Elucidating Pancreatic Ductal Adenocarcinoma Carcinogenesis at Single-Cell Resolution and Identifying Subtype Specific Drug Candidates
Abstract
1. Introduction
2. Results
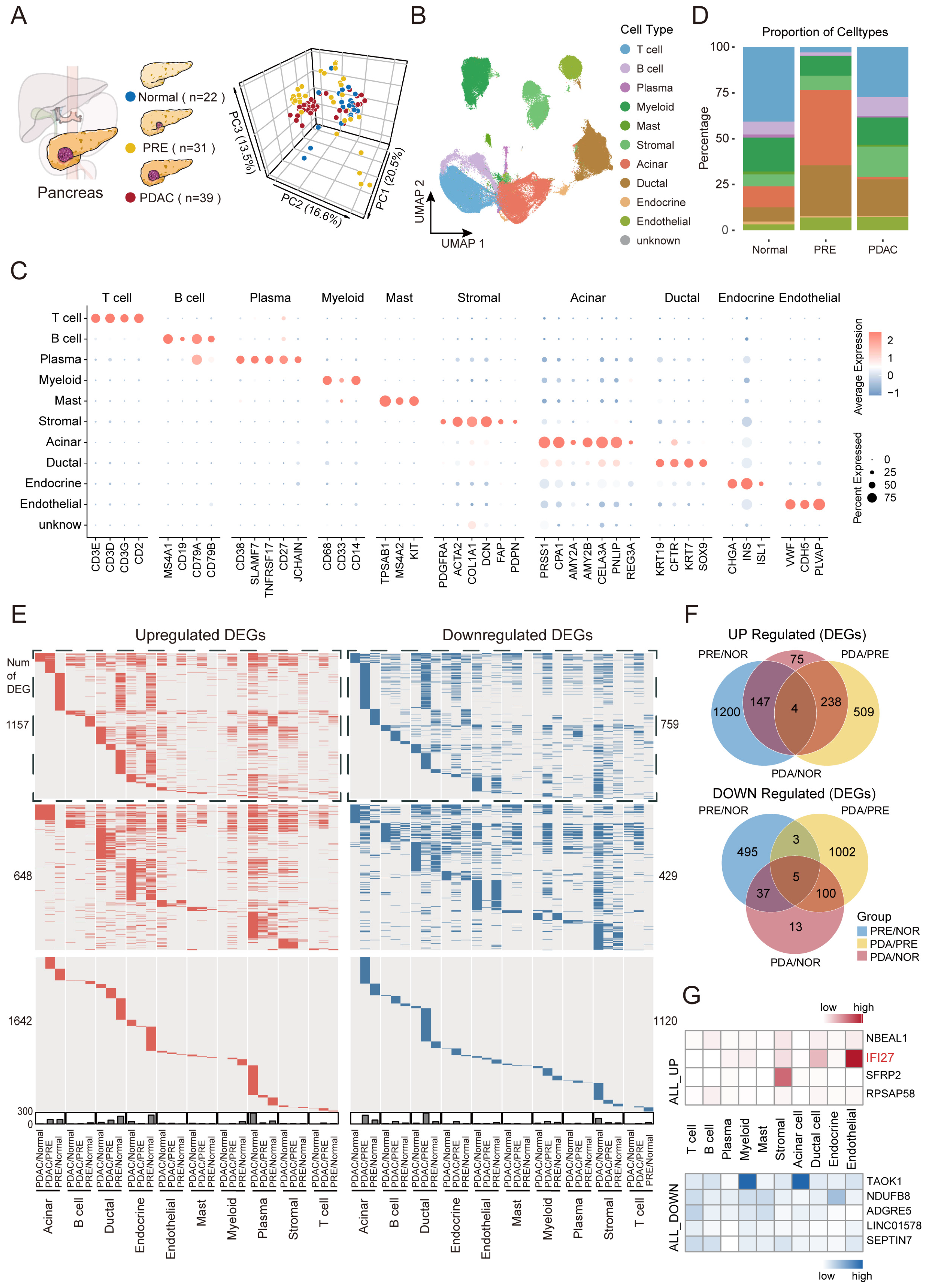
2.1. Dynamic Landscape Uncovers Persistently Dysregulated Genes Throughout PDAC Carcinogenesis
2.2. Identification and Characterization of Malignant Preneoplastic Clusters in Ductal and Acinar Cells
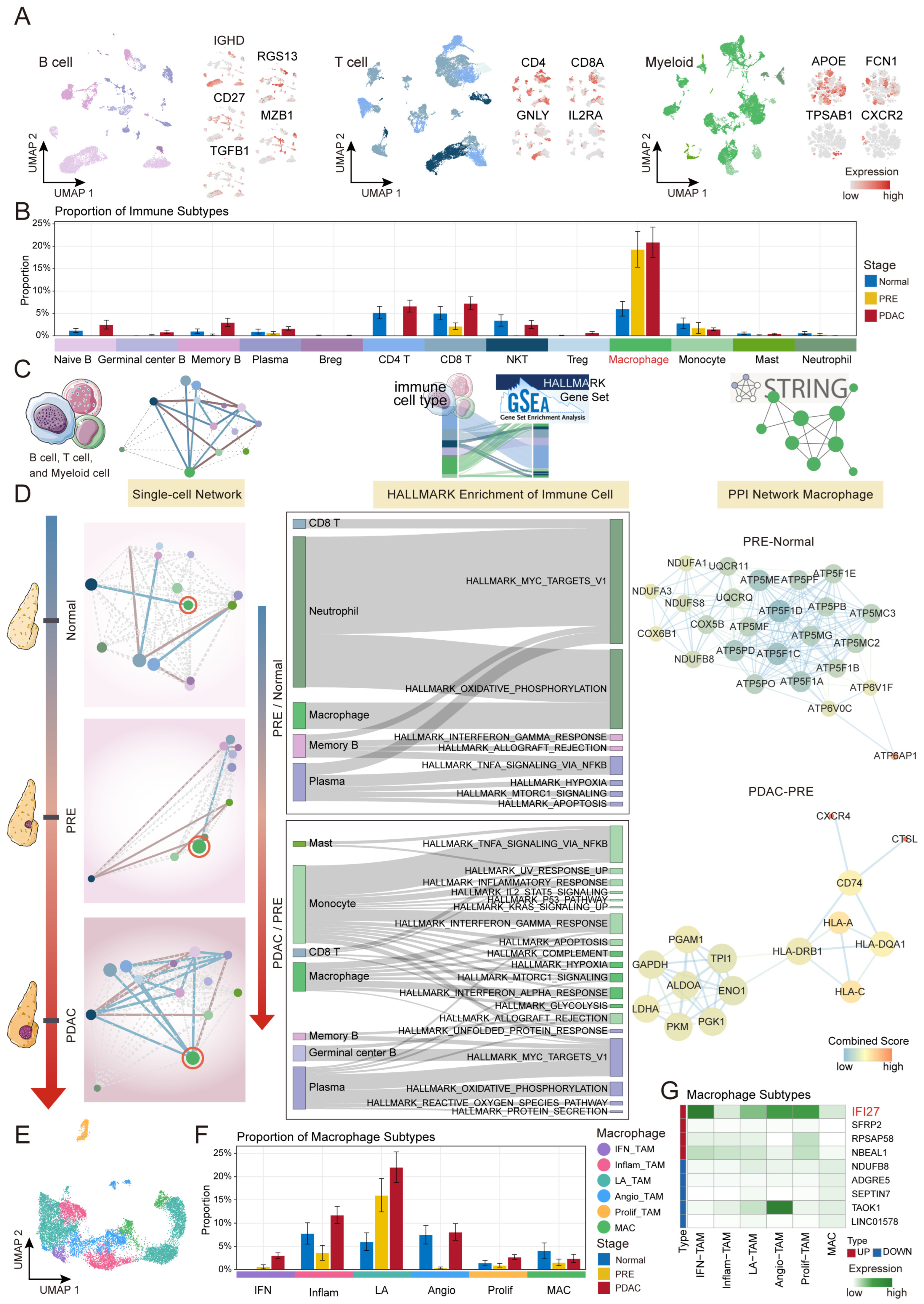
2.3. Macrophage-Driven Immune Microenvironment Remodeling During PDAC Carcinogenesis
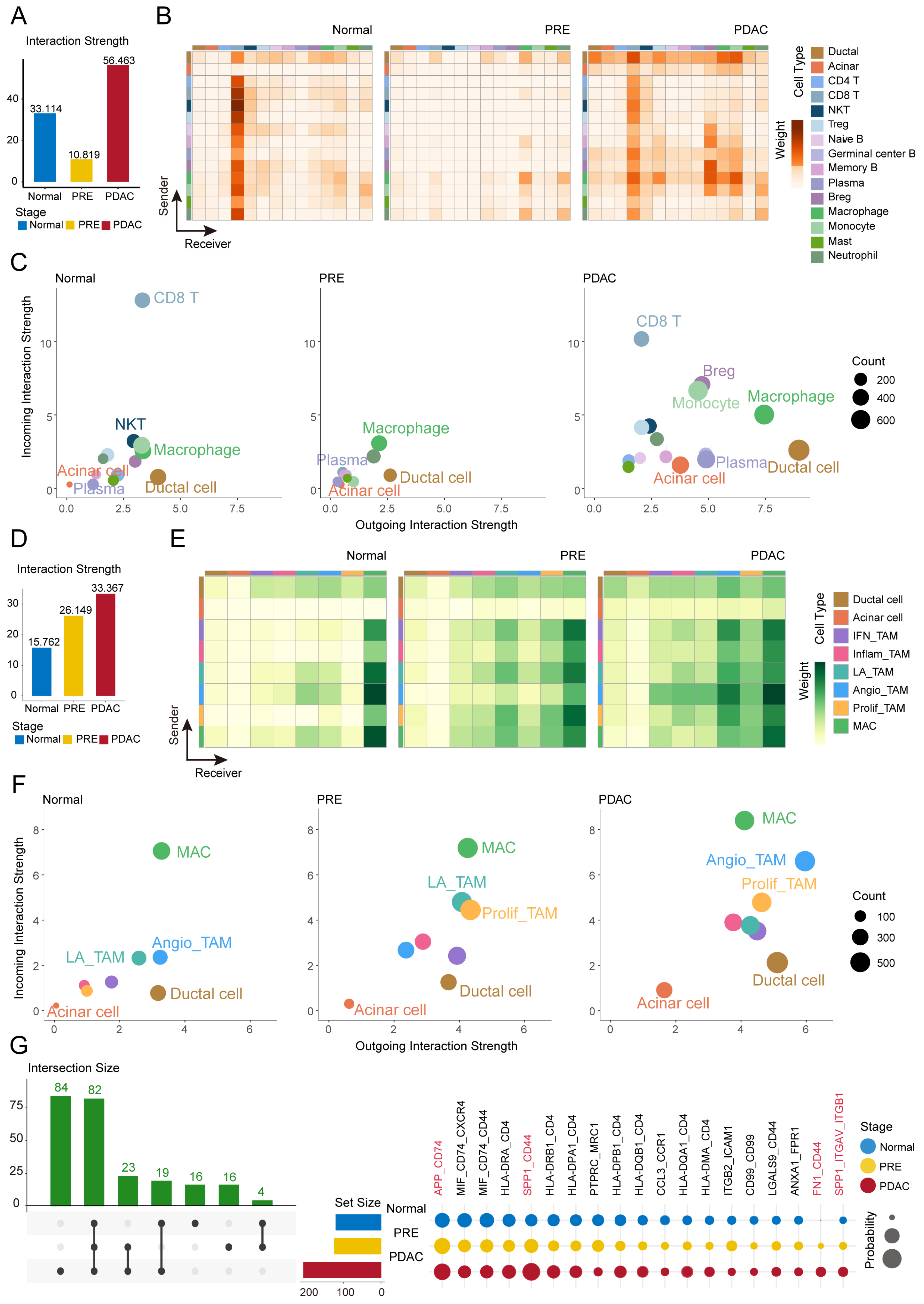
2.4. Dynamic Intensification of Macrophage-Ductal and Acinar Crosstalk During PDAC Carcinogenesis
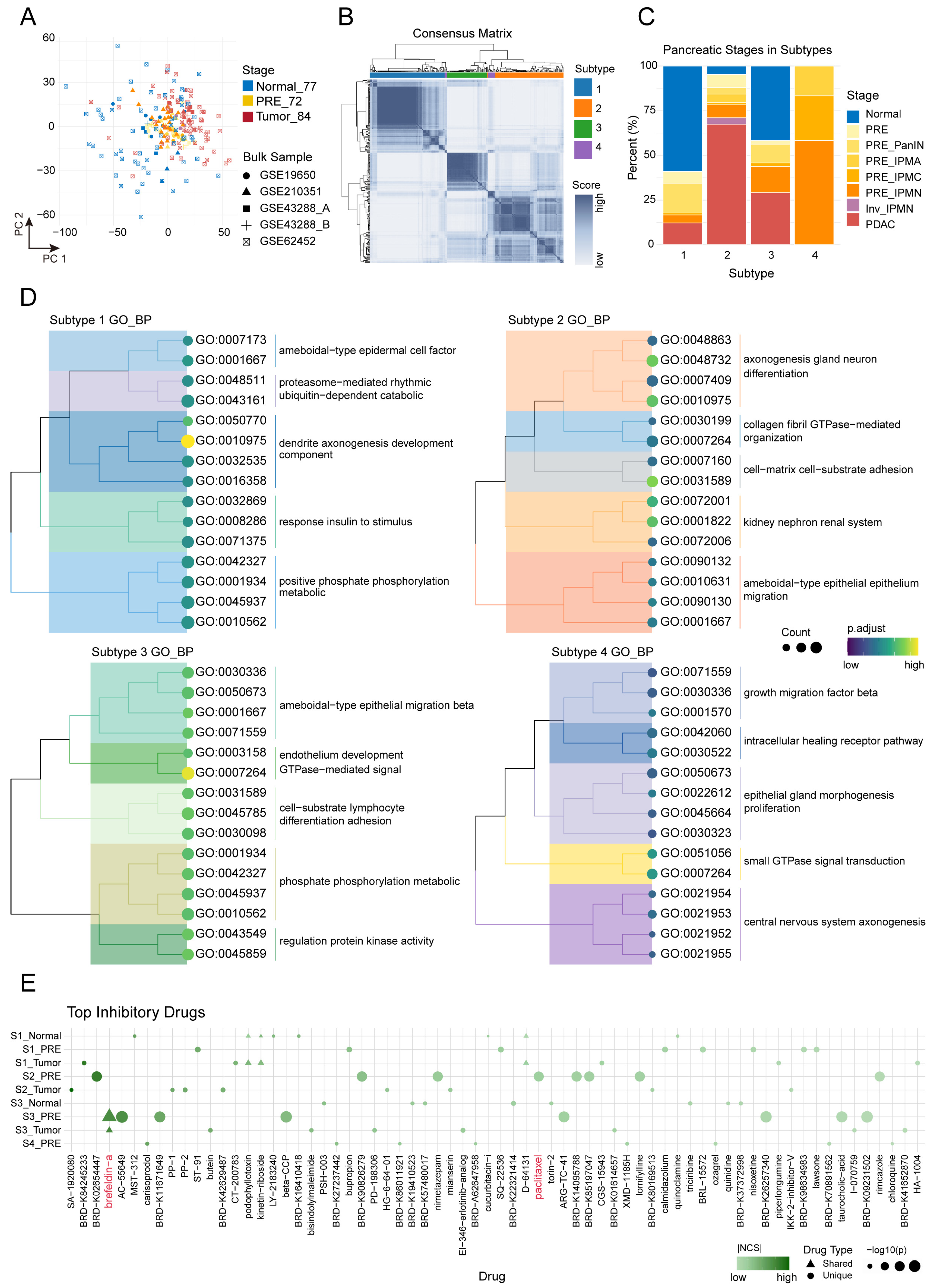
2.5. Consensus Subtypes and Drug Reversal Profiling Inform Precision Strategies in Pancreatic Carcinogenesis
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Single-Cell RNA-Seq Data Quality Control and Analysis
4.3. Clustering and Cell Type Annotation
4.4. Identification of Malignant Cells and Malignant Preneoplastic Clusters
4.5. Pseudotime Trajectory Inference
4.6. Construction of Stage-Specific Immune Networks and Correlation Analysis
4.7. Protein–Protein Interaction (PPI) Network Analysis
4.8. Gene Set Enrichment and Visualization
4.9. Cellchat Analysis
4.10. Consensus Clustering
4.11. Differential Expression Analysis
4.12. Drug Reversal Analysis
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Niksic, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Esteve, J.; et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.L.; Canto, M.I.; Klein, A.P.; Hruban, R.H.; Goggins, M. Recent Trends in the Incidence and Survival of Stage 1A Pancreatic Cancer: A Surveillance, Epidemiology, and End Results Analysis. JNCI J. Natl. Cancer Inst. 2020, 112, 1162–1169, Correction in JNCI J. Natl. Cancer Inst. 2021, 113, 216. [Google Scholar] [CrossRef] [PubMed]
- Halbrook, C.J.; Lyssiotis, C.A.; Pasca di Magliano, M.; Maitra, A. Pancreatic cancer: Advances and challenges. Cell 2023, 186, 1729–1754. [Google Scholar] [CrossRef] [PubMed]
- Sciano, F.; Terrana, F.; Pecoraro, C.; Parrino, B.; Cascioferro, S.; Diana, P.; Giovannetti, E.; Carbone, D. Exploring the therapeutic potential of focal adhesion kinase inhibition in overcoming chemoresistance in pancreatic ductal adenocarcinoma. Future Med. Chem. 2024, 16, 271–289. [Google Scholar] [CrossRef]
- Chung, V.; Mizrahi, J.D.; Pant, S. Novel Therapies for Pancreatic Cancer. JCO Oncol. Pract. 2025, 21, 613–619. [Google Scholar] [CrossRef]
- Randazzo, O.; Cascioferro, S.M.; Pecoraro, C.; Iddouch, W.A.; Avan, A.; Parrino, B.; Carbone, D.; Perricone, U.; Peters, G.J.; Diana, P.; et al. SF3B1 modulators affect key genes in metastasis and drug influx: A new approach to fight pancreatic cancer chemoresistance. Cancer Drug Resist. 2021, 4, 904–922. [Google Scholar] [CrossRef]
- Than, M.T.; O’Hara, M.; Stanger, B.Z.; Reiss, K.A. KRAS-Driven Tumorigenesis and KRAS-Driven Therapy in Pancreatic Adenocarcinoma. Mol. Cancer Ther. 2024, 23, 1378–1388. [Google Scholar] [CrossRef]
- Bandi, D.S.R.; Nagaraju, G.P.; Sarvesh, S.; Carstens, J.L.; Foote, J.B.; Graff, E.C.; Fang, Y.D.; Keeton, A.B.; Chen, X.; Valiyaveettil, J.; et al. ADT-1004: A first-in-class, oral pan-RAS inhibitor with robust antitumor activity in preclinical models of pancreatic ductal adenocarcinoma. Mol. Cancer 2025, 24, 76. [Google Scholar] [CrossRef]
- Wagner, A.; Regev, A.; Yosef, N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 2016, 34, 1145–1160. [Google Scholar] [CrossRef]
- Peng, J.; Sun, B.F.; Chen, C.Y.; Zhou, J.Y.; Chen, Y.S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.S.; et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738, Correction in Cell Res. 2019, 29, 777. [Google Scholar] [CrossRef]
- Raghavan, S.; Winter, P.S.; Navia, A.W.; Williams, H.L.; DenAdel, A.; Lowder, K.E.; Galvez-Reyes, J.; Kalekar, R.L.; Mulugeta, N.; Kapner, K.S.; et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell 2021, 184, 6119–6137. e26. [Google Scholar] [CrossRef] [PubMed]
- Flowers, B.M.; Xu, H.; Mulligan, A.S.; Hanson, K.J.; Seoane, J.A.; Vogel, H.; Curtis, C.; Wood, L.D.; Attardi, L.D. Cell of Origin Influences Pancreatic Cancer Subtype. Cancer Discov. 2021, 11, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Guerra, C.; Schuhmacher, A.J.; Canamero, M.; Grippo, P.J.; Verdaguer, L.; Perez-Gallego, L.; Dubus, P.; Sandgren, E.P.; Barbacid, M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 2007, 11, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Baerthel, S.; Falcomata, C.; Rad, R.; Theis, F.J.; Saur, D. Single-cell profiling to explore pancreatic cancer heterogeneity, plasticity and response to therapy. Nat. Cancer 2023, 4, 454–467. [Google Scholar] [CrossRef]
- Yousuf, S.; Qiu, M.; von Voithenberg, L.V.; Hulkkonen, J.; Macinkovic, I.; Schulz, A.R.; Hartmann, D.; Mueller, F.; Mijatovic, M.; Ibberson, D.; et al. Spatially Resolved Multi-Omics Single-Cell Analyses Inform Mechanisms of Immune Dysfunction in Pancreatic Cancer. Gastroenterology 2023, 165, 891–908. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Q.; Li, M.; Guo, H.; Liu, W.; Wang, F.; Tian, X.; Yang, Y. Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression. EBioMedicine 2021, 66, 103315. [Google Scholar] [CrossRef]
- Shojaei, M.; McLean, A.S. Interferon-stimulated gene IFI27 as a multifaceted candidate target in precision medicine. Trends Immunol. 2025, 46, 219–228. [Google Scholar] [CrossRef]
- Huang, S.; Zhao, J.; Song, J.; Li, Y.; Zuo, R.; Sa, Y.; Ma, Z.; OuYang, H. Interferon alpha-inducible protein 27 (IFI27) is a prognostic marker for pancreatic cancer based on comprehensive bioinformatics analysis. Bioengineered 2021, 12, 8515–8528. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, Z.; Ma, X.; Sun, K.; Fan, L.; Fang, J.; Pan, J.; Wang, X.; An, H.; Zhou, J. TAOK1 negatively regulates IL-17-mediated signaling and inflammation. Cell Mol. Immunol. 2018, 15, 794–802, Correction in Cell Mol. Immunol. 2018, 15, 940. [Google Scholar] [CrossRef]
- Alasiri, G.; Alrfaei, B.; Alaseem, A.M.; AlKhamees, O.A.; Aldali, J.A.; Aljehani, A.M.; Alfahed, A.; Aziz, M.A.; Almuhaini, G.; Alshehri, M.M. The role of TAOK3 in cancer progression and development as a prognostic marker: A pan-cancer analysis study. Saudi Pharm. J. 2024, 32, 101942. [Google Scholar] [CrossRef]
- Shi, G.; Zhu, L.; Sun, Y.; Bettencourt, R.; Damsz, B.; Hruban, R.H.; Konieczny, S.F. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology 2009, 136, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Kopp, J.L.; von Figura, G.; Mayes, E.; Liu, F.F.; Dubois, C.L.; Morris, J.P.T.; Pan, F.C.; Akiyama, H.; Wright, C.V.; Jensen, K.; et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 2012, 22, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Storz, P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Ying, H.; Kimmelman, A.C.; Lyssiotis, C.A.; Hua, S.; Chu, G.C.; Fletcher-Sananikone, E.; Locasale, J.W.; Son, J.; Zhang, H.; Coloff, J.L.; et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012, 149, 656–670. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Yang, K.; Yang, T.; Yu, J.; Li, F.; Zhao, X. Integrated transcriptional analysis reveals macrophage heterogeneity and macrophage-tumor cell interactions in the progression of pancreatic ductal adenocarcinoma. BMC Cancer 2023, 23, 199. [Google Scholar] [CrossRef]
- Biasci, D.; Smoragiewicz, M.; Connell, C.M.; Wang, Z.; Gao, Y.; Thaventhiran, J.E.D.; Basu, B.; Magiera, L.; Johnson, T.I.; Bax, L.; et al. CXCR4 inhibition in human pancreatic and colorectal cancers induces an integrated immune response. Proc. Natl. Acad. Sci. USA 2020, 117, 28960–28970. [Google Scholar] [CrossRef]
- Gocheva, V.; Zeng, W.; Ke, D.; Klimstra, D.; Reinheckel, T.; Peters, C.; Hanahan, D.; Joyce, J.A. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006, 20, 543–556. [Google Scholar] [CrossRef]
- Yang, Y.; Li, S.; To, K.K.W.; Zhu, S.; Wang, F.; Fu, L. Tumor-associated macrophages remodel the suppressive tumor immune microenvironment and targeted therapy for immunotherapy. J. Exp. Clin. Cancer Res. 2025, 44, 145. [Google Scholar] [CrossRef]
- Azizi, E.; Carr, A.J.; Plitas, G.; Cornish, A.E.; Konopacki, C.; Prabhakaran, S.; Nainys, J.; Wu, K.; Kiseliovas, V.; Setty, M.; et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018, 174, 1293–1308. e36. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Crawford, H.C.; Pasca di Magliano, M.; Banerjee, S. Signaling Networks That Control Cellular Plasticity in Pancreatic Tumorigenesis, Progression, and Metastasis. Gastroenterology 2019, 156, 2073–2084. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chen, J.; Zhu, Z.; Hu, H.; Zhang, Q.; Jia, R.; Wang, N.; Xiang, S.; Zhou, Y.; Wang, Y.; et al. CD74 Blockade Disrupts Endothelial Migrasome Signaling to Prevent Inflammatory Macrophage Differentiation and Inhibit Atherosclerotic Progression. Adv. Sci. 2025, 12, e02838. [Google Scholar] [CrossRef]
- Nallasamy, P.; Nimmakayala, R.K.; Karmakar, S.; Leon, F.; Seshacharyulu, P.; Lakshmanan, I.; Rachagani, S.; Mallya, K.; Zhang, C.; Ly, Q.P.; et al. Pancreatic Tumor Microenvironment Factor Promotes Cancer Stemness via SPP1-CD44 Axis. Gastroenterology 2021, 161, 1998–2013 e1997. [Google Scholar] [CrossRef]
- Su, W.; Ye, Z.; Liu, J.; Deng, K.; Liu, J.; Zhu, H.; Duan, L.; Shi, C.; Wang, L.; Zhao, Y.; et al. Single-cell and spatial transcriptome analyses reveal tumor heterogeneity and immune remodeling involved in pituitary neuroendocrine tumor progression. Nat. Commun. 2025, 16, 5007. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. New Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Yang, J.; Cui, Y. TM9SF3 is a mammalian Golgiphagy receptor that safeguards Golgi integrity and glycosylation fidelity. Autophagy 2025, 21, 2526–2527. [Google Scholar] [CrossRef]
- Citterio, C.; Vichi, A.; Pacheco-Rodriguez, G.; Aponte, A.M.; Moss, J.; Vaughan, M. Unfolded protein response and cell death after depletion of brefeldin A-inhibited guanine nucleotide-exchange protein GBF1. Proc. Natl. Acad. Sci. USA 2008, 105, 2877–2882. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Sun, L.L.; Fu, B.Q.; Deng, J.; Jia, C.L.; Miao, M.X.; Yang, F.; Cao, Y.B.; Yan, T.H. Aneuploidy underlies brefeldin A-induced antifungal drug resistance in Cryptococcus neoformans. Front. Cell. Infect. Microbiol. 2024, 14, 1397724. [Google Scholar] [CrossRef]
- Verovski, V.N.; Van den Berge, D.L.; Delvaeye, M.M.; Scheper, R.J.; De Neve, W.J.; Storme, G.A. Low-level doxorubicin resistance in P-glycoprotein-negative human pancreatic tumour PSN1/ADR cells implicates a brefeldin A-sensitive mechanism of drug extrusion. Br. J. Cancer 1996, 73, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Togawa, A.; Ito, H.; Kimura, F.; Shimizu, H.; Ohtsuka, M.; Shimamura, F.; Yoshidome, H.; Katoh, A.; Miyazaki, M. Establishment of gemcitabine-resistant human pancreatic cancer cells and effect of brefeldin-a on the resistant cell line. Pancreas 2003, 27, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zeh, H.J.; Kang, R.; Kroemer, G.; Tang, D. Cell death in pancreatic cancer: From pathogenesis to therapy. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 804–823. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.G.; Shimizu, T.; Pommier, Y. Brefeldin A is a potent inducer of apoptosis in human cancer cells independently of p53. Exp. Cell Res. 1996, 227, 190–196. [Google Scholar] [CrossRef]
- Anadu, N.O.; Davisson, V.J.; Cushman, M. Synthesis and anticancer activity of brefeldin A ester derivatives. J. Med. Chem. 2006, 49, 3897–3905. [Google Scholar] [CrossRef]
- Tian, K.; Xu, F.; Gao, X.; Han, T.; Li, J.; Pan, H.; Zang, L.; Li, D.; Li, Z.; Uchita, T.; et al. Nitric oxide-releasing derivatives of brefeldin A as potent and highly selective anticancer agents. Eur. J. Med. Chem. 2017, 136, 131–143. [Google Scholar] [CrossRef]
- Paek, S.M. Recent Synthesis and Discovery of Brefeldin A Analogs. Mar. Drugs 2018, 16, 133. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902. e21. [Google Scholar] [CrossRef]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Li, B.; Gould, J.; Yang, Y.; Sarkizova, S.; Tabaka, M.; Ashenberg, O.; Rosen, Y.; Slyper, M.; Kowalczyk, M.S.; Villani, A.C.; et al. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat. Methods 2020, 17, 793–798. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.A.; Trapnell, C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 2017, 14, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452. e17. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Jiang, H.; Chen, H.; Yang, K.; Yang, K.; Sun, M.; Lv, N.; Ren, B.; Lin, X.; Li, X.; et al. Elucidating Pancreatic Ductal Adenocarcinoma Carcinogenesis at Single-Cell Resolution and Identifying Subtype Specific Drug Candidates. Int. J. Mol. Sci. 2025, 26, 12031. https://doi.org/10.3390/ijms262412031
Chen J, Jiang H, Chen H, Yang K, Yang K, Sun M, Lv N, Ren B, Lin X, Li X, et al. Elucidating Pancreatic Ductal Adenocarcinoma Carcinogenesis at Single-Cell Resolution and Identifying Subtype Specific Drug Candidates. International Journal of Molecular Sciences. 2025; 26(24):12031. https://doi.org/10.3390/ijms262412031
Chicago/Turabian StyleChen, Jing, Hui Jiang, Hui Chen, Kuan Yang, Kaiyue Yang, Mingyao Sun, Na Lv, Bolin Ren, Xinyi Lin, Xia Li, and et al. 2025. "Elucidating Pancreatic Ductal Adenocarcinoma Carcinogenesis at Single-Cell Resolution and Identifying Subtype Specific Drug Candidates" International Journal of Molecular Sciences 26, no. 24: 12031. https://doi.org/10.3390/ijms262412031
APA StyleChen, J., Jiang, H., Chen, H., Yang, K., Yang, K., Sun, M., Lv, N., Ren, B., Lin, X., Li, X., Zhang, Y., & Hu, C. (2025). Elucidating Pancreatic Ductal Adenocarcinoma Carcinogenesis at Single-Cell Resolution and Identifying Subtype Specific Drug Candidates. International Journal of Molecular Sciences, 26(24), 12031. https://doi.org/10.3390/ijms262412031






