Comprehensive Updated Genome-Wide Identification and Expression Patterns of the TaGeBP Gene Family in Wheat
Abstract
1. Introduction
2. Results
2.1. Identification of the GeBP Gene Family in Wheat
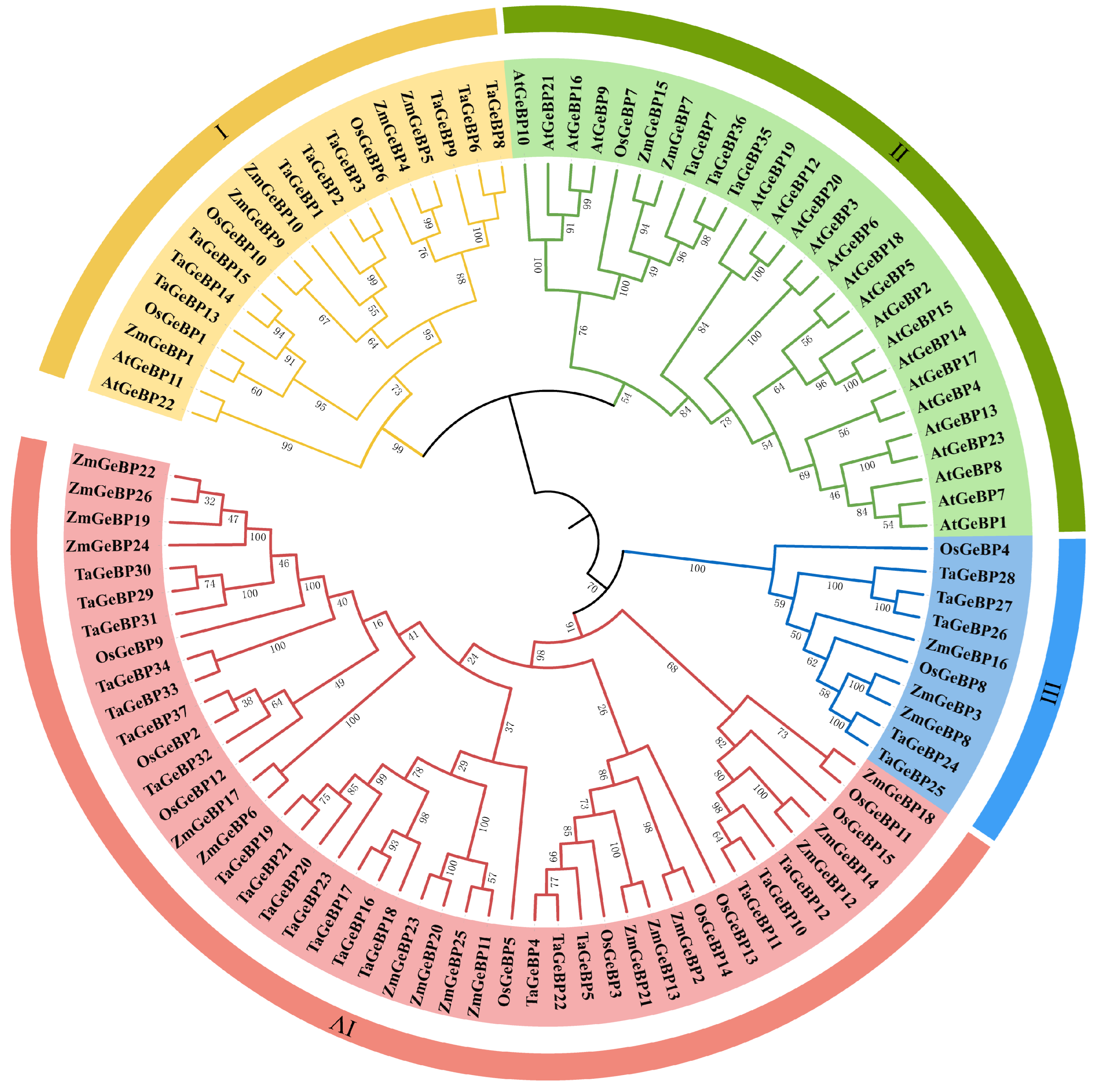
2.2. Phylogenetic Analysis of the TaGeBP Gene Family
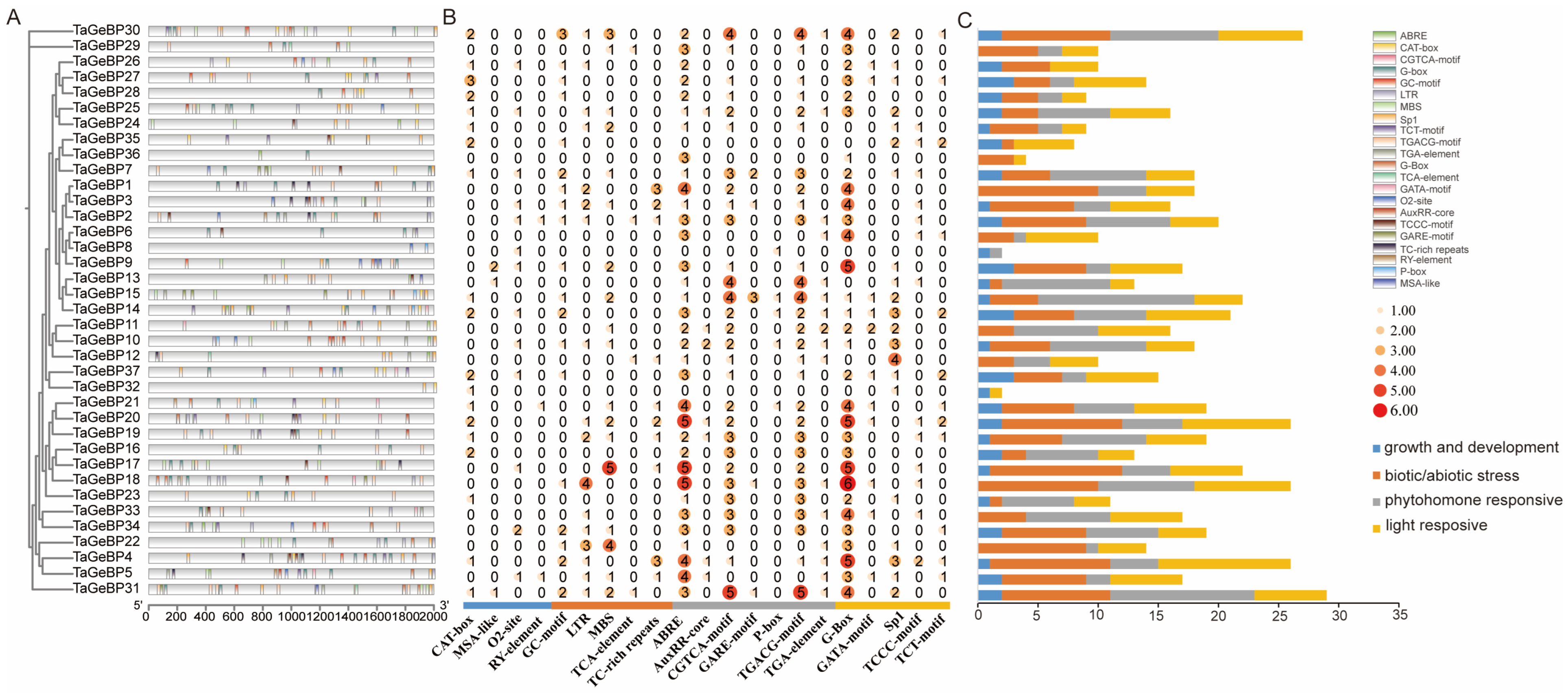
2.3. Analysis of Protein Conserved Motif and Gene Structure of GeBP in Wheat
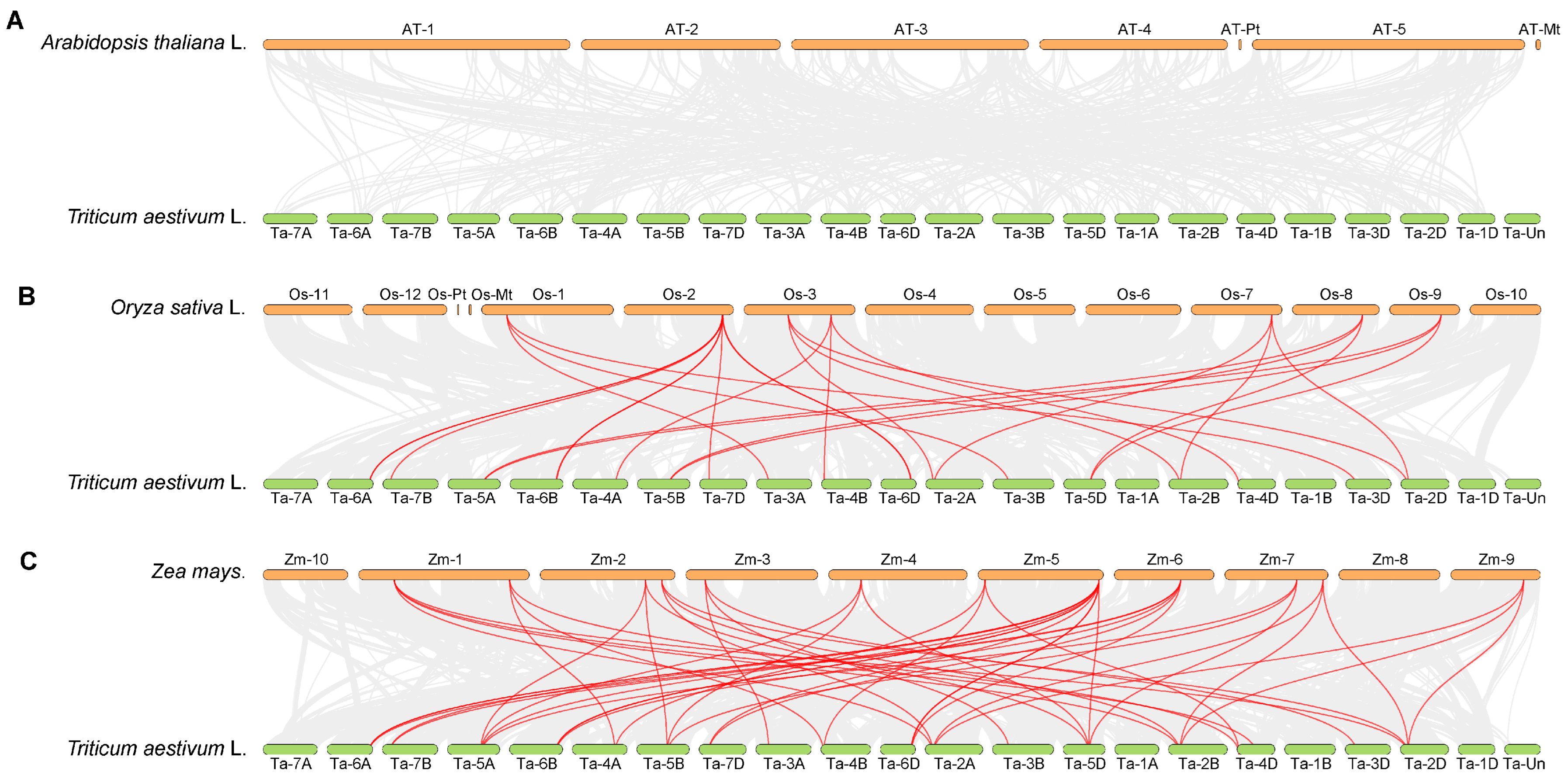
2.4. Chromosomal Localization and Collinearity Analysis of GeBP Genes in Wheat
2.5. Analysis of Cis-Elements in Promoters
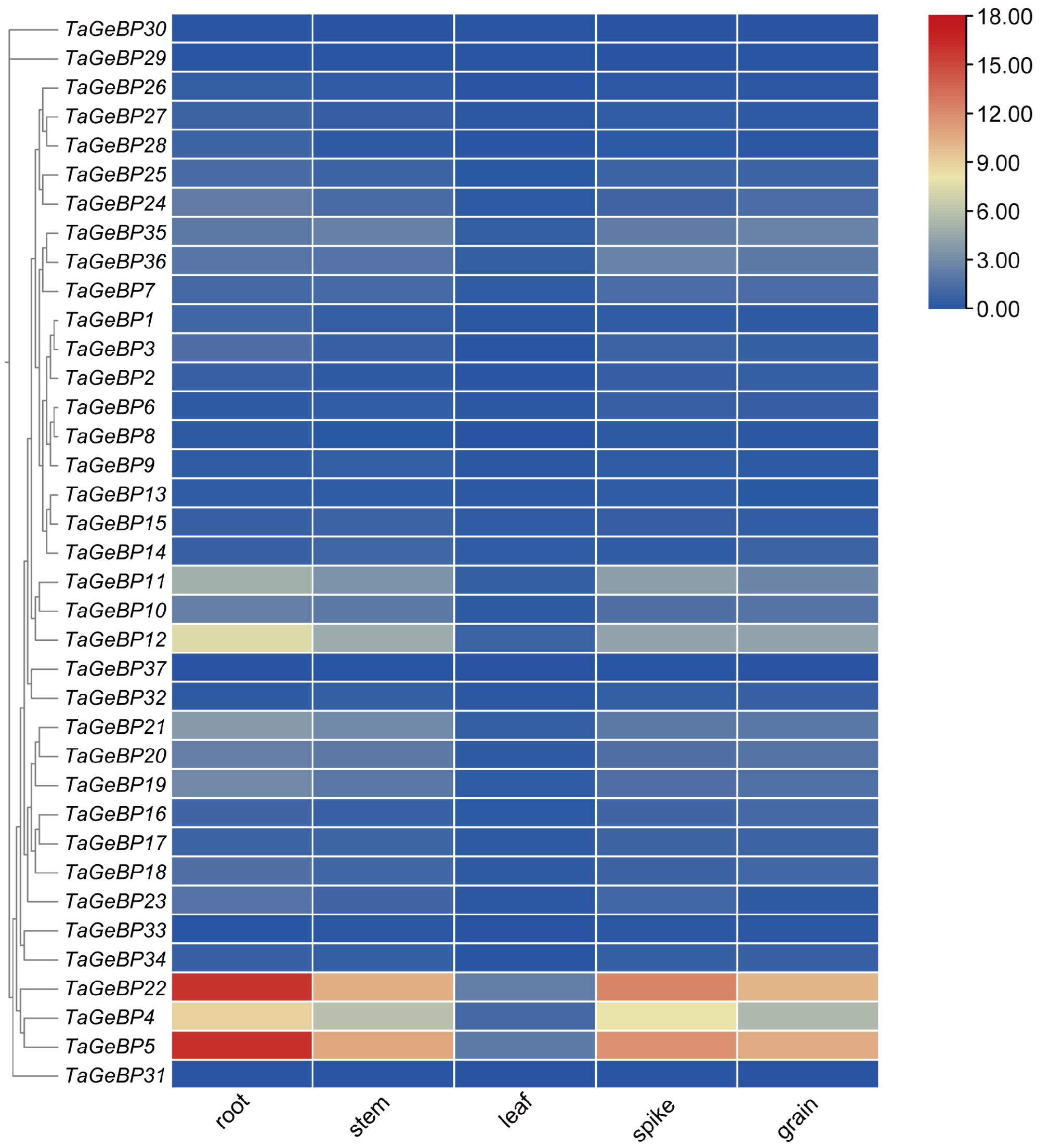
2.6. Expression Profile of the GeBP Gene Family in Wheat
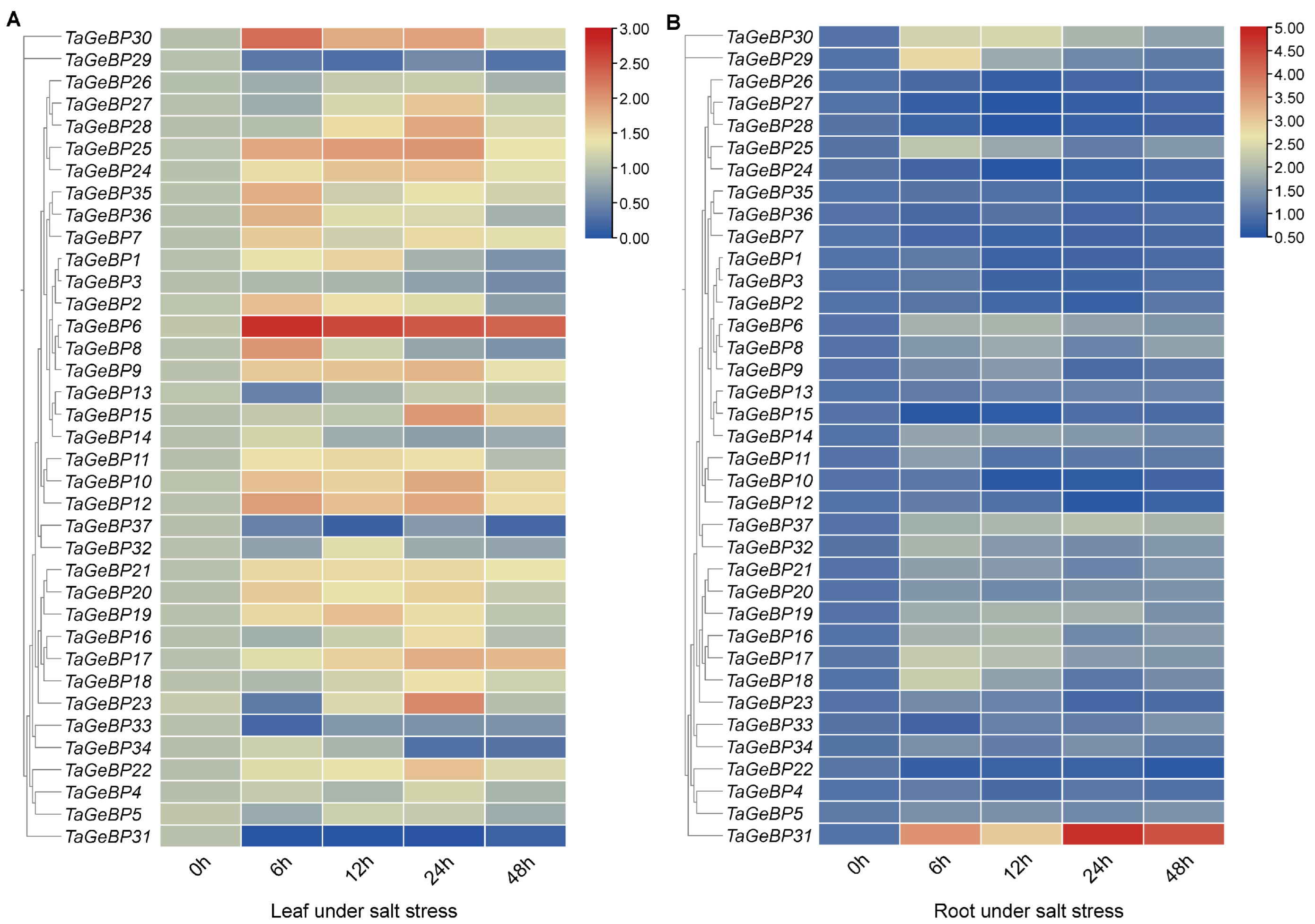
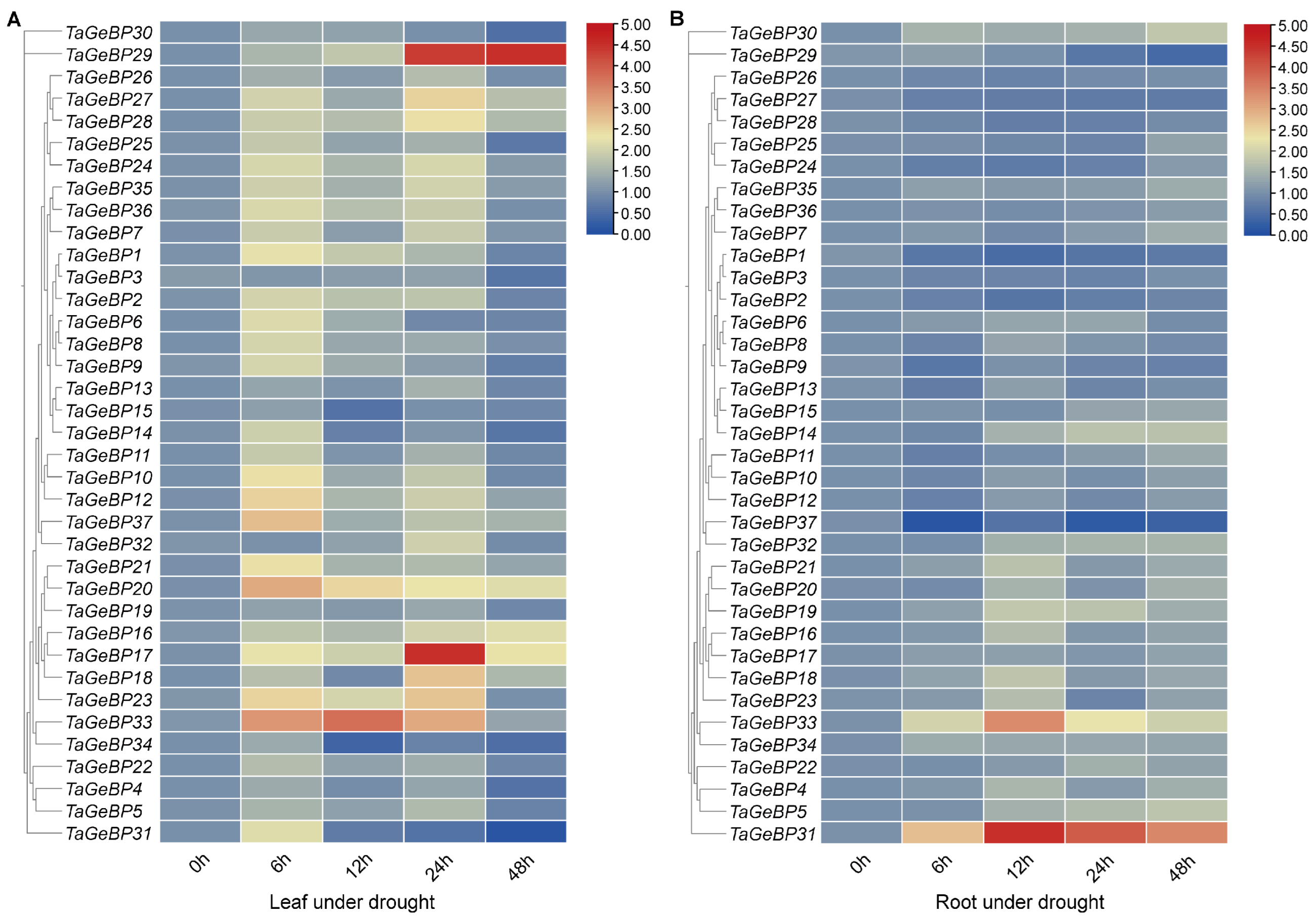
2.7. Expression Patterns of TaGeBP Genes Under Stress Conditions Using qRT-PCR Analyses
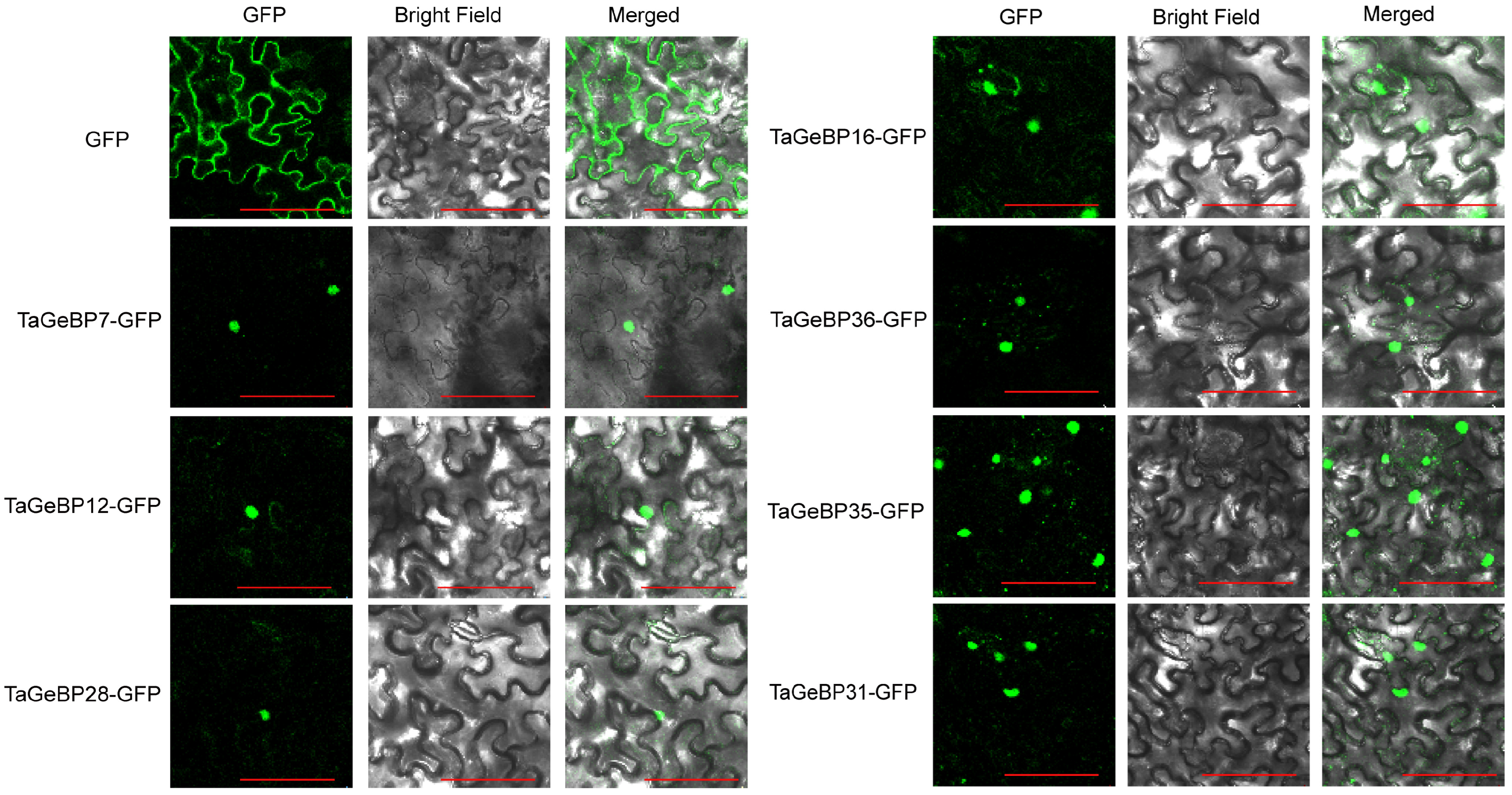
2.8. Subcellular Localization Analysis of TaGeBP Proteins
3. Discussion
4. Materials and Methods
4.1. Identification and Classification of TaGeBPs
4.2. Phylogenetic Analysis of the Wheat TaGeBP Gene Family
4.3. Analysis of Chromosomal Localization and Gene Collinearity
4.4. Analysis of Cis-Elements in Wheat TaGeBP Gene Promoters
4.5. Transcriptome Analysis of TaGeBPs in Different Tissues
4.6. TaGeBP Expression Profiles and qRT-PCR Analysis
4.7. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miransari, M.; Smith, D. Sustainable wheat (Triticum aestivum L.) production in saline fields: A review. Crit. Rev. Biotechnol. 2019, 39, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Skalicky, M.; Brestic, M.; Maitra, S.; Ashraful, A.M.; Syed, M.A.; Hossain, J.; Sarkar, S.; Saha, S.; Bhadra, P. Consequences and mitigation strategies of abiotic stresses in wheat (Triticum aestivum L.) under the changing climate. Agronomy 2021, 11, 241. [Google Scholar] [CrossRef]
- Singh, A.K.; Dhanapal, S.; Yadav, B.S. The dynamic responses of plant physiology and metabolism during environmental stress progression. Mol. Biol. Rep. 2020, 47, 1459–1470. [Google Scholar] [CrossRef]
- Cao, H.; Wang, D.; Li, X.; Zhang, Y.; Su, D.; Lu, W.; Xu, K.; Li, Z. Genome-Wide Identification and Expression Profiling of SlGeBP Gene Family in Response to Hormone and Abiotic Stresses in Solanum lycopersicum L. Int. J. Mol. Sci. 2025, 26, 6008. [Google Scholar] [CrossRef]
- Higgins, I.J.; Choudury, S.G.; Husbands, A.Y. Mechanisms driving functional divergence of transcription factor paralogs. New Phytol. 2025, 247, 2022–2033. [Google Scholar] [CrossRef]
- Curaba, J.; Herzog, M.; Vachon, G. GeBP, the first member of a new gene family in Arabidopsis, encodes a nuclear protein with DNA-binding activity and is regulated by KNAT1. Plant J. 2003, 33, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, F.; Perazza, D.; Laporte, F.; Le Hénanff, G.; Hornitschek, P.; Bonneville, J.M.; Vachon, G. GeBP and GeBP-like proteins are noncanonical leucine-zipper transcription factors that regulate cytokinin response in Arabidopsis. Plant Physiol. 2008, 146, 1142–1154. [Google Scholar] [CrossRef][Green Version]
- Liu, L.L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants: Functional domains. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F.; bZIP Research Group. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, Q.; He, Y.; Liu, W.; Xu, Y.; Liu, K.; Xian, F.; Li, J.; Hu, J. Genome-wide identification, expansion mechanism and expression profiling analysis of GLABROUS1 enhancer-binding protein (GeBP) gene family in Gramineae crops. Int. J. Mol. Sci. 2021, 22, 8758. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Liu, C.; Zhang, F.; Wei, J.; Li, B. Genome-wide characterization and expression analysis of GeBP family genes in soybean. Plants 2022, 11, 1848. [Google Scholar] [CrossRef]
- Wu, J.; Liu, R.; Xie, Y.; Zhao, S.; Yan, M.; Sun, N.; Zhan, Y.; Li, F.; Yu, S.; Feng, Z.; et al. Association of GhGeBP genes with fiber quality and early maturity related traits in upland cotton. BMC Genom. 2024, 25, 1058. [Google Scholar] [CrossRef]
- Wang, R.; Wu, X.; Wang, Z.; Zhang, X.; Chen, L.; Duan, Q.; Huang, J. Genome-wide identification and expression analysis of BrGeBP genes reveal their potential roles in cold and drought stress tolerance in Brassica rapa. Int. J. Mol. Sci. 2023, 24, 13597. [Google Scholar] [CrossRef]
- Liu, R.-X.; Li, H.-L.; Qiao, Z.-W.; Liu, H.-F.; Zhao, L.-L.; Wang, X.-F.; Zhang, Z.; Zhang, S.; Song, L.-Q.; You, C.-X. Genome-wide analysis of MdGeBP family and functional identification of MdGeBP3 in Malus domestica. Environ. Exp. Bot. 2023, 208, 105262. [Google Scholar] [CrossRef]
- Perazza, D.; Laporte, F.; Balagué, C.; Chevalier, F.; Remo, S.; Bourge, M.; Larkin, J.; Herzog, M.; Vachon, G. GeBP/GPL transcription factors regulate a subset of CPR5-dependent processes. Plant Physiol. 2011, 157, 1232–1242. [Google Scholar] [CrossRef]
- Zubo, Y.O.; Schaller, G.E. Role of the cytokinin-activated type-B response regulators in hormone crosstalk. Plants 2020, 9, 166. [Google Scholar] [CrossRef]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef]
- Curaba, J.; Moritz, T.; Blervaque, R.; Parcy, F.; Raz, V.; Herzog, M.; Vachon, G. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004, 136, 3660–3669. [Google Scholar] [CrossRef]
- Wang, H.; Umer, M.J.; Liu, F.; Cai, X.; Zheng, J.; Xu, Y.; Hou, Y.; Zhou, Z. Genome-wide identification and characterization of CPR5 genes in Gossypium reveals their potential role in trichome development. Front. Plant Sci. 2022, 13, 921096. [Google Scholar] [CrossRef] [PubMed]
- Khare, D.; Mitsuda, N.; Lee, S.; Song, W.-Y.; Hwang, D.; Ohme-Takagi, M.; Martinoia, E.; Lee, Y.; Hwang, J.-U. Root avoidance of toxic metals requires the GeBP-LIKE 4 transcription factor in Arabidopsis thaliana. New Phytol. 2017, 213, 1257–1273. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, M.; Zhang, L.; Wu, N.; Tang, X.; Yang, X.; Ghanem, H.; Wu, M.; Wu, G.; Qing, L. Insights into geminiviral pathogenesis: Interaction between βC1 protein and GLABROUS1 enhancer binding protein (GeBP) in Solanaceae. Phytopathol. Res. 2025, 7, 30. [Google Scholar] [CrossRef]
- Hu, Y.; Han, X.; Yang, M.; Zhang, M.; Pan, J.; Yu, D. The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2019, 31, 1520–1538. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Iqbal, J.; Naseer, S.; Shaukat, M.; Abbasi, B.A.; Yaseen, T.; Zahra, S.A.; Mahmood, T. Unfolding molecular switches in plant heat stress resistance: A comprehensive review. Plant Cell Rep. 2022, 41, 775–798. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhou, W.; Yao, X.; Zhao, Q.; Lu, L. Genome-wide investigation and functional analysis reveal that csgebp4 is required for tea plant trichome formation. Int. J. Mol. Sci. 2023, 24, 5207. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Genome evolution due to allopolyploidization in wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef]
- Evans, C.E.B.; Arunkumar, R.; Borrill, P. Transcription factor retention through multiple polyploidization steps in wheat. G3-Genes Genom. Genet. 2022, 12, jkac147. [Google Scholar] [CrossRef]
- Ma, J.; Yuan, M.; Sun, B.; Zhang, D.; Zhang, J.; Li, C.; Shao, Y.; Liu, W.; Jiang, L. Evolutionary divergence and biased expression of NAC transcription factors in hexaploid bread wheat (Triticum aestivum L.). Plants 2021, 10, 382. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef]
- Reams, A.B.; Neidle, E.L. Selection for gene clustering by tandem duplication. Annu. Rev. Microbiol. 2004, 58, 119–142. [Google Scholar] [CrossRef]
- Sethi, S.; Saini, J.S.; Mohan, A.; Brar, N.K.; Verma, S.; Sarao, N.K.; Gill, K.S. Comparative and evolutionary analysis of α-amylase gene across monocots and dicots. Funct. Integr. Genom. 2016, 16, 545–555. [Google Scholar] [CrossRef]
- Anantharaman, V.; Balaji, S.; Aravind, L. The signaling helix: A common functional theme in diverse signaling proteins. Biol. Direct. 2006, 1, 25. [Google Scholar] [CrossRef]
- Ho, C.-L.; Geisler, M. Genome-wide computational identification of biologically significant cis-regulatory elements and associated transcription factors from rice. Plants 2019, 8, 441. [Google Scholar] [CrossRef]
- Kong, W.; Ding, L.; Cheng, J.; Wang, B. Identification and expression analysis of genes with pathogen-inducible cis-regulatory elements in the promoter regions in Oryza sativa. Rice 2018, 11, 52. [Google Scholar] [CrossRef]
- Sarwar, R.; Jiang, T.; Ding, P.; Gao, Y.; Tan, X.; Zhu, K. Genome-wide analysis and functional characterization of the DELLA gene family associated with stress tolerance in B. napus. BMC Plant Biol. 2021, 21, 286. [Google Scholar] [CrossRef]
- Hahn, A.; Kilian, J.; Mohrholz, A.; Ladwig, F.; Peschke, F.; Dautel, R.; Harter, K.; Berendzen, K.W.; Wanke, D. Plant core environmental stress response genes are systemically coordinated during abiotic stresses. Int. J. Mol. Sci. 2013, 14, 7617–7641. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Shaikhali, J.; Davoine, C.; Brännström, K.; Rouhier, N.; Bygdell, J.; Björklund, S.; Wingsle, G. Biochemical and redox characterization of the mediator complex and its associated transcription factor GeBPL, a GLABROUS1 enhancer binding protein. Biochem. J. 2015, 468, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Boer, D.; Hayes, S.; Testerink, C. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, E.; Millas, R.; McNeal, W.; Faris, J.; Taheri, A. Variation analysis of root system development in wheat seedlings using root phenotyping system. Agronomy 2020, 10, 206. [Google Scholar] [CrossRef]
- Onyemaobi, O.; Sangma, H.; Garg, G.; Wallace, X.; Kleven, S.; Suwanchaikasem, P.; Roessner, U.; Dolferus, R. Reproductive stage drought tolerance in wheat: Importance of stomatal conductance and plant growth regulators. Genes 2021, 12, 1742. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Wani, S.H.; Bhardwaj, S.C.; Rani, K.; Bishnoi, S.K.; Singh, G.P. Wheat spike blast: Genetic interventions for effective management. Mol. Biol. Rep. 2022, 49, 5483–5494. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Chen, J.; Yue, M.; Xu, C.; Jian, W.; Liu, Y.; Song, B.; Gao, Y.; Cheng, Y.; Li, Z. Tomato transcriptional repressor MYB70 directly regulates ethylene-dependent fruit ripening. Plant J. 2020, 104, 1568–1581. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Ding, J.; Li, T.; Zhao, Y.; Xu, D.; Zeng, J.; Liu, W.; Qu, M.; Ma, W.; Dai, X. Comprehensive Updated Genome-Wide Identification and Expression Patterns of the TaGeBP Gene Family in Wheat. Int. J. Mol. Sci. 2025, 26, 11972. https://doi.org/10.3390/ijms262411972
Zhang S, Ding J, Li T, Zhao Y, Xu D, Zeng J, Liu W, Qu M, Ma W, Dai X. Comprehensive Updated Genome-Wide Identification and Expression Patterns of the TaGeBP Gene Family in Wheat. International Journal of Molecular Sciences. 2025; 26(24):11972. https://doi.org/10.3390/ijms262411972
Chicago/Turabian StyleZhang, Shuqing, Jianwen Ding, Tianao Li, Yuxuan Zhao, Dengan Xu, Jianbin Zeng, Wenxing Liu, Mei Qu, Wujun Ma, and Xuehuan Dai. 2025. "Comprehensive Updated Genome-Wide Identification and Expression Patterns of the TaGeBP Gene Family in Wheat" International Journal of Molecular Sciences 26, no. 24: 11972. https://doi.org/10.3390/ijms262411972
APA StyleZhang, S., Ding, J., Li, T., Zhao, Y., Xu, D., Zeng, J., Liu, W., Qu, M., Ma, W., & Dai, X. (2025). Comprehensive Updated Genome-Wide Identification and Expression Patterns of the TaGeBP Gene Family in Wheat. International Journal of Molecular Sciences, 26(24), 11972. https://doi.org/10.3390/ijms262411972






