Whole-Genome Analysis of PGP Endophytic Bacillus subtilis 10-4: Unraveling Molecular Insights into Plant Growth and Stress Resilience
Abstract
1. Introduction
2. Results
2.1. Genome Overview and General Features of B. subtilis 10-4
2.1.1. Assembly and Annotation
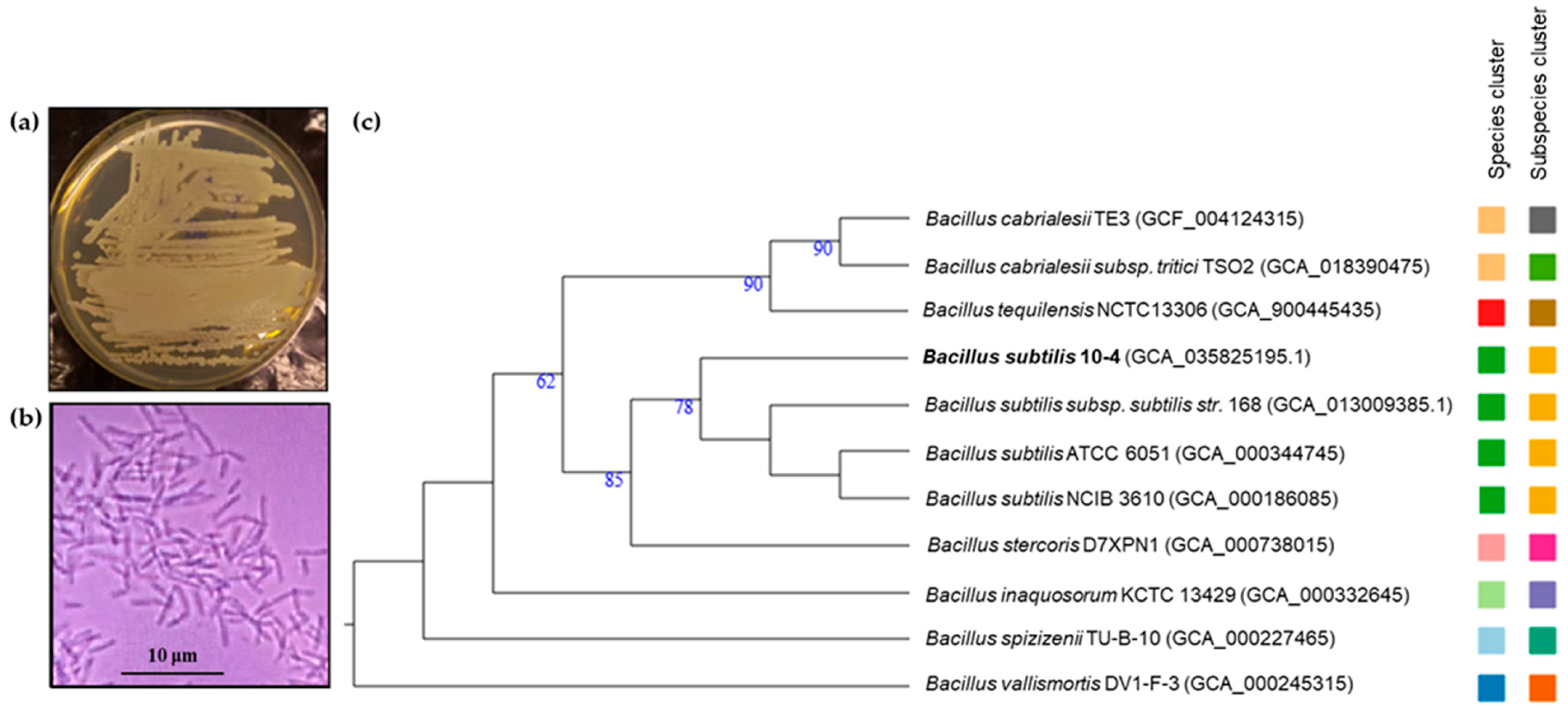
2.1.2. Taxonomic Assignment and Phylogenetic Position
2.2. Functional Annotation of B. subtilis 10-4 Genome and Identification Plant-Beneficial Genes
2.2.1. Genes Involved in Colonization and Interactions with Plants
2.2.2. Metabolic Pathways of B. subtilis 10-4 Underlying PGP Capabilities and Stress Resistance
Functional Genes Associated with Plant Growth and Mineral Nutrition
Functional Genes Associated with Stress Tolerance
Secondary Metabolite Biosynthetic Genes in B. subtilis 10-4 Genome
2.3. Growth-Simulating Impact of B. subtilis 10-4 on Plants Under Laboratory and Field Conditions
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain, Cultivation, and Inoculum Preparation
4.2. Genomic DNA Extraction, Whole Genome Sequencing, and Assembly
4.3. Genome Annotation and Phylogenetic Tree Construction
4.4. Functional Annotation and Genomic Properties of Strain 10-4
4.5. Plant Growth Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PGP | Plant Growth-Promoting |
| WGS | Whole-Genome Sequencing |
| BGCs | Biosynthetic Gene Clusters |
| SA | Salicylic Acid |
| IAA | Indole-3-Acetic Acid |
| LB | Luria–Bertani |
| CFU | Colony Forming Units |
| GABA | Gamma-Aminobutyric Acid |
| SOD | Superoxide Dismutase |
| ICS | Isochorismate Synthase |
| VOCs | Volatile Organic Compounds |
| EPS | Exopolysaccharide |
References
- NASA. National Aeronautics and Space Administration. 2021. Available online: https://earthobservatory.nasa.gov/Features/WorldOfChange/decadaltemp.php (accessed on 18 July 2025).
- FAO. Food and Agriculture Organization of the United Nations, Statistical Yearbook 2022. Rome. Available online: https://doi.org/10.4060/cc2211en (accessed on 18 July 2025). [CrossRef]
- Asseng, S.; Martre, P.; Maiorano, A.; Rötter, R.P.; O’Leary, G.J.; Fitzgerald, G.J.; Ewert, F. Climate change impact and adaptation for wheat protein. Glob. Change Biol. 2019, 25, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.; Waraic, E.A.; Akhtar, S.; Anjum, S.; Ahmad, T.; Mahboob, W.; Hafeez, O.B.A.; Tapera, T.; Labuschagne, M.; Rizwan, M. Physiological responses of wheat to drought stress and its mitigation approaches. Acta Physiol. Plant. 2018, 40, 80. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Fritschi, F.B.; Mittler, R. Global warming, climate change, and environmental pollution: Recipe for a multifactorial stress combination disaster. Trends Plant Sci. 2021, 26, 588–599. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Ali, S.; Mir, R.A.; Haque, M.A.; Danishuddin Almalki, M.A.; Alfredan, M.; Mir, Z.A. Exploring physiological and molecular dynamics of drought stress responses in plants: Challenges and future directions. Front. Plant Sci. 2025, 16, 1565635. [Google Scholar] [CrossRef]
- Sinha, R.; Irulappan, V.; Mohan-Raju, B.; Suganthi, A.; Senthil-Kumar, M. Impact of drought stress on simultaneously occurring pathogen infection in field-grown chickpea. Sci. Rep. 2019, 9, 5577. [Google Scholar] [CrossRef]
- Priya, R.S.; Yuvaraj, M.; Sharmila, R.; Jagathjothi, N.; Saranya, M.; Suganthi, N.; Sivaji, M. Effects of climate change on plant diseases. In Plant Quarantine Challenges Under Climate Change Anxiety; Abd-Elsalam, K.A., Abdel-Momen, S.M., Eds.; Springer Nature: Cham, Switzerland, 2024; pp. 183–225. [Google Scholar]
- SCIP Database. Stress Combinations and Their Interactions in Plants (SCIP) Database. Available online: http://223.31.159.3/plant_complete/index_orangesunset.php (accessed on 18 July 2025).
- Sena, L.; Mica, E.; Valè, G.; Vaccino, P.; Pecchioni, N. Exploring the potential of endophyte-plant interactions for improving crop sustainable yields in a changing climate. Front. Plant Sci. 2024, 15, 1349401. [Google Scholar] [CrossRef]
- Ali, S.; Glick, B. Bacterial alleviation of drought stress in plants: Recent advances and future challenges. Sustain. Agric. Under Drought Stress 2025, 367–383. [Google Scholar]
- Guo, S.; Pan, R.; Zhang, Y.; Gu, Q.; Shen, Q.; Yang, J.; Li, R. Plant-microbe interactions influence plant performance via boosting beneficial root-endophytic bacteria. Environ. Microbiome 2025, 20, 18. [Google Scholar] [CrossRef]
- Singh, Y.; Likhita, J.; Sinha, S.; Arya, M.; Garg, P. Emerging Role of beneficial microbes in plant stress management under climate change. In Plant-Microbiome Interactions for Climate-Resilient Agriculture; Pankaj, U., Babele, P., Singh, A.K., Eds.; Springer Nature: Singapore, 2025; pp. 121–147. [Google Scholar]
- Sorokan, A.V.; Gabdrakhmanova, V.F.; Mardanshin, I.S.; Maksimov, I.V. Effect of endophytic bacteria Bacillus subtilis 26D and Bacillus velezensis M66 on resistance of potato plants to the causative agent of early blight Alternaria solani. Appl. Biochem. Microbiol. 2024, 60, 1313–1320. [Google Scholar] [CrossRef]
- König, F.; Sandri, M.R.; Russi, A.; Granada, C.E.; Schwambach, J. Biocontrol of tomato pathogens by Bacillus subtilis F62 and its synergistic action in plant growth promotion with Rhizobium sp. L5. Biocontrol Sci. Technol. 2024, 34, 551–565. [Google Scholar] [CrossRef]
- Yadav, U.; Anand, V.; Kumar, S.; Verma, I.; Anshu, A.; Pandey, I.A.; Singh, P.C. Bacillus subtilis NBRI-W9 simultaneously activates SAR and ISR against Fusarium chlamydosporum NBRI-FOL7 to increase wilt resistance in tomato. J. Appl. Microbiol. 2024, 135, lxae013. [Google Scholar] [CrossRef]
- Mannaa, M.; Park, A.R.; Kim, J.C.; Seo, Y.S. Microbial allies recruited by Bacillus subtilis JCK-1398 to defend pine trees against pinewood nematode. Sci. Rep. 2025, 15, 9670. [Google Scholar] [CrossRef]
- Vasantha-Srinivasan, P.; Park, K.B.; Kim, K.Y.; Jung, W.J.; Han, Y.S. The role of Bacillus species in the management of plant-parasitic nematodes. Front. Microbiol. 2025, 15, 1510036. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, M.; Ali, Q.; Zhao, W.; Chi, Y.K.; Ali, F.; Rashid, K.A.; Qi, R.D. Exploring plant growth promoting traits and biocontrol potential of new isolated Bacillus subtilis BS-2301 strain in suppressing Sclerotinia sclerotiorum through various mechanisms. Front. Plant Sci. 2024, 15, 1444328. [Google Scholar] [CrossRef]
- Rumyantsev, S.D.; Alekseev, V.Y.; Sorokan, A.V.; Burkhanova, G.F.; Cherepanova, E.A.; Maksimov, I.V.; Veselova, S.V. Search for biocontrol agents among endophytic lipopeptide-synthesizing bacteria Bacillus spp. to protect wheat plants against Greenbug aphid (Schizaphis graminum). Vavilov J. Genet. Breed. 2024, 28, 276. [Google Scholar] [CrossRef]
- Janaki, M.; Sivadasan Unni, P.K.; Stanley-Raja, V.; Senthil-Nathan, S.; Almutairi, B.O.; Abdel-Megeed, A. Biocontrol effect of Bacillus subtilis against Cnaphalocrocis medinalis (Guenèe) (Lepidoptera: Pyralidae): A sustainable approach to rice pest management. Agronomy 2024, 14, 310. [Google Scholar] [CrossRef]
- Gagné-Bourque, F.; Bertrand, A.; Claessens, A.; Aliferis, K.A.; Jabaji, S. Alleviation of drought stress and metabolic changes in timothy (Phleum pratense L.) colonized with Bacillus subtilis B26. Front. Plant Sci. 2016, 7, 584. [Google Scholar] [CrossRef]
- Puthiyottil, P.; Akkara, Y. Pretreatment with Bacillus subtilis mitigates drought induced photo-oxidative damages in okra by modulating antioxidant system and photochemical activity. Physiol. Mol. Biol. Plants 2021, 27, 945–957. [Google Scholar] [CrossRef]
- Fonseca, M.C.D.; Bossolani, J.W.; de Oliveira, S.L.; Moretti, L.G.; Portugal, J.R.; Scudeletti, D.; de Oliveira, E.F.; Crusciol, C.A.C. Bacillus subtilis inoculation improves nutrient uptake and physiological activity in sugarcane under drought stress. Microorganisms 2022, 10, 809. [Google Scholar] [CrossRef]
- Opadokun, E.O.; Wichai, T.; Kotchaplai, P. Effect of γ-polyglutamic acid produced by drought tolerant Bacillus subtilis FSO3 on soil moisture retention. J. Basic Microbiol. 2025, 65, e70047. [Google Scholar] [CrossRef]
- Ji, C.; Tian, H.; Wang, X.; Song, X.; Ju, R.; Li, H.; Gao, Q.; Li, C.; Zhang, P.; Li, J.; et al. Bacillus subtilis HG-15, a halotolerant rhizoplane bacterium, promotes growth and salinity tolerance in wheat (Triticum aestivum). BioMed Res. Int. 2022, 2022, 9506227. [Google Scholar] [CrossRef]
- Patel, M.; Islam, S.; Husain, F.M.; Yadav, V.K.; Park, H.K.; Yadav, K.K.; Bagatharia, S.; Joshi, M.; Jeon, B.H.; Patel, A. Bacillus subtilis ER-08, a multifunctional plant growth-promoting rhizobacterium, promotes the growth of fenugreek (Trigonella foenum-graecum L.) plants under salt and drought stress. Front. Microbiol. 2023, 14, 1208743. [Google Scholar] [CrossRef] [PubMed]
- da Silva, D.M.M.; Santos, C.C.; Wagner, F.E.; Martins, L.O.M.; Ozório, J.P.A.; da Silva, O.A.; Ribeiro, D.M.; Scalon, S.P.Q. Seed biopriming with Parachlorella, Bacillus subtilis, and Trichoderma harzianum alleviates the effects of salinity in soybean. BMC Plant Biol. 2024, 24, 1149. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Rehman, A.; Akram, W.; Anjum, T.; Yasin, N.A.; Aftab, Z.E.; Munir, B.; Khan, W.U.; Li, G. Unlocking salinity stress resilience in turnip (Brassica rapa subsp. rapa) plants using Bacillus subtilis Z-12 and Bacillus aryabhattai Z-48. Microorganisms 2025, 13, 359. [Google Scholar] [CrossRef] [PubMed]
- Harwood, C.R. Bacillus subtilis and its relatives: Molecular biological and industrial workhorses. Trends Biotechnol. 1992, 10, 247–256. [Google Scholar] [CrossRef]
- Su, Y.; Liu, C.; Fang, H.; Zhang, D. Bacillus subtilis: A universal cell factory for industry, agriculture, biomaterials and medicine. Microb. Cell Factories 2020, 19, 173. [Google Scholar] [CrossRef]
- Kumar, A.; Sangwan, P.; Kumar, V.; Pandey, A.K.; Pooja Kumar, A.; Chauhan, P.; Koubouris, G.; Fanourakis, D.; Parmar, K. Physio-biochemical insights of endophytic microbial community for crop stress resilience: An updated overview. J. Plant Growth Regul. 2025, 44, 2641–2664. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Y.; Jia, B.; Tao, L.; Li, H.; Wang, J.; Yao, Y. Four decades of Bacillus biofertilizers: Advances and future prospects in agriculture. Microorganisms 2025, 13, 187. [Google Scholar] [CrossRef]
- Lastochkina, O.V. Adaptation and tolerance of wheat plants to drought mediated by natural growth regulators Bacillus spp.: Mechanisms and practical importance. Agric. Biol. 2021, 56, 843–867. [Google Scholar] [CrossRef]
- Lastochkina, O.; Bosacchi, M. Potential roles for endophytic plant growth-promoting microbes in wheat adaptation and tolerance under herbicide and drought stresses combination. Turk. J. Agric. For. 2023, 47, 688–712. [Google Scholar] [CrossRef]
- Lastochkina, O.; Aliniaeifard, S.; Garshina, D.; Garipova, S.; Pusenkova, L.; Allagulova, C.; Fedorova, K.; Baymiev, A.; Koryakov, I.; Sobhani, M. Seed priming with endophytic Bacillus subtilis strain-specifically improves growth of Phaseolus vulgaris plants under normal and salinity conditions and exerts anti-stress effect through induced lignin deposition in roots and decreased oxidative and osmotic damages. J. Plant Physiol. 2021, 263, 153462. [Google Scholar]
- Lastochkina, O.V.; Garipova, S.R.; Pusenkova, L.I.; Garshina, D.Y.; Baymiev, A.K.; Koryakov, I.S. Effect of endophytic bacteria Bacillus subtilis on seedling growth and root lignification of Pisum sativum L. under normal and sodium chloride salt conditions. Russ. J. Plant Physiol. 2023, 70, 97. [Google Scholar] [CrossRef]
- Pusenkova, L.; Lastochkina, O.; Ercişli, S. The Potential of hydroponic seed minituber enrichment with the endophyte Bacillus subtilis for improving the yield components and quality of potato (Solanum tuberosum L.). Agriculture 2023, 13, 1626. [Google Scholar] [CrossRef]
- Lastochkina, O.; Pusenkova, L.; Yuldashev, R.; Babaev, M.; Garipova, S.; Blagova, D.; Khairullin, R.; Aliniaeifard, S. Effects of Bacillus subtilis on some physiological and biochemical parameters of Triticum aestivum L. (wheat) under salinity. Plant Physiol. Biochem. 2017, 121, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Lastochkina, O.; Garshina, D.; Ivanov, S.; Yuldashev, R.; Khafizova, R.; Allagulova, C.; Fedorova, K.; Avalbaev, A.; Maslennikova, D.; Bosacchi, M. Seed priming with endophytic Bacillus subtilis modulates physiological responses of two different Triticum aestivum L. cultivars under drought stress. Plants 2020, 9, 1810. [Google Scholar] [CrossRef]
- Maslennikova, D.; Koryakov, I.; Yuldashev, R.; Avtushenko, I.; Yakupova, A.; Lastochkina, O. Endophytic plant growth-promoting bacterium Bacillus subtilis reduces the toxic effect of cadmium on wheat plants. Microorganisms 2023, 11, 1653. [Google Scholar] [CrossRef]
- Lastochkina, O.; Baymiev, A.; Shayahmetova, A.; Garshina, D.; Koryakov, I.; Shpirnaya, I.; Pusenkova, L.; Mardanshin, I.; Kasnak, C.; Palamutoglu, R. Effects of endophytic Bacillus subtilis and salicylic acid on postharvest diseases (Phytophthora infestans, Fusarium oxysporum) development in stored potato tubers. Plants 2020, 9, 76. [Google Scholar] [CrossRef]
- Lastochkina, O.; Garshina, D.; Allagulova, C.; Fedorova, K.; Koryakov, I.; Vladimirova, A. Application of endophytic Bacillus subtilis and salicylic acid to improve wheat growth and tolerance under combined drought and fusarium root rot stresses. Agronomy 2020, 10, 1343. [Google Scholar] [CrossRef]
- Lastochkina, O.V.; Pusenkova, L.I.; Il’yasova, E.Y.; Aliniaeifard, S. Effect of Bacillus subtilis based biologicals on physiological and biochemical parameters of sugar beat (Beta vulgaris L.) plants infected with Alternaria alternata. Agrobiology 2018, 53, 958–968. [Google Scholar]
- Lastochkina, O.; Garshina, D.; Ibragimov, A. Enhanced performance of endophytic Bacillus subtilis in conjunction with salicylic acid meliorated simultaneous drought and fusarium root rot stresses in Triticum aestivum L. Biol. Life Sci. Forum 2021, 4, 20. [Google Scholar]
- Lastochkina, O.; Yakupova, A.; Avtushenko, I.; Lastochkin, A.; Yuldashev, R. Effect of seed priming with endophytic Bacillus subtilis on some physio-biochemical parameters of two wheat varieties exposed to drought after selective herbicide application. Plants 2023, 12, 1724. [Google Scholar] [CrossRef]
- Markova, O.; Garipova, S.; Chistoedova, A.; Matyunina, V.; Lubyanova, A.; Lastochkina, O.; Garipov, A.; Shpirnaya, I.; Pusenkova, L. Predicting field effectiveness of endophytic Bacillus subtilis inoculants for common bean using morphometric and biochemical markers. Plants 2024, 13, 1769. [Google Scholar] [CrossRef]
- Pusenkova, L.I.; Garipova, S.R.; Lastochkina, O.V.; Valieva, V.A.; Chistoedova, A.V.; Matyunina, V.D. Pre-planting inoculation with endophytes as a way to manage seed productivity and quality of healthy tubers of potato. Agrochem. Her. 2024, 5, 78–86. [Google Scholar]
- Lastochkina, O.; Yuldashev, R.; Avalbaev, A.; Allagulova, C.; Veselova, S. The contribution of hormonal changes to the protective effect of endophytic bacterium Bacillus subtilis on two wheat genotypes with contrasting drought sensitivities under osmotic stress. Microorganisms 2023, 11, 2955. [Google Scholar] [CrossRef]
- Lastochkina, O.; Ivanov, S.; Petrova, S.; Garshina, D.; Lubyanova, A.; Yuldashev, R.; Kuluev, B.; Zaikina, E.; Maslennikova, D.; Allagulova, C.; et al. Role of endogenous salicylic acid as a hormonal intermediate in the bacterial endophyte Bacillus subtilis-induced protection of wheat genotypes contrasting in drought susceptibility under dehydration. Plants 2022, 11, 3365. [Google Scholar] [CrossRef] [PubMed]
- Lubyanova, A.R.; Allagulova, C.R.; Lastochkina, O.V. The effects of seed pretreatment with endophytic bacteria Bacillus subtilis on the water balance of spring and winter wheat seedlings under short-time water deficit. Plants 2023, 12, 2684. [Google Scholar] [CrossRef]
- Maslennikova, D.; Lastochkina, O. Contribution of ascorbate and glutathione in endobacteria Bacillus subtilis-mediated drought tolerance in two Triticum aestivum L. genotypes contrasting in drought sensitivity. Plants 2021, 10, 2557. [Google Scholar] [CrossRef] [PubMed]
- Trotel-Aziz, P.; Couderchet, M.; Biagianti, S.; Aziz, A. Characterization of new bacterial biocontrol agents Acinetobacter, Bacillus, Pantoea and Pseudomonas spp. mediating grapevine resistance against Botrytis cinerea. Environ. Exp. Bot. 2008, 64, 21–32. [Google Scholar] [CrossRef]
- Trotel-Aziz, P.; Abou-Mansour, E.; Courteaux, B.; Rabenoelina, F.; Clément, C.; Fontaine, F.; Aziz, A. Bacillus subtilis PTA-271 counteracts Botryosphaeria dieback in grapevine, triggering immune responses and detoxification of fungal phytotoxins. Front. Plant Sci. 2019, 10, 25. [Google Scholar] [CrossRef]
- Thiruvengadam, R.; Gandhi, K.; Vaithiyanathan, S.; Sankarasubramanian, H.; Loganathan, K.; Lingan, R.; Rajagopalan, V.R.; Muthurajan, R.; Ebenezer Iyadurai, J.; Kuppusami, P. Complete genome sequence analysis of Bacillus subtilis Bbv57, a promising biocontrol agent against phytopathogens. Int. J. Mol. Sci. 2022, 23, 9732. [Google Scholar] [CrossRef] [PubMed]
- Hazarika, D.J.; Bora, S.S.; Naorem, R.S.; Sharma, D.; Boro, R.C.; Barooah, M. Genomic insights into Bacillus subtilis MBB3B9 mediated aluminium stress mitigation for enhanced rice growth. Sci. Rep. 2023, 13, 16467. [Google Scholar] [CrossRef] [PubMed]
- Douka, D.; Spantidos, T.N.; Tsalgatidou, P.C.; Katinakis, P.; Venieraki, A. Whole-genome profiling of endophytic strain BL Ns. 14 from Nigella sativa reveals potential for agricultural bioenhancement. Microorganisms 2024, 12, 2604. [Google Scholar] [CrossRef]
- Iqbal, S.; Qasim, M.; Rahman, H.; Khan, N.; Paracha, R.Z.; Bhatti, M.F.; Javed, A.; Janjua, H.A. Genome mining, antimicrobial and plant growth-promoting potentials of halotolerant Bacillus paralicheniformis ES-1 isolated from salt mine. Mol. Genet. Genom. 2023, 298, 79–93. [Google Scholar] [CrossRef]
- Belaouni, H.; Compant, S.; Antonielli, L.; Nikolic, B.; Zitouni, A.; Sessitsch, A. In-depth genome analysis of Bacillus sp. BH32, a salt stress-tolerant endophyte obtained from a halophyte in a semiarid region. Appl. Microbiol. Biotechnol. 2022, 106, 3113–3137. [Google Scholar] [CrossRef]
- Narayanasamy, S.; Thankappan, S.; Kumaravel, S.; Ragupathi, S.; Uthandi, S. Complete genome sequence analysis of a plant growth-promoting phylloplane Bacillus altitudinis FD48 offers mechanistic insights into priming drought stress tolerance in rice. Genomics 2023, 115, 110550. [Google Scholar] [CrossRef]
- Su, Z.; Liu, G.; Liu, X.; Li, S.; Lu, X.; Wang, P.; Zhao, W.; Zhang, X.; Dong, L.; Qu, Y.; et al. Functional analyses of the Bacillus velezensis HMB26553 genome provide evidence that its genes are potentially related to the promotion of plant growth and prevention of cotton rhizoctonia damping-off. Cells 2023, 12, 1301. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Z.; Zhao, Q.; Yang, X.; Li, Y.; Zhou, H.; Zheng, H. Whole-genome analysis revealed the growth-promoting and biological control mechanism of the endophytic bacterial strain Bacillus halotolerans Q2H2, with strong antagonistic activity in potato plants. Front. Microbiol. 2024, 14, 1287921. [Google Scholar] [CrossRef]
- Etesami, H.; Glick, B.R. Bacterial indole-3-acetic acid: A key regulator for plant growth, plant-microbe interactions, and agricultural adaptive resilience. Microbiol. Res. 2024, 281, 127602. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Li, Y.; Zhang, L.; Mu, J.; Jiang, Y.; Fu, H.; Zhang, Y.; Cui, H.; Yu, X.; Ye, Z. Biosynthetic pathways and functions of indole-3-acetic acid in microorganisms. Microorganisms 2023, 11, 2077. [Google Scholar] [CrossRef]
- Duca, D.R.; Glick, B.R. Indole-3-acetic acid biosynthesis and its regulation in plant-associated bacteria. Appl. Microbiol. Biotechnol. 2020, 104, 8607–8619. [Google Scholar] [CrossRef]
- Pahari, A.; Pradhan, A.; Nayak, S.K.; Mishra, B.B. Bacterial siderophore as a plant growth promoter. In Microbial Biotechnology: Applications in Agriculture and Environment; Patra, J., Vishnuprasad, C., Das, G., Eds.; Springer: Singapore, 2018; Volume 1, pp. 163–180. [Google Scholar]
- Qi, Y.; Kobayashi, Y.; Hulett, F.M. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J. Bacteriol. 1997, 179, 2534–2539. [Google Scholar] [CrossRef]
- Sessitsch, A.; Hardoim, P.; Döring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant-Microbe Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genomics 2016, 17, 882. [Google Scholar] [CrossRef] [PubMed]
- Hanif, A.; Zhang, F.; Li, P.; Li, C.; Xu, Y.; Zubair, M.; Zhang, M.; Jia, D.; Zhao, X.; Liang, J.; et al. Fengycin produced by Bacillus amyloliquefaciens FZB42 inhibits Fusarium graminearum growth and mycotoxins biosynthesis. Toxins 2019, 11, 295. [Google Scholar] [CrossRef]
- Ma, L.; Lu, Y.; Yan, H.; Wang, X.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Screening of cellulolytic bacteria from rotten wood of Qinling (China) for biomass degradation and cloning of cellulases from Bacillus methylotrophicus. BMC Biotechnol. 2020, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, H.; Huang, Q.; Zhang, Y.; Zhao, L.; Liu, F.; Wang, G. Four superoxide dismutases of Bacillus cereus 0–9 are non-redundant and perform different functions in diverse living conditions. World J. Microbiol. Biotechnol. 2020, 36, 12. [Google Scholar] [CrossRef]
- Kappes, R.M.; Kempf, B.; Bremer, E. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: Haracterization of OpuD. J. Bacteriol. 1996, 178, 5071–5079. [Google Scholar] [CrossRef]
- Xie, S.S.; Wu, H.J.; Zang, H.Y.; Wu, L.M.; Zhu, Q.Q.; Gao, X.W. Plant growth promotion by spermidine-producing Bacillus subtilis OKB105. Mol. Plant Microbe Interact. 2014, 27, 655–663. [Google Scholar] [CrossRef]
- Gong, Y.; Liu, J.Q.; Xu, M.J.; Zhang, C.M.; Gao, J.; Li, C.G.; Xing, K.; Qin, S. Antifungal volatile organic compounds from Streptomyces setonii WY228 control black spot disease of sweet potato. Appl. Environ. Microbiol. 2022, 88, 0231721. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.E.Á.; de Brito, M.V.; Pimenta, A.T.Á.; da Silva, G.S.; Zocolo, G.J.; Muniz, C.R.; de Medeiros, S.C.; Grangeiro, T.B.; Lima, M.A.S.; da Silva, C.F.B. Antagonism of volatile organic compounds of the Bacillus sp. against Fusarium kalimantanense. World J. Microbiol. Biotechnol. 2022, 39, 60. [Google Scholar] [CrossRef]
- Park, Y.; Solhtalab, M.; Thongsomboon, W.; Aristilde, L. Strategies of organic phosphorus recycling by soil bacteria: Acquisition, metabolism, and regulation. Environ. Microbiol. Rep. 2022, 14, 3–24. [Google Scholar] [CrossRef]
- Xie, F.; Li, G.; Wang, Y.; Zhang, Y.; Zhou, L.; Wang, C.; Liu, S.; Liu, S.; Wang, C. Pyridoxal phosphate synthases PdxS/PdxT are required for Actinobacillus pleuropneumoniae viability, stress tolerance and virulence. PLoS ONE 2017, 12, e0176374. [Google Scholar] [CrossRef]
- Decsi, K.; Ahmed, M.; Abdul-Hamid, D.; Tóth, Z. The role of salicylic acid in activating plant stress responses—Results of the past decade and future perspectives. Int. J. Mol. Sci. 2025, 26, 4447. [Google Scholar] [CrossRef]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic Acid Biosynthesis in Plants. Front. Plant Sci. 2020, 11, 338, Erratum in Front. Plant Sci. 2025, 16, 1697849. https://doi.org/10.3389/fpls.2025.1697849. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Sardà Carbasse, J.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid. Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 106, 645–667. [Google Scholar] [CrossRef]
- Kreft, L.; Botzki, A.; Coppens, F.; Vandepoele, K.; Van Bel, M. PhyD3: A phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 2017, 33, 2946–2947. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. Antismash 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. Antismash 7.0: New and improved predictions for detection, regulation, chemical structures and visualization. Nucleic Acid. Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- GOST 33777-2016; Method for Determination of Phytotoxicity for Higher Plants Seeds. Standartinform: Moscow, Russia, 2016. Available online: https://files.stroyinf.ru/Data2/1/4293754/4293754192.pdf (accessed on 12 September 2025).
- Dospekhov, M.B. Methods of Field Experiment; Kolos: Moscow, Russia, 1979; 416p. [Google Scholar]
- GOST 26213-91; Soils. Methods for Determination of Organic Matter. Izdatel’stvo Standartov: Moscow, Russia, 1992. Available online: https://docs.cntd.ru/document/1200023481 (accessed on 21 April 2025).
- GOST 26483-85; Soils. Preparation of Salt Extract and Determination of Its pH by CINAO Method. Izdatel’stvo Standartov: Moscow, Russia, 1985. Available online: https://files.stroyinf.ru/Data/292/29278.pdf (accessed on 21 April 2025).
- GOST 26205-91; Soils. Determination of Mobile Compounds of Phosphorus and Potassium by Machigin Method Modified by CINAO. Izdatel’stvo Standartov: Moscow, Russia, 1993. Available online: https://files.stroyinf.ru/Data2/1/4294828/4294828275.html (accessed on 21 April 2025).
- GOST 26951; Soils. Determination of Nitrates by Ionometric Method. Izdatel’stvo Standartov: Moscow, Russia, 1993. Available online: https://docs.cntd.ru/document/1200023499 (accessed on 21 April 2025).
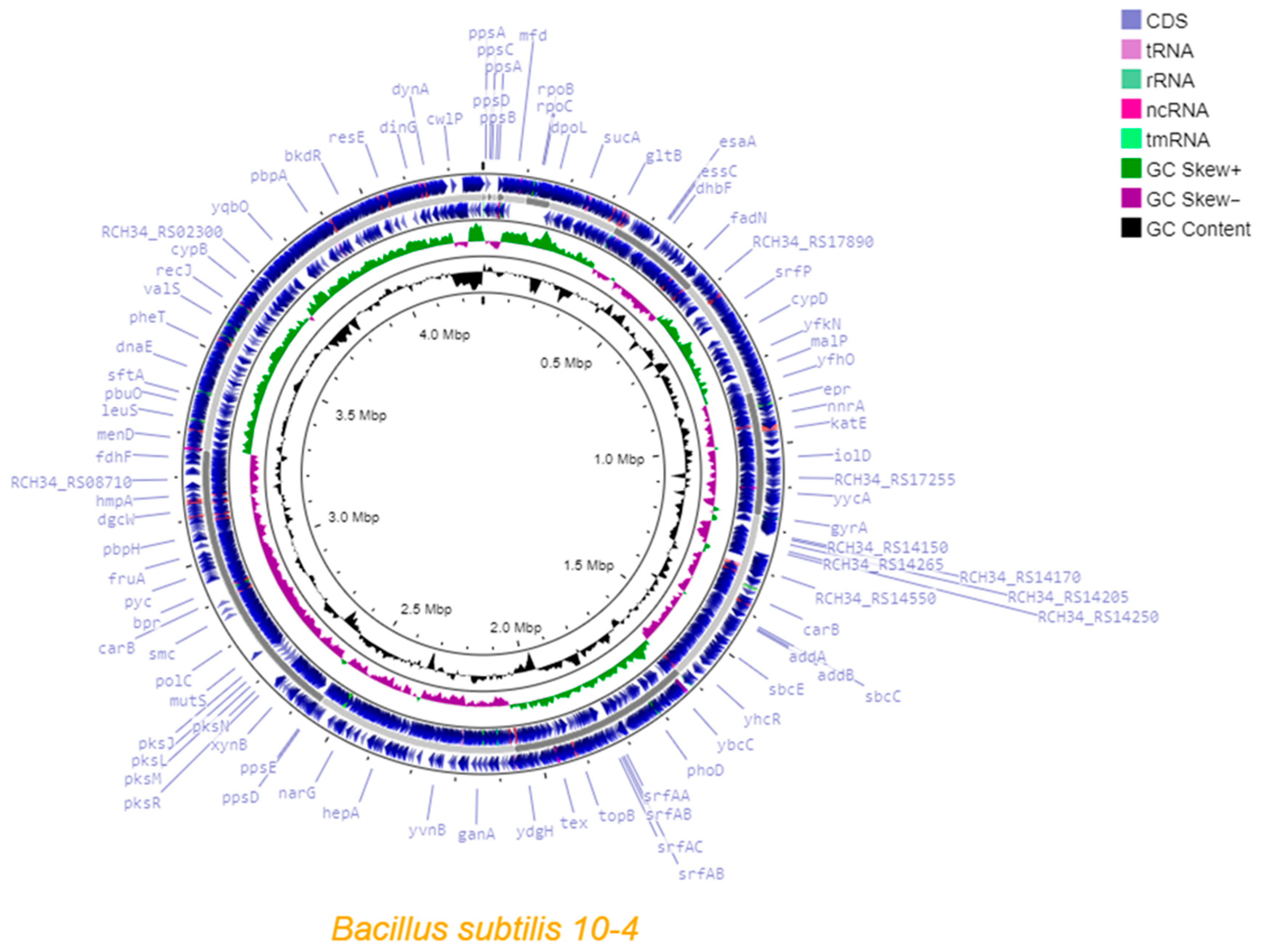




| Genome Size (bp) | 4,278,582 (4.3 Mb) |
| Number of Contigs | 19 |
| Contig N50 | 496.9 kb |
| Contig L50 | 3 |
| G+C Content (%) | 43.5 |
| Genome Coverage | 99.0× |
| Assembly level | contig |
| Genes (total) | 4476 |
| CDSs (total) | 4473 |
| Genes (coding) | 4314 |
| CDSs (with protein) | 4314 |
| Genes (RNA) | 53 |
| rRNAs | 1, 3, 2 (5S, 16S, 23S) |
| Complete rRNAs | 1, 1 (5S, 16S) |
| Partial rRNAs | 2, 2 (16S, 23S) |
| tRNAs | 42 |
| ncRNAs | 5 |
| Pseudo Genes (total) | 109 |
| Pseudo Genes (without protein) | 109 |
| Genome | Strains | |||||
|---|---|---|---|---|---|---|
| B. subtilis 10-4 | B. subtilis 26D | B. subtilis PTA-271 | B. subtilis Bbv57 | B. subtilis MBB3B9_DBT-NECAB | B. subtilis subsp. subtilis str. 168 | |
| NCBI number | PRJNA1008864 | PRJNA1182114 | RJNA646528 | PRJNA794929 | PRJNA786394 | SAMEA3138188 |
| Genome size (bp) | 4,278,582 | 4,160,174 | 4,190,000 | 4,302,465 | 4,149,783 | 4,215,606 |
| Genes (total) | 4476 | 4342 | 4141 | 4363 | 4354 | 4135 |
| CDSs (with protein) | 4314 | 4133 | 3940 | 4281 | 4163 | 4120 |
| rRNAs | 1, 3, 2 (5S, 16S, 23S) | 9, 7, 4 (5S, 16S, 23S) | 9, 7, 4 (5S, 16S, 23S) | 5 | 5, 1, 1 (5S, 16S, 23S) | 30 (5S, 16S, 23S) |
| tRNAs | 42 | 82 | 81 | 76 | 85 | 86 |
| ncRNAs | 5 | 5 | 5 | 5 | 1 | |
| Pseudo Genes (total) | 109 | 102 | 95 | 27 | 94 | 13 |
| Source | Soil | Cotton Leaves | Gravepine Rhizospheric Soil | Soil | Soil | Soil |
| Properties | Biocontrol, growth promotion | Biocontrol, growth promotion | Biocontrol | Biocontrol | Biocontrol, growth promotion | Reference Lab Domesticated |
| PGP Activities | Gene Name | Function |
|---|---|---|
| Auxin biosynthesis | trpA, trpB, trpC, trpP, trpD, trpE, trpF, TRPs, TSa, TSb | Tryptophan biosynthesis |
| Nitrogen metabolism | glnG, nrgB | Regulation of nitrogen metabolism |
| NarG, NorD, NarH, NarJ, NorQ, NarI | Denitrification | |
| Amt, NsrR | Assimilation | |
| NarH, NarJ, NarG, NiR1b, NarI, NiR1a | Ammonification of nitrates and nitrites | |
| gltA, gltB, glnA, fpgS | Glutamate/glutamine metabolism | |
| narT, nasA, nasB, nasC, nasD, nasE, narX, narG, narH | Nitrate/nitrite assimilation and metabolism | |
| Phosphorus metabolism | pstA, pstC, pstS | Phosphate-binding and transport |
| pstB | Phosphate transport system energetics | |
| pitA | Alternative phosphate transport | |
| NaPi, PhoH, PhoP, PhoH, PpaX2, PhoR | Phosphate metabolism | |
| Iron acquisition and metabolism | dhbC, FeuA, FeuB, dhbB, FeuC, dhbE, dhbF, yuiI, dhbA, Hyp1, Fe-ABC1, X-ABC3, X-ABC2 | Siderophores |
| X-ABC1, X-ABC3, HtsA, S, A, HtsB, ZnH, Hyp, HtsC, X-ABC2, R4 | Heme, hemin uptake and utilization systems in GramPositives | |
| EfeB, EfeU, EfeO | Ferrous iron transporter EfeUOB, low-pH-induced | |
| Potassium metabolism | KefA | Potassium homeostasis |
| Sulfur metabolism | AS | Galactosylceramide and Sulfatide metabolism |
| Tpx, AhpC-like, TrxR, Bcp | Thioredoxin-disulfide reductase |
| Trait | Gene Name | Function |
|---|---|---|
| General stress | RsbWUSVRT, SigB, ysnF, yhdN CstA, Csr RseP | Stress response regulation Carbon starvation Periplasmic stress |
| Drought, salt stress | opuCA, opuCB, opuCC, opuCD, opuD | Glycine betaine/choline transporter |
| Oxidative stresses | gpx GloA sodA, sodB, sodC, sodMn, PerR, Osmcl, OsmclR, AhpC, NsrR, fur | Glutathione: redox cycle Glutathione: non-redox reactions Protection against ROS Oxidative stress protection |
| Osmotic stress | OpuD, OpuBA, GbsB, OpuAC, OpuAA, OpuBC, OpuBB, OpuBD, OpuAB, BetB glpF | Choline and betaine uptake and betaine biosynthesis Osmoregulation |
| Parameter | B. subtilis Strain 10-4 Cells Concentration (CFU mL−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 (H2O) | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | |
| Oat (Avena sativa L.) | |||||||||
| Root Leight (cm) | 1.6 ± 0.5 | 1.6 ± 0.6 | 1.7 ± 0.6 | 1.8 ± 0.6 | 2 ± 0.7 | 2.1 ± 0.6 | 1.9 ± 0.8 | 2 ± 0.6 | 1.8 ± 0.6 |
| % of Control | 100 e | 100 e | 106 d | 113 cd | 125 b | 131 a | 119 c | 125 b | 113 cd |
| Radish (Raphanus sativus L.) | |||||||||
| Root Leight (cm) | 2.1 ± 1.1 | 2.1 ± 1.3 | 2.1 ± 0.8 | 2.7 ± 1.3 | 2.8 ± 1.0 | 2.7 ± 1.4 | 2.9 ± 1.4 | 2.3 ± 1.0 | 2.1 ± 1.1 |
| % of Control | 100 e | 100 e | 100 f | 127 c | 127 c | 132 b | 136 a | 105 d | 100 e |
| Variant | Parameter | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (cm) | Fresh Biomass (g) | Dry Biomass (g) | Number of Productive Stems Plant−1 | ||||||||||
| Roots | Shoots | Roots | Shoots | Roots | Shoots | ||||||||
| dpi | dpi | dpi | dpi | dpi | dpi | dpi | |||||||
| 21 | 54 | 21 | 54 | 21 | 54 | 21 | 54 | 21 | 54 | 21 | 54 | 54 | |
| Control | 6.4 bD | 11.0 bC | 34.7 bD | 79.4 bB | 0.12 bD | 2.93 aB | 1.97 bD | 25.0 bB | 0.09 bD | 2.33 bB | 0.90 bD | 11.06 bB | 1.0 b |
| 10-4 | 12.7 aB | 13.4 aA | 37.8 aC | 81.1 aA | 0.18 aC | 3.56 aA | 3.50 aC | 30.0 aA | 0.15 aC | 2.78 aA | 1.49 aC | 13.98 aA | 1.26 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lastochkina, O.; Pusenkova, L. Whole-Genome Analysis of PGP Endophytic Bacillus subtilis 10-4: Unraveling Molecular Insights into Plant Growth and Stress Resilience. Int. J. Mol. Sci. 2025, 26, 11904. https://doi.org/10.3390/ijms262411904
Lastochkina O, Pusenkova L. Whole-Genome Analysis of PGP Endophytic Bacillus subtilis 10-4: Unraveling Molecular Insights into Plant Growth and Stress Resilience. International Journal of Molecular Sciences. 2025; 26(24):11904. https://doi.org/10.3390/ijms262411904
Chicago/Turabian StyleLastochkina, Oksana, and Liudmila Pusenkova. 2025. "Whole-Genome Analysis of PGP Endophytic Bacillus subtilis 10-4: Unraveling Molecular Insights into Plant Growth and Stress Resilience" International Journal of Molecular Sciences 26, no. 24: 11904. https://doi.org/10.3390/ijms262411904
APA StyleLastochkina, O., & Pusenkova, L. (2025). Whole-Genome Analysis of PGP Endophytic Bacillus subtilis 10-4: Unraveling Molecular Insights into Plant Growth and Stress Resilience. International Journal of Molecular Sciences, 26(24), 11904. https://doi.org/10.3390/ijms262411904









