Nodal Spread Prediction in Human Oral Tongue Squamous Cell Carcinoma Using a Cancer-Testis Antigen Genes Signature
Abstract
1. Introduction
2. Results
2.1. Patient Cohort and Clinical Characteristics (Table 1)
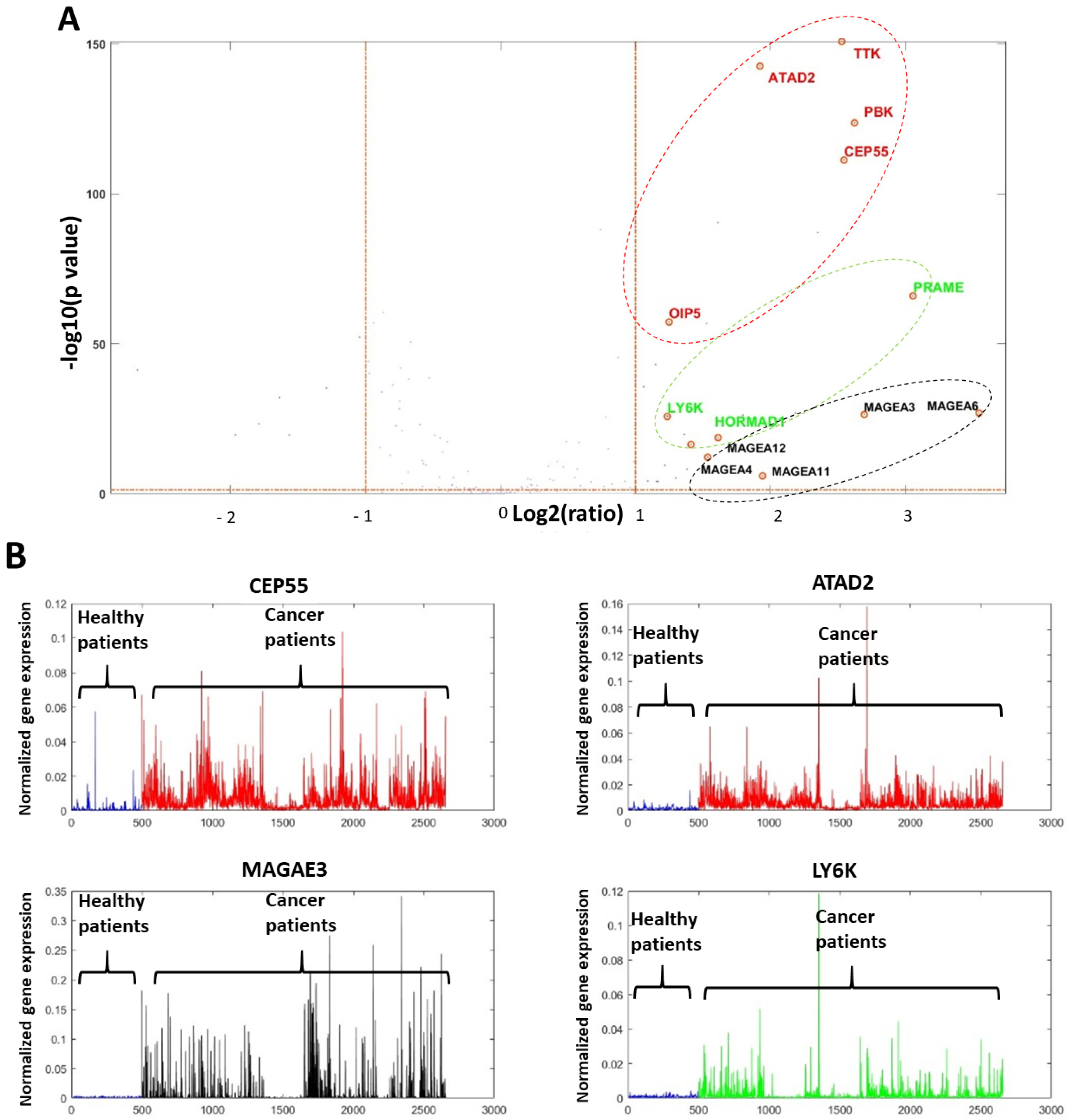
2.2. Genomic Computational Analysis
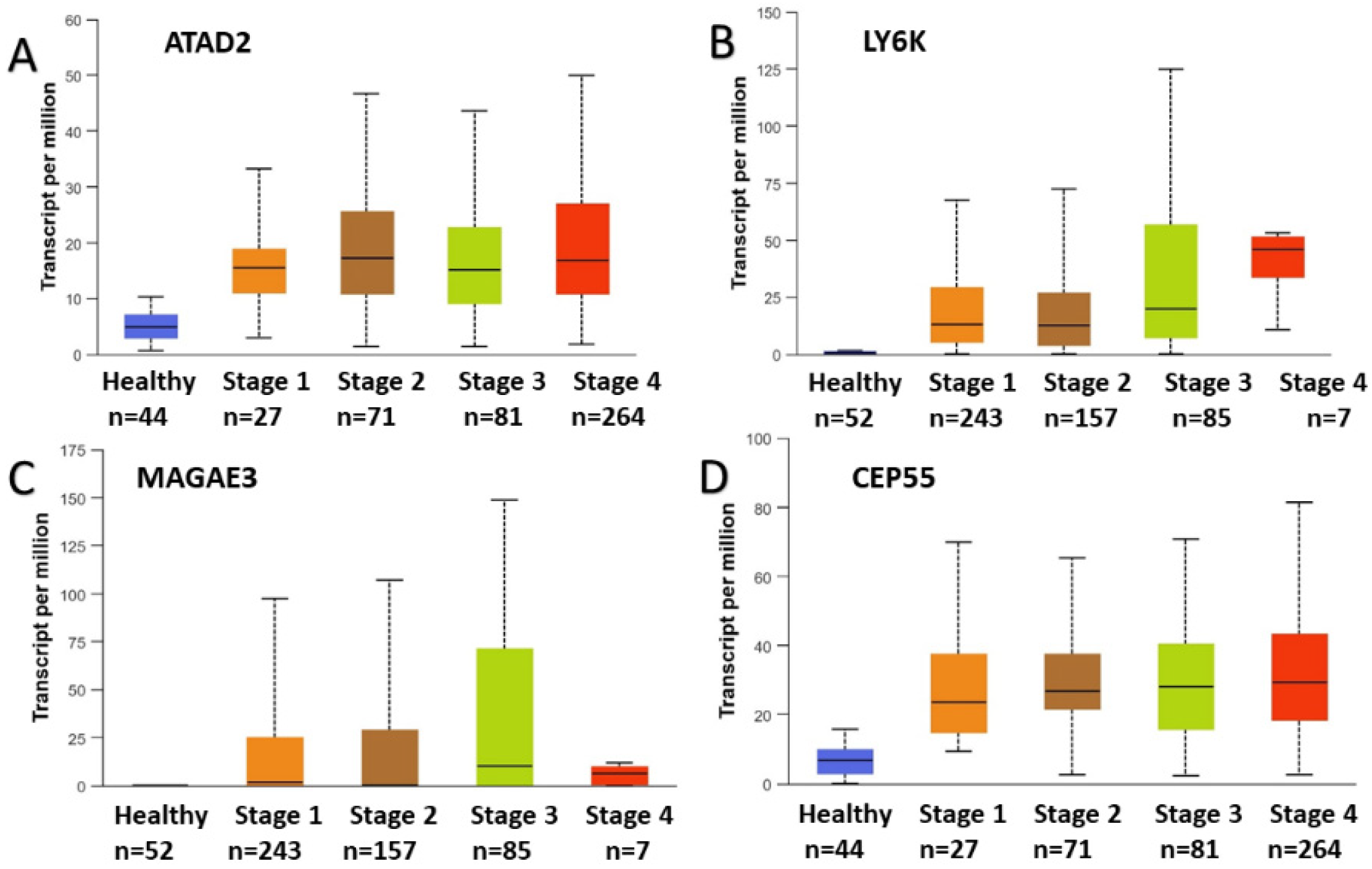
2.3. Focused Computational Analysis on Tongue Cancer Datasets
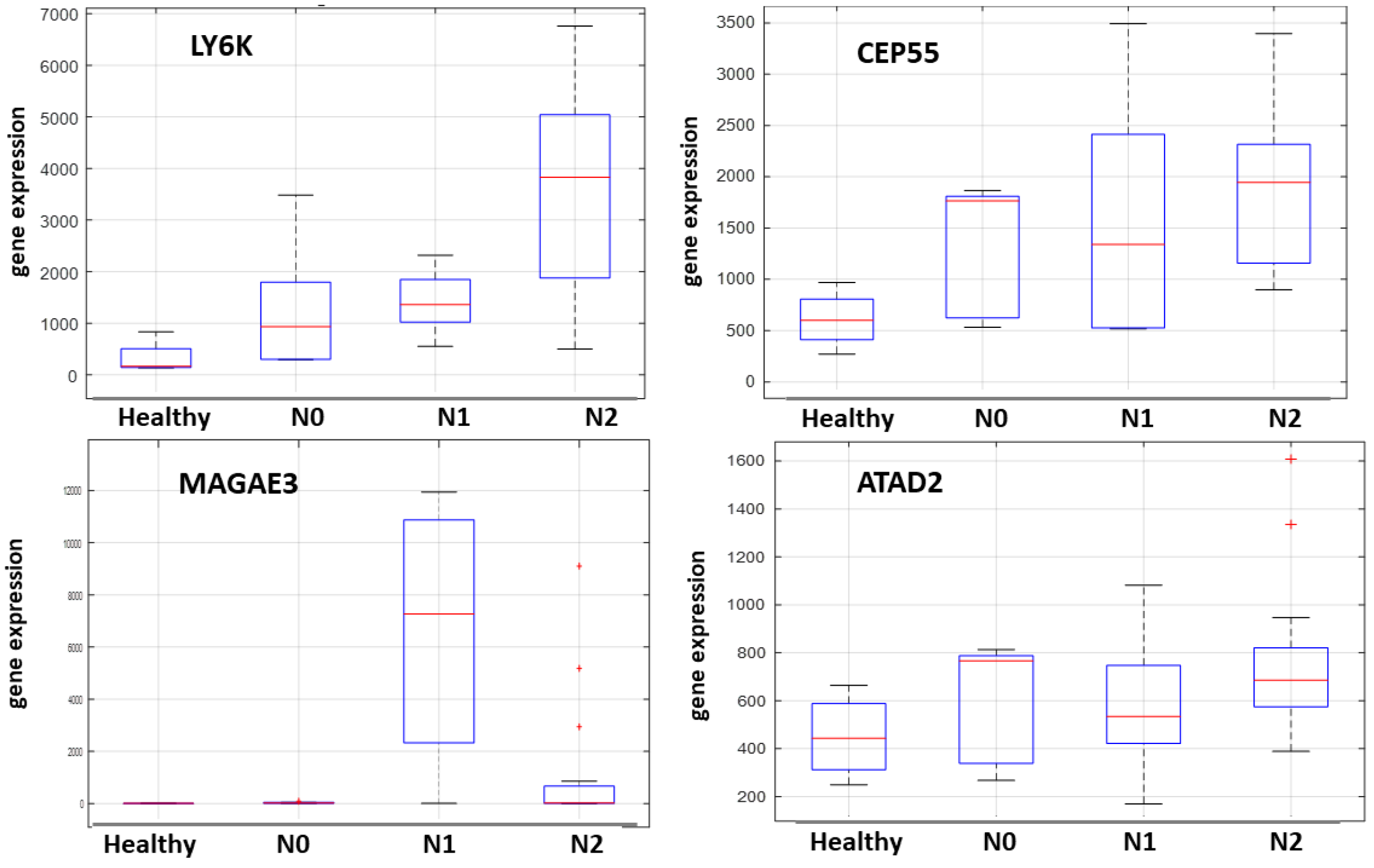
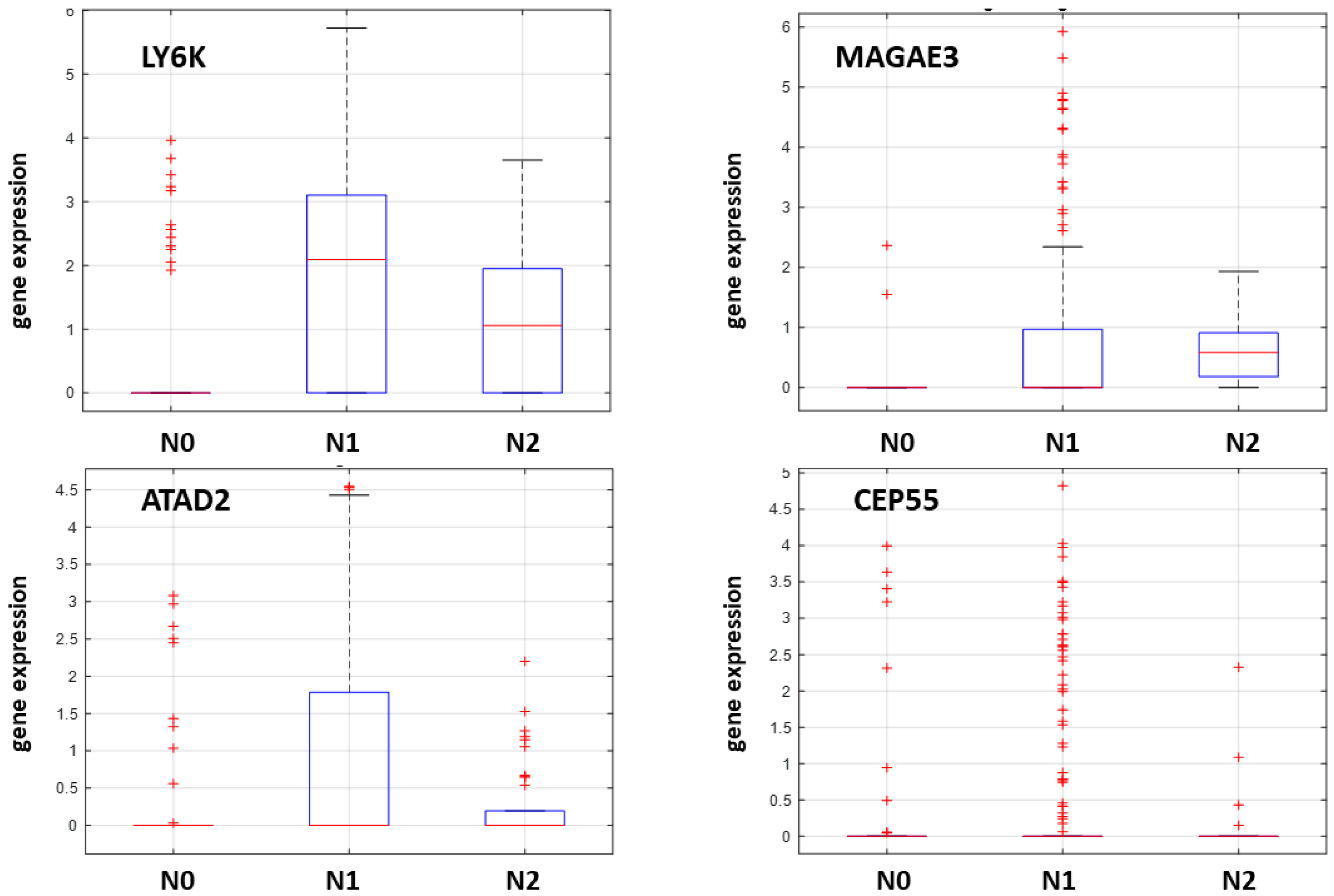
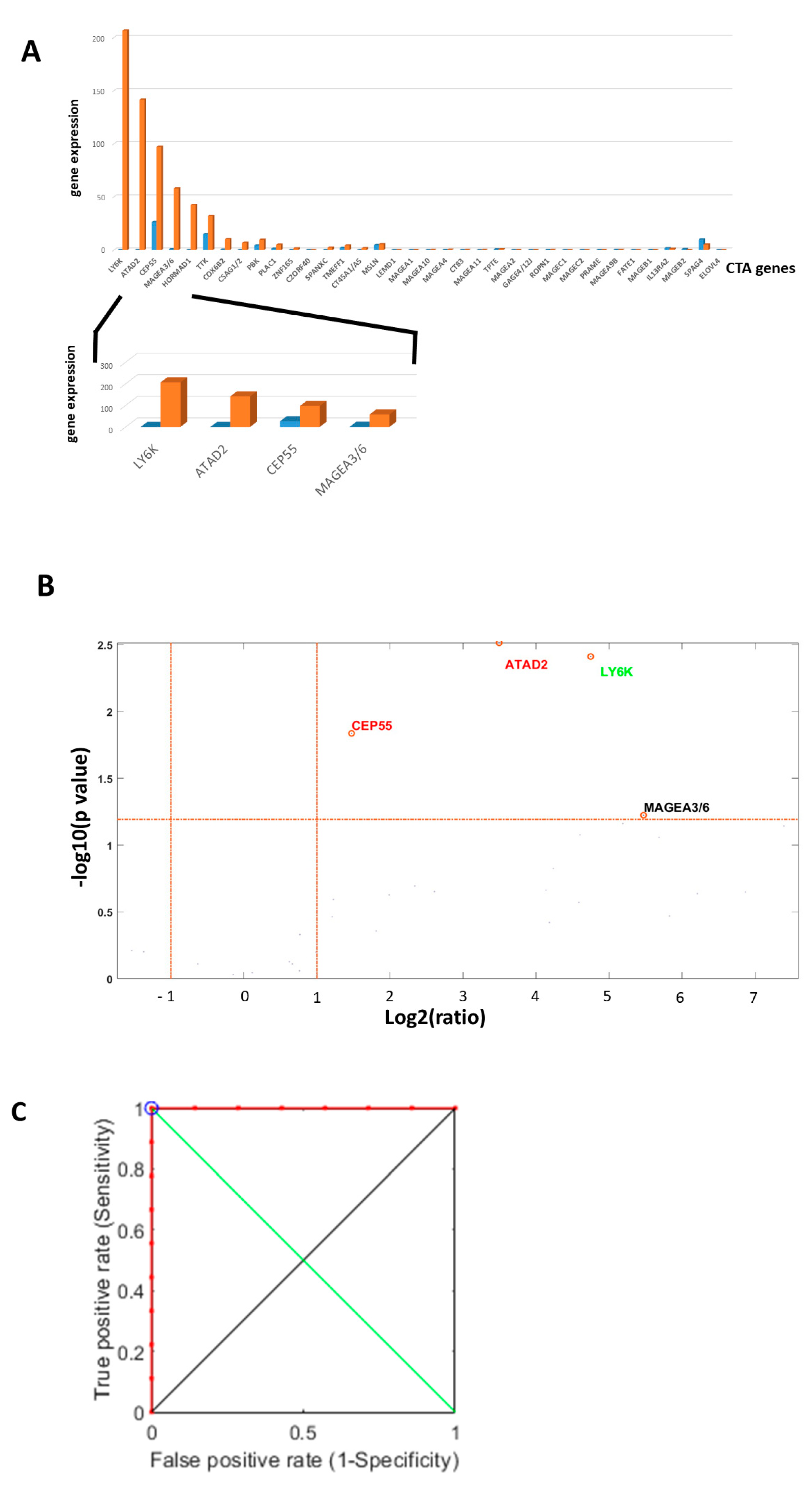
2.4. NanoString nCounter Validation and CTA Gene Expression in Oral Tongue Cancer
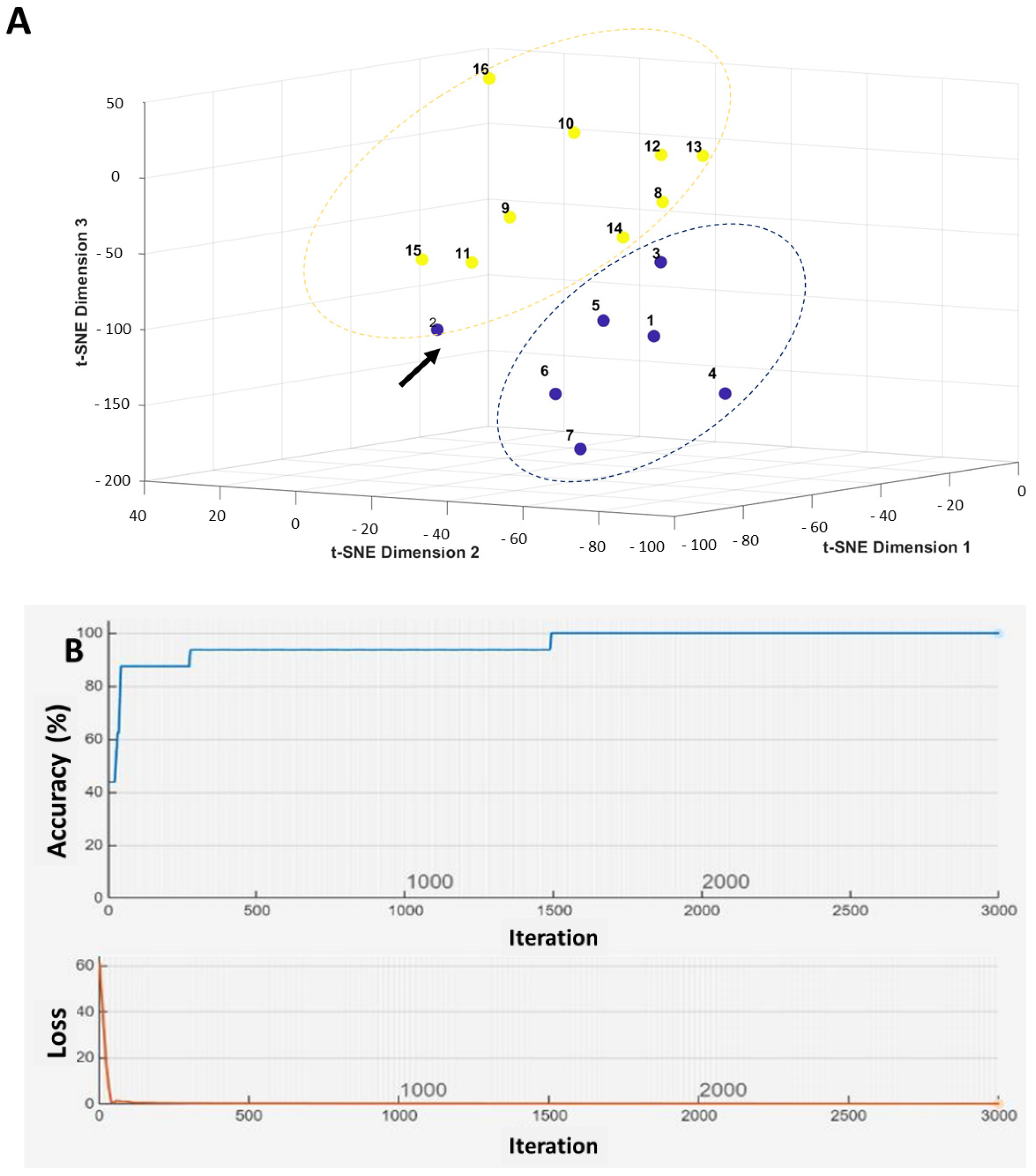
2.5. Machine Learning-Based Prediction of Neck Nodal Spread
3. Discussion
4. Materials and Methods
4.1. Primary Endpoints
4.2. Patients
4.3. Main Inclusion Criteria
4.4. Exclusion Criteria
4.5. RNA Extraction Quality Control
4.6. Demographic and Clinical Data
4.7. Genomic Computational Analysis
4.8. Tissue Analysis
4.9. RNA Extraction
4.10. NanoString nCounter Analysis
4.11. Oral Tongue Cancer CTA Gene Analysis, Expression and Accuracy
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
List of Abbreviations
| SCC | Squamous cell carcinoma |
| HNC | Head and neck cancer |
| AJCC | American Joint Committee on Cancer |
| CNN | Convolutional Neural Network |
| CTA | Cancer-Testis Antigen |
| DOI | Depth of Invasion |
| ENE | Extra-nodal Extension |
| EXPO | Expression Project for Oncology |
| GEO | Gene Expression Omnibus |
| PORT | Postoperative Radiotherapy |
| CRT | Chemoradiotherapy |
| TCGA | The Cancer Genome Atlas |
| t-SNE | t-distributed Stochastic Neighbor Embedding |
| LOOCV | Leave-One-Out Cross-Validation |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Stepan, K.O.; Mazul, A.L.; Larson, J.; Shah, P.; Jackson, R.S.; Pipkorn, P.; Kang, S.Y.; Puram, S.V. Changing Epidemiology of Oral Cavity Cancer in the United States. Otolaryngol. Head Neck Surg. 2023, 168, 761–768. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424, Erratum in CA Cancer J. Clin. 2020, 70, 313. https://doi.org/10.3322/caac.21609. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wu, J.; Wang, J.; Huang, R. Tobacco and oral squamous cell carcinoma: A review of carcinogenic pathways. Tob. Induc. Dis. 2019, 17, 29. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sim, Y.C.; Hwang, J.H.; Ahn, K.M. Overall and disease-specific survival outcomes following primary surgery for oral squamous cell carcinoma: Analysis of consecutive 67 patients. J. Korean Assoc. Oral. Maxillofac. Surg. 2019, 45, 83–90. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- D’Cruz, A.K.; Vaish, R.; Kapre, N.; Dandekar, M.; Gupta, S.; Hawaldar, R.; Agarwal, J.P.; Pantvaidya, G.; Chaukar, D.; Deshmukh, A.; et al. Elective versus Therapeutic Neck Dissection in Node-Negative Oral Cancer. N. Engl. J. Med. 2015, 373, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Sparano, A.; Weinstein, G.; Chalian, A.; Yodul, M.; Weber, R. Multivariate predictors of occult neck metastasis in early oral tongue cancer. Otolaryngol. Head Neck Surg. 2004, 131, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Ghai, S.; Mhaske, S.; Singh, R. Elective Neck Dissection Versus Therapeutic Neck Dissection in Clinically Node-Negative Early Stage Oral Cancer: A Meta-analysis of Randomized Controlled Trials. J. Maxillofac. Oral Surg. 2022, 21, 340–349. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McDonald, C.; Kent, S.; Schache, A.; Rogers, S.; Shaw, R. Health-related quality of life, functional outcomes, and complications after sentinel lymph node biopsy and elective neck dissection in early oral cancer: A systematic review. Head Neck 2023, 45, 2754–2779. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wan, S.Q.; Huang, L.; Zhong, W.J.; Zhang, B.L.; Song, J.; Ma, Y.H.; Hu, M. Analysis of hospitalization costs and length of stay for oral cancer patients undergoing surgery: Evidence from Hunan, China. Oral Oncol. 2021, 119, 105363. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, C.; Forker, K.; Zhang, X.; Wu, D.; Zhou, P.; Bowser, J.L. Pathological modulation of genome maintenance by cancer/testes antigens (CTAs). DNA Repair 2025, 147, 103818. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nin, D.S.; Deng, L.W. Biology of Cancer-Testis Antigens and Their Therapeutic Implications in Cancer. Cells 2023, 12, 926. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Seager, R.J.; Senosain, M.F.; Van Roey, E.; Gao, S.; DePietro, P.; Nesline, M.K.; Dash, D.P.; Zhang, S.; Ko, H.; Hastings, S.B.; et al. Cancer testis antigen burden (CTAB): A novel biomarker of tumor-associated antigens in lung cancer. J. Transl. Med. 2024, 22, 141. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Naik, A.; Thomas, R.; Al-Khadairi, G.; Bacha, R.; Hendrickx, W.; Decock, J. Cancer testis antigen PRAME: An anti-cancer target with immunomodulatory potential. J. Cell Mol. Med. 2021, 25, 10376–10388. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Smith, Y. Methods of Diagnosing Cancer Using Cancer Testis Antigens. USA Patent 12,018,331, 25 June 2024. [Google Scholar]
- Faisal, M.; Dhanani, R.; Ullah, S.; Bakar, M.A.; Irfan, N.; Malik, K.I.; Loya, A.; Boban, E.M.; Hussain, R.; Jamshed, A. Prognostic outcomes of treatment naïve oral tongue squamous cell carcinoma (OTSCC): A comprehensive analysis of 14 years. Eur. Arch. 2021, 278, 3045–3053, Erratum in Eur. Arch. Oto-Rhino-Laryngol. 2021, 278, 3055. https://doi.org/10.1007/s00405-021-06617-8. [Google Scholar] [CrossRef] [PubMed]
- Ghorbanpour, M.; Salarvand, S.; Shahsavari, F.; Shirkhoda, M.; Shakib, P.A.; Ghalehtaki, R. Depth of invasion and extranodal extension: The influential factors to predict survival rate of patients with oral tongue squamous cell carcinoma. BMC Cancer 2024, 24, 1256. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guo, C.B.; Feng, Z.; Zhang, J.G.; Peng, X.; Cai, Z.G.; Mao, C.; Zhang, Y.; Yu, G.Y.; Li, J.N.; Niu, L.X. Supraomohyoid neck dissection and modified radical neck dissection for clinically node-negative oral squamous cell carcinoma: A prospective study of prognosis, complications and quality of life. J. Cranio-Maxillofac. Surg. 2014, 42, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Kao, H.J.; Weng, T.H.; Chen, C.H.; Chen, Y.C.; Chi, Y.H.; Huang, K.Y.; Weng, S.L. Integrating In Silico and In Vitro Approaches to Identify Natural Peptides with Selective Cytotoxicity against Cancer Cells. Int. J. Mol. Sci. 2024, 25, 6848. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fu, J.; Zhang, J.; Chen, X.; Liu, Z.; Yang, X.; He, Z.; Hao, Y.; Liu, B.; Yao, D. ATPase family AAA domain-containing protein 2 (ATAD2): From an epigenetic modulator to cancer therapeutic target. Theranostics 2023, 13, 787–809. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guruvaiah, P.; Chava, S.; Sun, C.W.; Singh, N.; Penn, C.A.; Gupta, R. ATAD2 is a driver and a therapeutic target in ovarian cancer that functions by upregulating CENPE. Cell Death Dis. 2023, 14, 456. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Biju, T.; Venkatesh, C.; Honnasiddappa, D.B.; Sajjan, M.; Mahadeva, N.K.; Dinesh, B.G.H.; Kumar, B.S.; Ganjipete, S.; Ramar, M.; Kunjiappan, S.; et al. ATAD2 bromodomain in cancer therapy: Current status and future perspectives. Int. J. Biol. Macromol. 2025, 311 Pt 3, 143948. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Senapati, S. Immunological and functional aspects of MAGEA3 cancer/testis antigen. Adv. Protein Chem. Struct. Biol. 2021, 125, 121–147. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Kang, Y.; Zhong, R.; You, J.; Chen, J.; Li, L.; Chen, J.; Chen, L. Cancer testis antigen MAGEA3 in serum and serum-derived exosomes serves as a promising biomarker in lung adenocarcinoma. Sci. Rep. 2024, 14, 7573. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gao, X.; Chen, G.; Cai, H.; Wang, X.; Song, K.; Liu, L.; Qiu, T.; He, Y. Aberrantly enhanced melanoma-associated antigen (MAGE)-A3 expression facilitates cervical cancer cell proliferation and metastasis via actuating Wnt signaling pathway. Biomed. Pharmacother. 2020, 122, 109710. [Google Scholar] [CrossRef] [PubMed]
- Andrys-Olek, J.; Selvanesan, B.C.; Varghese, S.; Arriaza, R.H.; Tiwari, P.B.; Chruszcz, M.; Borowski, T.; Upadhyay, G. Experimental and Computational Studies Reveal Novel Interaction of Lymphocytes Antigen 6K to TGF-β Receptor Complex. Int. J. Mol. Sci. 2023, 24, 12779. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sastry, N.G.; Wan, X.; Huang, T.; Alvarez, A.A.; Pangeni, R.P.; Song, X.; James, C.D.; Horbinski, C.M.; Brennan, C.W.; Nakano, I.; et al. LY6K promotes glioblastoma tumorigenicity via CAV-1-mediated ERK1/2 signaling enhancement. Neuro-Oncology 2020, 22, 1315–1326, Erratum in Neuro-Oncology 2024, 26, 1947. https://doi.org/10.1093/neuonc/noae151. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Park, S.; Park, D.; Han, S.; Chung, G.E.; Soh, S.; Ka, H.I.; Joo, H.J.; Yang, Y. LY6K depletion modulates TGF-β and EGF signaling. Cancer Med. 2023, 12, 12593–12607. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, L.; Zhu, Y.; Guo, F.; Long, H.; Yin, M. Pan-cancer analysis of oncogenic role of CEP55 and experiment validation in clear cell renal cell carcinoma. Sci. Rep. 2024, 14, 28279. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yan, S.-M.; Liu, L.; Gu, W.-Y.; Huang, L.-Y.; Yang, Y.; Huang, Y.-H.; Luo, R.-Z.; Zheng, D. CEP55 Positively Affects Tumorigenesis of Esophageal Squamous Cell Carcinoma and Is Correlated with Poor Prognosis. J. Oncol. 2021, 2021, 8890715. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wangmo, D.; Gates, T.J.; Zhao, X.; Sun, R.; Subramanian, S. Centrosomal Protein 55 (CEP55) Drives Immune Exclusion and Resistance to Immune Checkpoint Inhibitors in Colorectal Cancer. Vaccines 2024, 12, 63. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
| Scheme | Female N (%) | 10 (62.5%) | |||
| Male N (%) | 6 (37.5%) | ||||
| Age, years | Mean | 60.6 | |||
| Median | 64.5 | ||||
| Range | 24–77 | ||||
| Disease stage | Early (stage I&II) N (%) | 6 (37.5%) | |||
| Advanced (stage III&IV) N (%) | 10 (62.5%) | ||||
| Nodal disease | Positive N (%) | 9 (56.25%) | Positive ENE | 5 (55.5%) | |
| Negative ENE | 4 (44.5%) | ||||
| Negative N (%) | 7 (43.75%) | ||||
| Adjuvant treatment | Radiation N (%) | 12 (63.15%) | |||
| Chemotherapy N (%) | 7 (43.75%) | ||||
| Survival | Death of disease N (%) | 4 (25%) | Time, months | Mean | 12 |
| Median | 12.5 | ||||
| Range | 8–15 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, Y.; Cohen, A.; Neuman, T.; Fleissig, Y.; Hirshoren, N. Nodal Spread Prediction in Human Oral Tongue Squamous Cell Carcinoma Using a Cancer-Testis Antigen Genes Signature. Int. J. Mol. Sci. 2025, 26, 9258. https://doi.org/10.3390/ijms26189258
Smith Y, Cohen A, Neuman T, Fleissig Y, Hirshoren N. Nodal Spread Prediction in Human Oral Tongue Squamous Cell Carcinoma Using a Cancer-Testis Antigen Genes Signature. International Journal of Molecular Sciences. 2025; 26(18):9258. https://doi.org/10.3390/ijms26189258
Chicago/Turabian StyleSmith, Yoav, Amit Cohen, Tzahi Neuman, Yoram Fleissig, and Nir Hirshoren. 2025. "Nodal Spread Prediction in Human Oral Tongue Squamous Cell Carcinoma Using a Cancer-Testis Antigen Genes Signature" International Journal of Molecular Sciences 26, no. 18: 9258. https://doi.org/10.3390/ijms26189258
APA StyleSmith, Y., Cohen, A., Neuman, T., Fleissig, Y., & Hirshoren, N. (2025). Nodal Spread Prediction in Human Oral Tongue Squamous Cell Carcinoma Using a Cancer-Testis Antigen Genes Signature. International Journal of Molecular Sciences, 26(18), 9258. https://doi.org/10.3390/ijms26189258






