Bioinformatics Analysis and Expression Profiling Under Abiotic Stress of the DREB Gene Family in Glycyrrhiza uralensis
Abstract
1. Introduction
2. Results
2.1. Impact of Drought Stress on ABA Accumulation in G. uralensis Tissues
2.2. Differential Expression Patterns of the GuDREB Gene Family in Response to Drought Stress
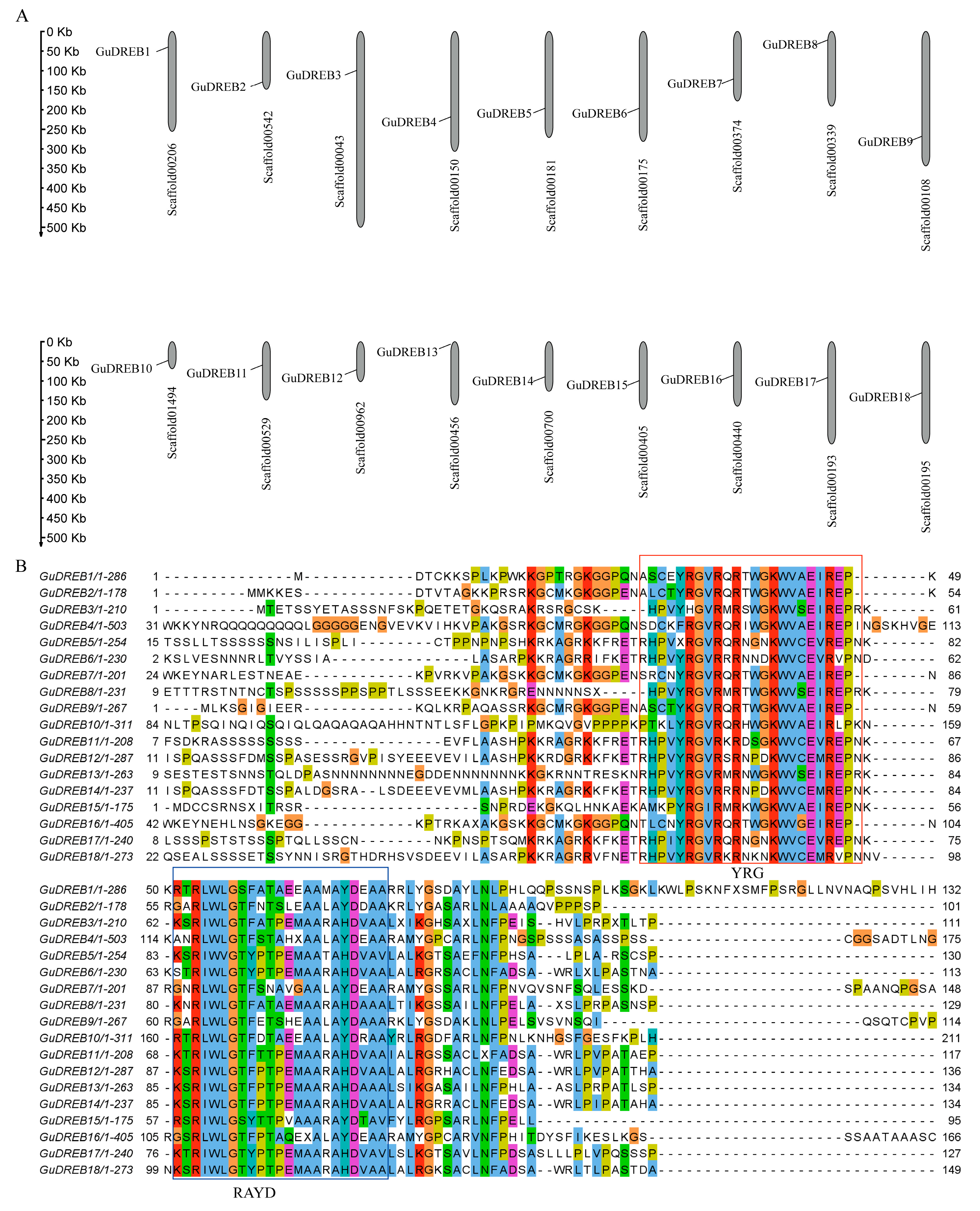
2.3. Genomic Scaffold Localization and Multiple Sequence Alignment of GuDREB Proteins
2.4. Physicochemical Characterization and Secondary Structure Prediction of GuDREB Proteins
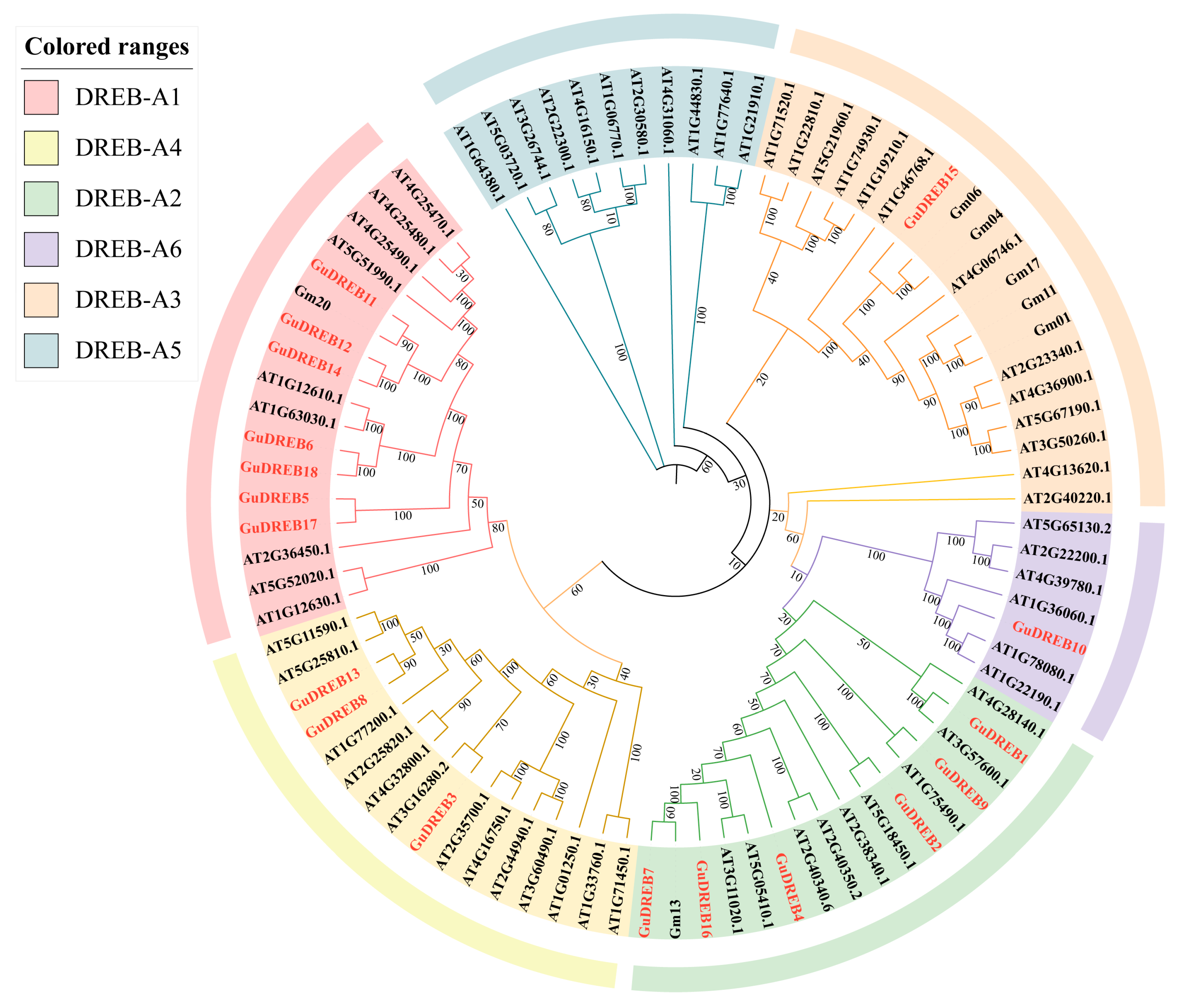
2.5. Phylogenetic Analysis of Plant DREB Family Members
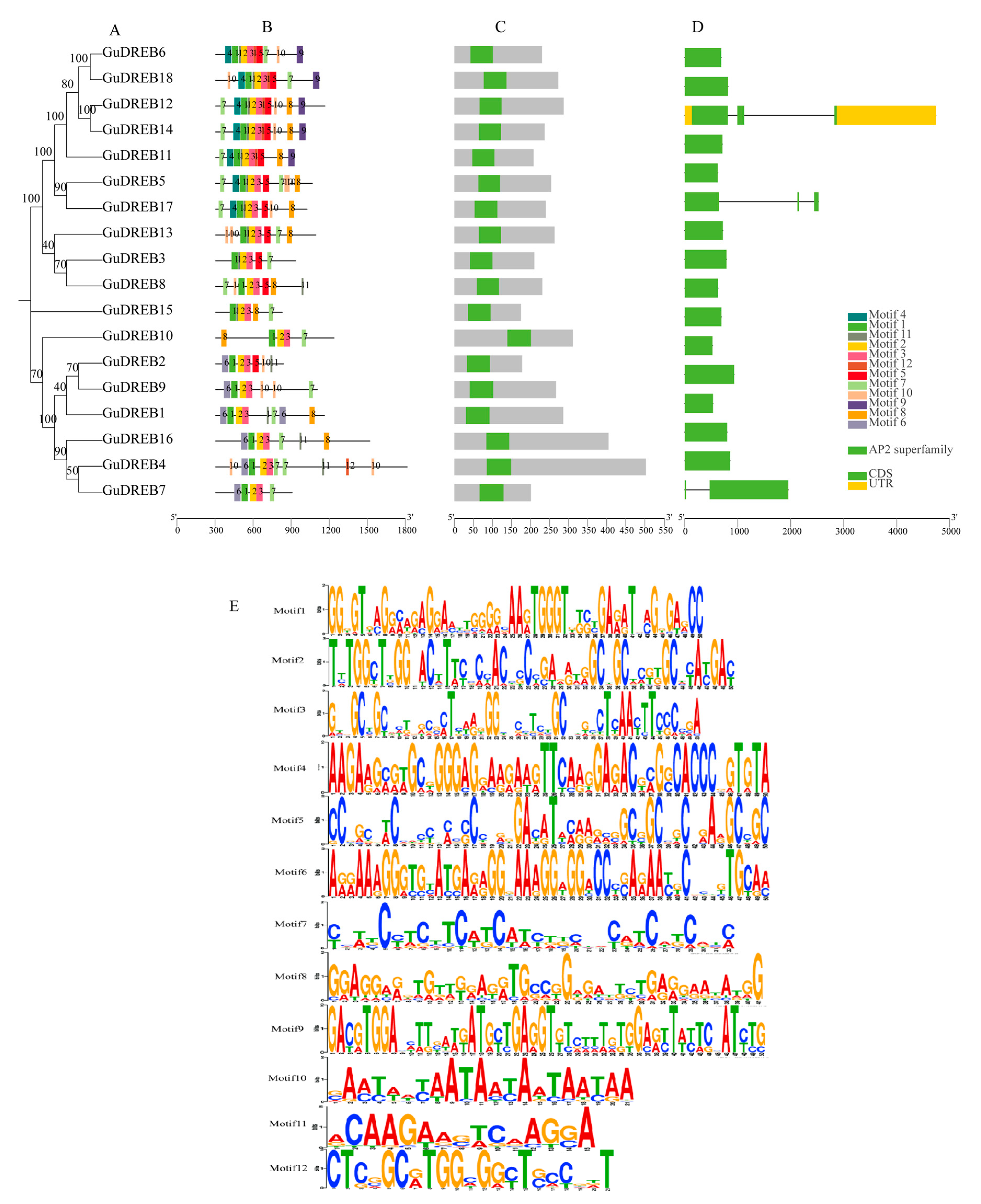
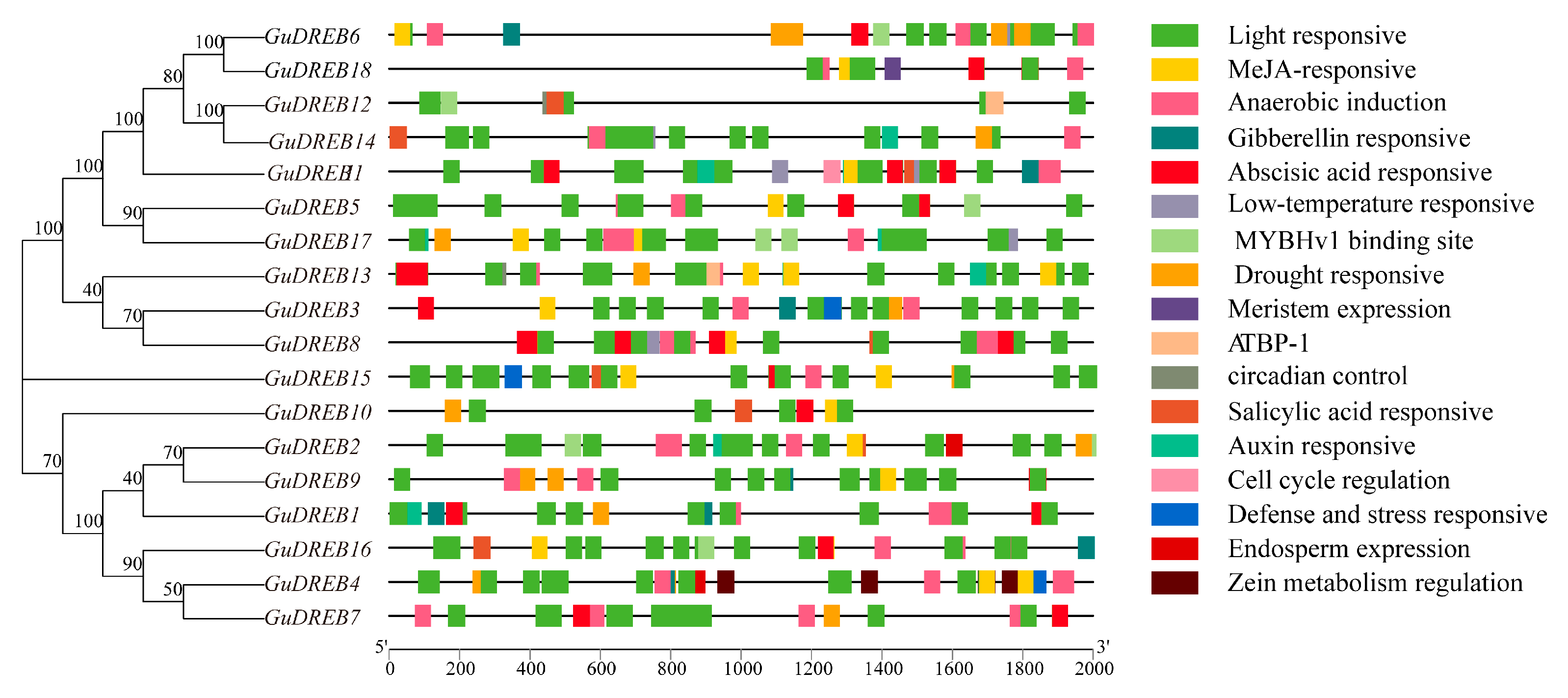
2.6. Integrated Analysis of Gene Structures, Conserved Domains, and Motifs in the G. uralensis DREB Gene Family
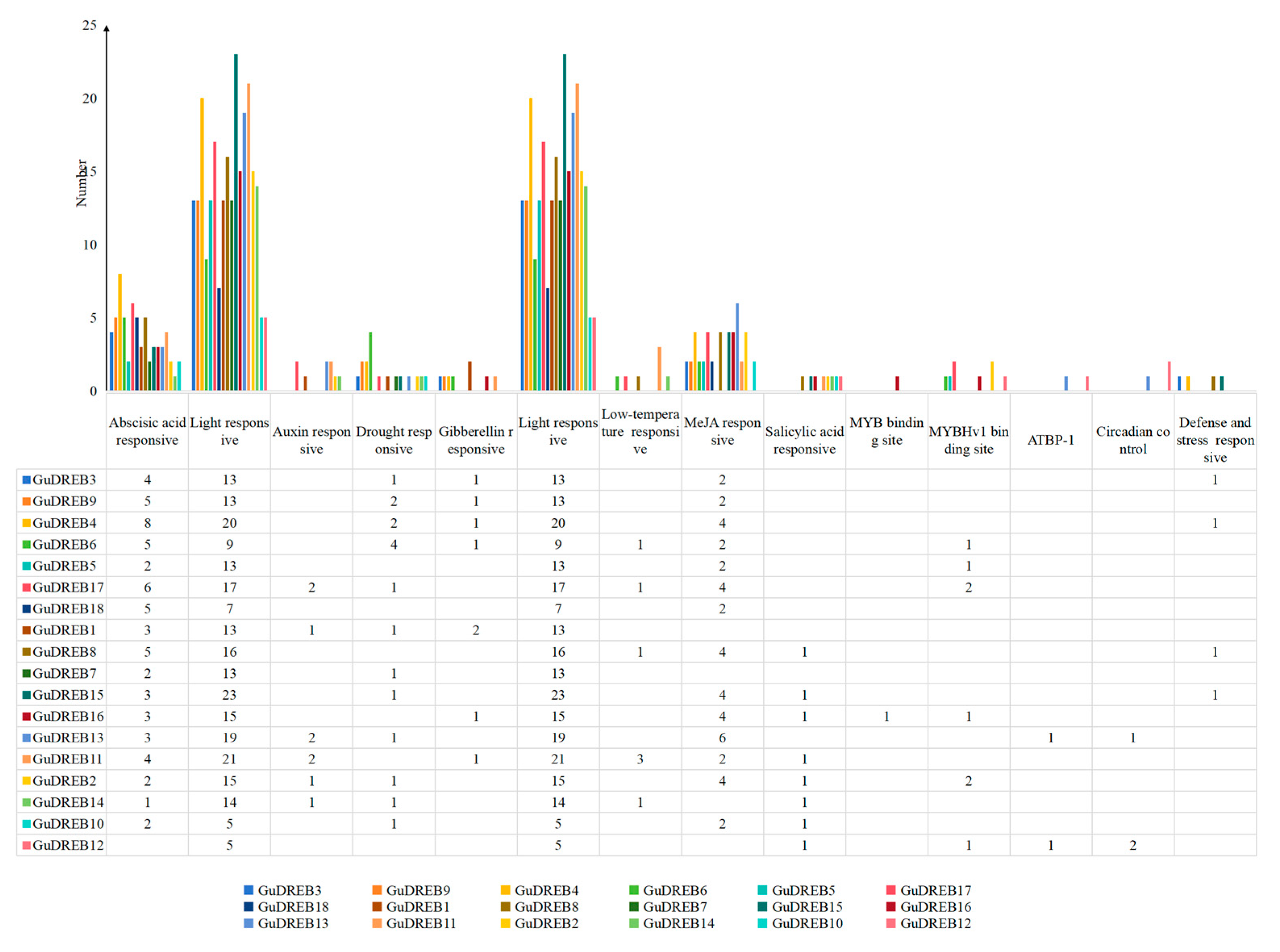
2.7. Analysis of Cis-Regulatory Elements in G. uralensis DREB Subfamily Members
3. Discussion
4. Materials and Methods
4.1. Plant Material and Data Sources
4.2. Bioinformatics Analysis of the GuDREB Gene Family
4.3. Plant Material Treatment
4.4. Sample Tissue Processing
4.5. Standard Curve Preparation
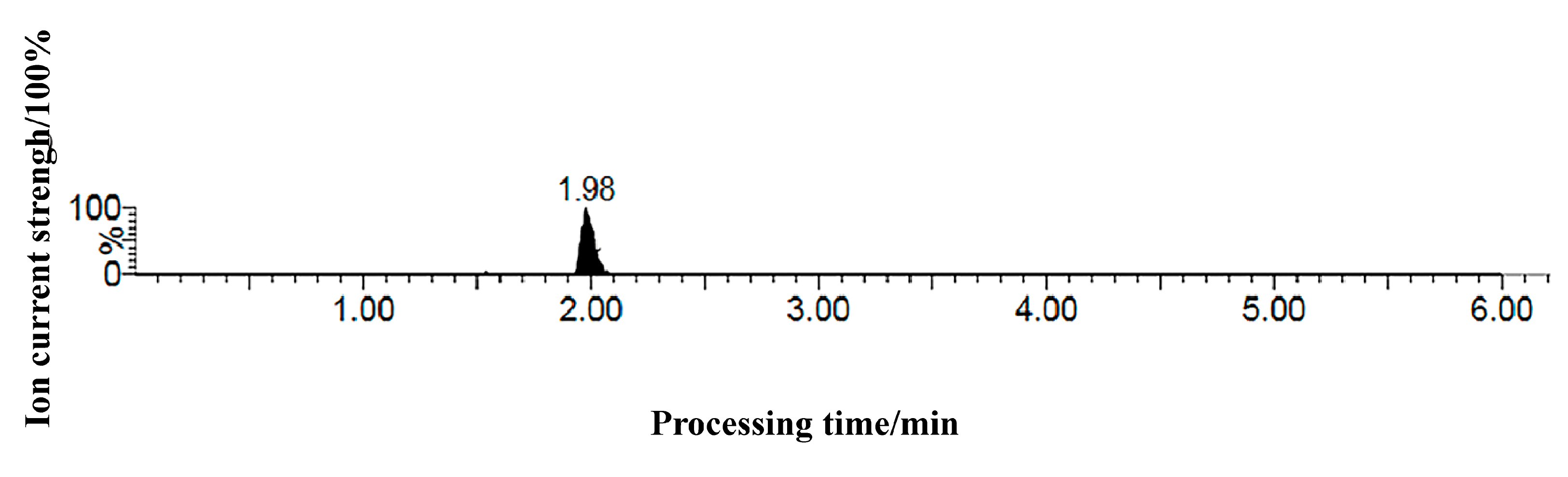
4.6. Chromatographic Conditions
4.7. Mass Spectrometry Conditions
4.8. Linearity Analysis for Abscisic Acid Quantification
4.9. RNA Extraction and Transcriptome Sequencing
4.10. Validation of RNA-Seq Data Using qRT-PCR
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zuo, J.; Meng, T.; Wang, Y.; Tang, W. A Review of the Antiviral Activities of Glycyrrhizic Acid, Glycyrrhetinic Acid and Glycyrrhetinic Acid Monoglucuronide. Pharmaceuticals 2023, 16, 641. [Google Scholar] [CrossRef]
- Jiang, L.; Akram, W.; Luo, B.; Hu, S.; Faruque, M.O.; Ahmad, S.; Yasin, N.A.; Khan, W.U.; Ahmad, A.; Shikov, A.N.; et al. Metabolomic and Pharmacologic Insights of Aerial and Underground Parts of Glycyrrhiza uralensis Fisch. ex DC. for Maximum Utilization of Medicinal Resources. Front. Pharmacol. 2021, 12, 658670. [Google Scholar] [CrossRef]
- Cheng, M.; Ding, L.; Kan, H.; Zhang, H.; Jiang, B.; Sun, Y.; Cao, S.; Li, W.; Koike, K.; Qiu, F. Isolation, structural elucidation and in vitro hepatoprotective activity of flavonoids from Glycyrrhiza uralensis. J. Nat. Med. 2019, 73, 847–854. [Google Scholar] [CrossRef]
- Marui, A.; Kotera, A.; Furukawa, Z.; Yasufuku, N.; Omine, K.; Nagano, T.; Tuvshintogtokh, I.; Mandakh, B. Monitoring the Growing Environment of Wild Licorice with Analysis of Satellite Data at a Semi-arid Area in Mongolia (DESERT TECHNOLOGY 11 INTERNATIONAL CONFERENCE). J. Arid. Land Stud. 2014, 24, 99–202. [Google Scholar]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Chai, M.; Cheng, H.; Yan, M.; Priyadarshani, S.; Zhang, M.; He, Q.; Huang, Y.; Chen, F.; Liu, L.; Huang, X.; et al. Identification and expression analysis of the DREB transcription factor family in pineapple (Ananas comosus (L.) Merr.). PeerJ 2020, 8, e9006. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Lu, Z.; Gao, L.; Guo, S.; Shen, Q. Is Nitrogen a Key Determinant of Water Transport and Photosynthesis in Higher Plants Upon Drought Stress? Front. Plant Sci. 2018, 9, 1143. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Mei, F.; Chen, B.; Du, L.; Li, S.; Zhu, D.; Chen, N.; Zhang, Y.; Li, F.; Wang, Z.; Cheng, X.; et al. A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat. Plant Cell 2022, 34, 4472–4494. [Google Scholar] [CrossRef]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; Van Montagu, M.; Jofuku, K.D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar] [CrossRef]
- Thomashow, M.F. PLANT COLD ACCLIMATION: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Donde, R.; Gupta, M.K.; Gouda, G.; Kumar, J.; Vadde, R.; Sahoo, K.K.; Dash, S.K.; Behera, L. Computational characterization of structural and functional roles of DREB1A, DREB1B and DREB1C in enhancing cold tolerance in rice plant. Amino Acids 2019, 51, 839–853. [Google Scholar] [CrossRef]
- Akbudak, M.A.; Filiz, E.; Kontbay, K. DREB2 (dehydration-responsive element-binding protein 2) type transcription factor in sorghum (Sorghum bicolor): Genome-wide identification, characterization and expression profiles under cadmium and salt stresses. 3 Biotech 2018, 8, 426. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, L.; Wang, L.; Wang, S.; Cheng, X. VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J. Plant Res. 2016, 129, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Xi, Y.; Chen, X.; Cheng, Z.M. Genome-wide identification of AP2/EREBP in Fragaria vesca and expression pattern analysis of the FvDREB subfamily under drought stress. BMC Plant Biol. 2021, 21, 295. [Google Scholar] [CrossRef]
- Niu, X.; Luo, T.; Zhao, H.; Su, Y.; Ji, W.; Li, H. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene 2020, 740, 144514. [Google Scholar] [CrossRef]
- Ghorbani, R.; Zakipour, Z.; Alemzadeh, A.; Razi, H. Genome-wide analysis of AP2/ERF transcription factors family in Brassica napus. Physiol. Mol. Biol. Plants 2020, 26, 1463–1476. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, N.; Munir, F.; Gul, A.; Amir, R.; Zafar Paracha, R. Genome-wide analysis, identification, evolution and genomic organization of dehydration responsive element-binding (DREB) gene family in Solanum tuberosum. PeerJ 2021, 9, e11647. [Google Scholar] [CrossRef]
- Wang, C.; Wu, D.; Jiang, L.; Liu, X.; Xie, T. Multi-Omics Elucidates Difference in Accumulation of Bioactive Constituents in Licorice (Glycyrrhiza uralensis) under Drought Stress. Molecules 2023, 28, 7042. [Google Scholar] [CrossRef]
- Su, J.; Song, S.; Wang, Y.; Zeng, Y.; Dong, T.; Ge, X.; Duan, H. Genome-wide identification and expression analysis of DREB family genes in cotton. BMC Plant Biol. 2023, 23, 169. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Gupta, K.; Lopato, S.; Agarwal, P. Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 2017, 68, 2135–2148. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.J.; Hwang, J.E.; Chen, H.; Hong, J.K.; Yang, K.A.; Choi, M.S.; Lee, K.O.; Chung, W.S.; Lee, S.Y.; Lim, C.O. Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem. Biophys. Res. Commun. 2007, 362, 431–436. [Google Scholar] [CrossRef]
- Qin, F.; Kakimoto, M.; Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Tran, L.S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007, 50, 54–69. [Google Scholar] [CrossRef]
- Mizoi, J.; Ohori, T.; Moriwaki, T.; Kidokoro, S.; Todaka, D.; Maruyama, K.; Kusakabe, K.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 2013, 161, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Peng, X.; Fan, W.; Tang, M.; Liu, J.; Shen, S. Functional analysis of BpDREB2 gene involved in salt and drought response from a woody plant Broussonetia papyrifera. Gene 2014, 535, 140–149. [Google Scholar] [CrossRef]
- Singh, K.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Lin, R.C.; Park, H.J.; Wang, H.Y. Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 2008, 1, 42–57. [Google Scholar] [CrossRef]
- Rae, L.; Lao, N.T.; Kavanagh, T.A. Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors. Planta 2011, 234, 429–444. [Google Scholar] [CrossRef]
- Feng, J.X.; Liu, D.; Pan, Y.; Gong, W.; Ma, L.G.; Luo, J.C.; Deng, X.W.; Zhu, Y.X. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 2005, 59, 853–868. [Google Scholar] [CrossRef]
- Xu, J.Y.; Lu, Y.; Sun, R.D.; Wang, X.S.; Ding, W.T.; Deng, S.S.; Wang, P.W.; Zhao, J.M.; Guo, N.; Xing, H. Functional analysis of GmDREB8 under drought stress in soybean. J. Nanjing Agric. Univ. 2023, 46, 226–236. [Google Scholar] [CrossRef]
- Kuromori, T.; Seo, M.; Shinozaki, K. ABA Transport and Plant Water Stress Responses. Trends Plant Sci. 2018, 23, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Christmann, A.; Moes, D.; Himmelbach, A.; Yang, Y.; Tang, Y.; Grill, E. Integration of abscisic acid signalling into plant responses. Plant Biol 2006, 8, 314–325. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Q.Y.; Cheng, X.G.; Xu, Z.S.; Li, L.C.; Ye, X.G.; Xia, L.Q.; Ma, Y.Z. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007, 353, 299–305. [Google Scholar] [CrossRef] [PubMed]

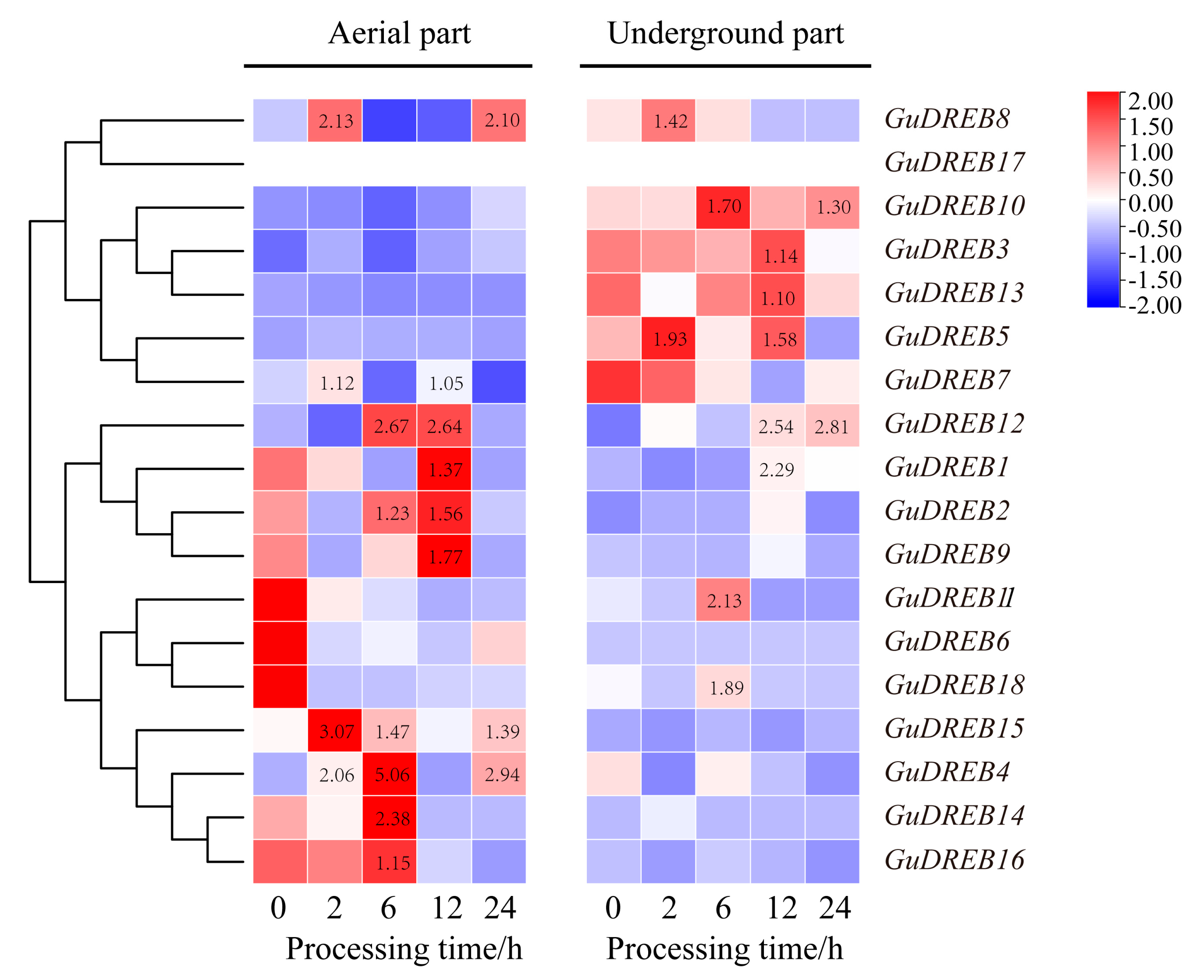
| Gene Name | Aerial Part | Underground Part | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 2 h | 6 h | 12 h | 24 h | 0 h | 2 h | 6 h | 12 h | 24 h | |
| GuDREB1 | 0.89 ± 0.12 | 0.56 ± 0.08 | 0.15 ± 0.03 | 1.22 ± 0.15 | 0.16 ± 0.02 | 0.21 ± 0.04 | 0.09 ± 0.01 | 0.14 ± 0.02 | 0.48 ± 0.06 | 0.43 ± 0.05 |
| GuDREB2 | 0.48 ± 0.07 | 0.08 ± 0.01 | 0.59 ± 0.08 | 0.75 ± 0.09 | 0.13 ± 0.02 | 0.00 ± 0.00 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.27 ± 0.03 | 0.00 ± 0.00 |
| GuDREB3 | 4.70 ± 0.62 | 8.78 ± 1.12 | 4.01 ± 0.52 | 8.02 ± 1.03 | 10.49 ± 1.35 | 23.53 ± 3.06 | 21.89 ± 2.84 | 19.96 ± 2.59 | 26.92 ± 3.50 | 13.89 ± 1.81 |
| GuDREB4 | 0.16 ± 0.02 | 0.33 ± 0.04 | 0.81 ± 0.10 | 0.13 ± 0.02 | 0.47 ± 0.06 | 0.36 ± 0.05 | 0.09 ± 0.01 | 0.33 ± 0.04 | 0.19 ± 0.02 | 0.11 ± 0.01 |
| GuDREB5 | 0.00 ± 0.00 | 0.07 ± 0.01 | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.00 ± 0.00 | 0.57 ± 0.07 | 1.10 ± 0.14 | 0.39 ± 0.05 | 0.90 ± 0.12 | 0.00 ± 0.00 |
| GuDREB6 | 2.46 ± 0.32 | 0.10 ± 0.01 | 0.26 ± 0.03 | 0.00 ± 0.00 | 0.66 ± 0.08 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| GuDREB7 | 20.31 ± 2.64 | 22.80 ± 2.96 | 16.98 ± 2.21 | 21.39 ± 2.78 | 16.22 ± 2.11 | 28.85 ± 3.75 | 27.27 ± 3.55 | 22.67 ± 2.95 | 18.76 ± 2.44 | 22.44 ± 2.92 |
| GuDREB8 | 4.46 ± 0.58 | 9.50 ± 1.24 | 1.44 ± 0.19 | 2.01 ± 0.26 | 9.38 ± 1.22 | 6.48 ± 0.84 | 9.23 ± 1.20 | 6.64 ± 0.87 | 4.23 ± 0.55 | 4.26 ± 0.56 |
| GuDREB9 | 0.77 ± 0.10 | 0.00 ± 0.00 | 0.47 ± 0.06 | 1.36 ± 0.18 | 0.00 ± 0.00 | 0.10 ± 0.01 | 0.06 ± 0.01 | 0.04 ± 0.01 | 0.27 ± 0.03 | 0.00 ± 0.00 |
| GuDREB10 | 28.90 ± 3.76 | 26.76 ± 3.48 | 17.16 ± 2.23 | 27.77 ± 3.61 | 44.55 ± 5.79 | 65.70 ± 8.54 | 64.83 ± 8.43 | 111.64 ± 14.51 | 76.35 ± 9.93 | 85.10 ± 11.06 |
| GuDREB11 | 8.21 ± 1.07 | 2.75 ± 0.36 | 1.61 ± 0.21 | 0.66 ± 0.09 | 0.97 ± 0.13 | 1.86 ± 0.24 | 1.17 ± 0.15 | 5.04 ± 0.65 | 0.37 ± 0.05 | 0.39 ± 0.05 |
| GuDREB12 | 0.39 ± 0.05 | 0.22 ± 0.03 | 1.04 ± 0.13 | 1.03 ± 0.13 | 0.37 ± 0.05 | 0.26 ± 0.03 | 0.58 ± 0.07 | 0.43 ± 0.05 | 0.66 ± 0.08 | 0.73 ± 0.09 |
| GuDREB13 | 1.41 ± 0.18 | 0.78 ± 0.10 | 0.00 ± 0.00 | 0.27 ± 0.03 | 0.48 ± 0.06 | 13.97 ± 1.82 | 5.70 ± 0.74 | 12.54 ± 1.63 | 15.38 ± 2.00 | 8.08 ± 1.05 |
| GuDREB14 | 0.26 ± 0.03 | 0.13 ± 0.02 | 0.62 ± 0.08 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.08 ± 0.01 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| GuDREB15 | 76.38 ± 9.93 | 234.19 ± 30.44 | 112.25 ± 14.60 | 65.57 ± 8.52 | 106.33 ± 13.82 | 26.10 ± 3.40 | 16.09 ± 2.10 | 33.96 ± 4.41 | 17.35 ± 2.26 | 32.92 ± 4.28 |
| GuDREB16 | 19.04 ± 2.48 | 16.86 ± 2.20 | 21.96 ± 2.85 | 5.36 ± 0.70 | 1.78 ± 0.23 | 4.12 ± 0.54 | 1.85 ± 0.24 | 4.81 ± 0.62 | 3.39 ± 0.44 | 1.38 ± 0.18 |
| GuDREB17 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| GuDREB18 | 3.33 ± 0.43 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.13 ± 0.02 | 0.16 ± 0.02 | 0.45 ± 0.06 | 0.03 ± 0.01 | 0.85 ± 0.11 | 0.06 ± 0.01 | 0.03 ± 0.01 |
| Gene Name | Genomic (bp) | CDS (bp) | Aliphatic Index | Average Hydrophilicity | Length (aa) | pI | Mw (Da) | Sublocation | Instability Index (II) |
|---|---|---|---|---|---|---|---|---|---|
| GuDREB1 | 858 | 858 | 65.75 | −0.860 | 285 | 6.47 | 32,149.59 | nucl | 53.48 |
| GuDREB2 | 534 | 534 | 60.79 | −0.672 | 177 | 8.44 | 19,521.95 | cyto, nucl | 35.04 |
| GuDREB3 | 630 | 630 | 68.18 | −0.445 | 209 | 5.38 | 23,142.96 | nucl | 63.95 |
| GuDREB4 | 1509 | 1509 | 58.86 | −0.897 | 502 | 6.64 | 55,106.82 | nucl | 53.82 |
| GuDREB5 | 762 | 762 | 59.41 | −0.841 | 253 | 8.61 | 28,264.57 | nucl | 47.20 |
| GuDREB6 | 690 | 690 | 70.74 | −0.507 | 229 | 5.32 | 25,572.11 | cyto, nucl | 56.38 |
| GuDREB7 | 1158 | 603 | 55.20 | −0.816 | 200 | 9.30 | 21,799.72 | cyto, nucl | 43.40 |
| GuDREB8 | 693 | 693 | 49.74 | −0.715 | 230 | 5.16 | 24,845.95 | nucl | 65.91 |
| GuDREB9 | 801 | 801 | 69.70 | −0.443 | 266 | 5.13 | 29,171.77 | cyto, nucl | 52.94 |
| GuDREB10 | 933 | 933 | 63.94 | −0.666 | 310 | 7.03 | 34,350.45 | nucl | 52.27 |
| GuDREB11 | 624 | 624 | 55.85 | −0.543 | 207 | 5.68 | 22,747.49 | nucl | 60.17 |
| GuDREB12 | 2864 | 861 | 58.04 | −0.698 | 286 | 6.12 | 32,225.61 | nucl | 64.33 |
| GuDREB13 | 789 | 789 | 56.68 | −0.774 | 262 | 5.59 | 28,224.83 | nucl | 53.50 |
| GuDREB14 | 711 | 711 | 58.39 | −0.766 | 236 | 5.37 | 26,557.51 | nucl | 65.84 |
| GuDREB15 | 525 | 525 | 60.11 | −0.923 | 174 | 9.32 | 19,567.12 | nucl | 51.94 |
| GuDREB16 | 1925 | 1215 | 55.57 | −0.713 | 404 | 4.97 | 43,894.36 | nucl | 38.60 |
| GuDREB17 | 720 | 720 | 64.06 | −0.628 | 239 | 5.12 | 26,568.61 | nucl | 52.37 |
| GuDREB18 | 819 | 819 | 64.96 | −0.626 | 272 | 5.21 | 30,668.00 | nucl | 70.13 |
| Gene Name | α-Helix | Extended Strand | β-Turn | Random Coil |
|---|---|---|---|---|
| GuDREB1 | 29.47% | 16.84% | 9.47% | 44.21% |
| GuDREB2 | 35.03% | 13.56% | 9.04% | 42.37% |
| GuDREB3 | 29.67% | 15.31% | 5.74% | 49.28% |
| GuDREB4 | 44.02% | 12.55% | 7.17% | 36.25% |
| GuDREB5 | 15.02% | 22.92% | 7.11% | 54.94% |
| GuDREB6 | 29.26% | 25.33% | 4.80% | 40.61% |
| GuDREB7 | 37.00% | 13.50% | 6.00% | 43.50% |
| GuDREB8 | 22.17% | 21.30% | 4.35% | 52.17% |
| GuDREB9 | 26.69% | 18.05% | 6.77% | 48.50% |
| GuDREB10 | 23.55% | 15.48% | 6.45% | 54.52% |
| GuDREB11 | 31.88% | 17.87% | 7.73% | 42.51% |
| GuDREB12 | 28.32% | 19.58% | 5.94% | 46.15% |
| GuDREB13 | 24.81% | 12.60% | 4.96% | 57.63% |
| GuDREB14 | 33.05% | 15.25% | 5.93% | 45.76% |
| GuDREB15 | 33.91% | 14.94% | 4.02% | 47.13% |
| GuDREB16 | 33.17% | 12.87% | 10.15% | 43.81% |
| GuDREB17 | 23.85% | 25.52% | 7.11% | 43.51% |
| GuDREB18 | 24.63% | 25.00% | 8.46% | 41.91% |
| Compounds | Retention Time (min) | Relative Molecular Weight/Da | Ionization Mode | MS (m/z) | MS2 (m/z) | Cone- Voltage/V | Collision Energy/eV |
|---|---|---|---|---|---|---|---|
| Abscisic acid | 2.00 | 264 | ESI- | 263.2 | 153.0 * | 22 | 12 |
| Component | Linear Equation | R2 | Linear Range (ng·mL−1) |
|---|---|---|---|
| Abscisic acid | Y = 103.72X − 99.05 | 0.9956 | 1.2~100 |
| Primer Name | Primer Sequences F (5′-3′) | Primer Sequences R (5′-3′) |
|---|---|---|
| GuDREB3 | GCAGCAAGCACCCAGTTTAC | CTGCCATTTCAGGGGTAGCA |
| GuDREB 7 | GAAAGGGAAAGGAGGACCCG | TGTTCGGCTCCCGAATTTCA |
| GuDREB 8 | AACCATCAAAGGCTCCTCCG | ACGAACTCGCTATTCGGGTC |
| GuDREB 15 | AGTTGGTGCCAGAGTCGATG | AGGTCAAGGCAATCTTCGGG |
| GuLectin | CTGATGCAGAGCTTCAAATCGAG | TTCGGAAGGAAGGTTGAGGTAAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, L.; Shi, N.; Du, X.; Huang, T.; Zhang, Y.; Zhao, C.; Zhao, K.; Lin, Z.; Ma, D.; Li, Q.; et al. Bioinformatics Analysis and Expression Profiling Under Abiotic Stress of the DREB Gene Family in Glycyrrhiza uralensis. Int. J. Mol. Sci. 2025, 26, 9235. https://doi.org/10.3390/ijms26189235
Cheng L, Shi N, Du X, Huang T, Zhang Y, Zhao C, Zhao K, Lin Z, Ma D, Li Q, et al. Bioinformatics Analysis and Expression Profiling Under Abiotic Stress of the DREB Gene Family in Glycyrrhiza uralensis. International Journal of Molecular Sciences. 2025; 26(18):9235. https://doi.org/10.3390/ijms26189235
Chicago/Turabian StyleCheng, Linyuan, Nana Shi, Xiangrong Du, Teng Huang, Yaxin Zhang, Chenjie Zhao, Kun Zhao, Zirun Lin, Denglin Ma, Qiuling Li, and et al. 2025. "Bioinformatics Analysis and Expression Profiling Under Abiotic Stress of the DREB Gene Family in Glycyrrhiza uralensis" International Journal of Molecular Sciences 26, no. 18: 9235. https://doi.org/10.3390/ijms26189235
APA StyleCheng, L., Shi, N., Du, X., Huang, T., Zhang, Y., Zhao, C., Zhao, K., Lin, Z., Ma, D., Li, Q., Wang, F., Yao, H., & Shen, H. (2025). Bioinformatics Analysis and Expression Profiling Under Abiotic Stress of the DREB Gene Family in Glycyrrhiza uralensis. International Journal of Molecular Sciences, 26(18), 9235. https://doi.org/10.3390/ijms26189235





