Synthesis of Novel Tricyclic N-Acylhydrazones as Tubulin Polymerization Inhibitors
Abstract
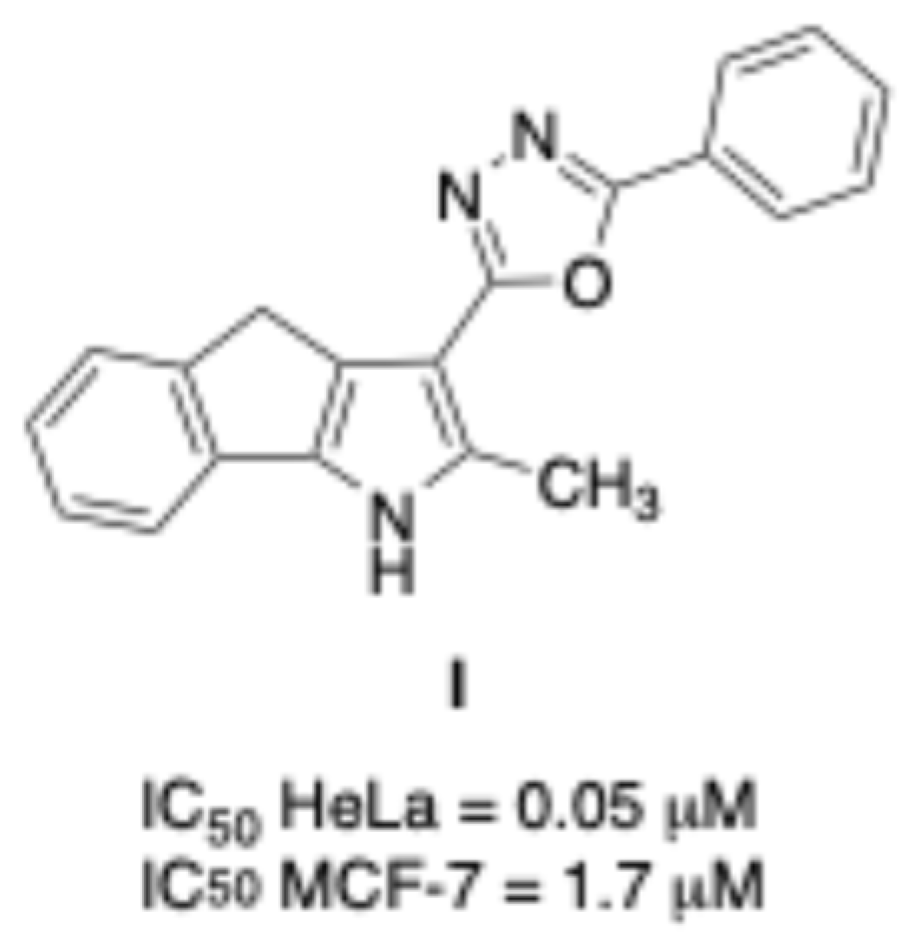
1. Introduction
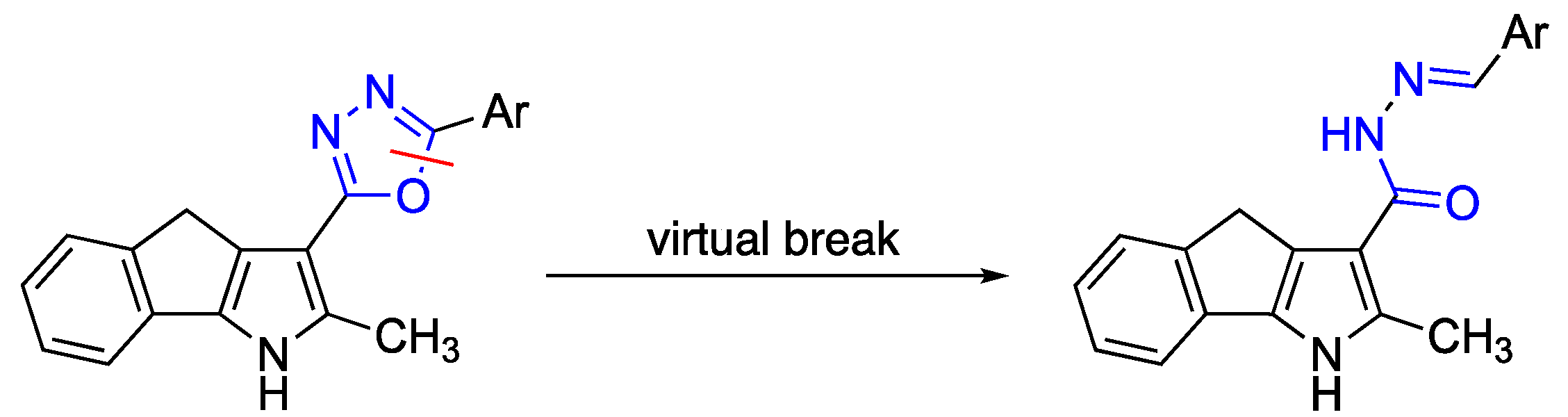
2. Results and Discussion
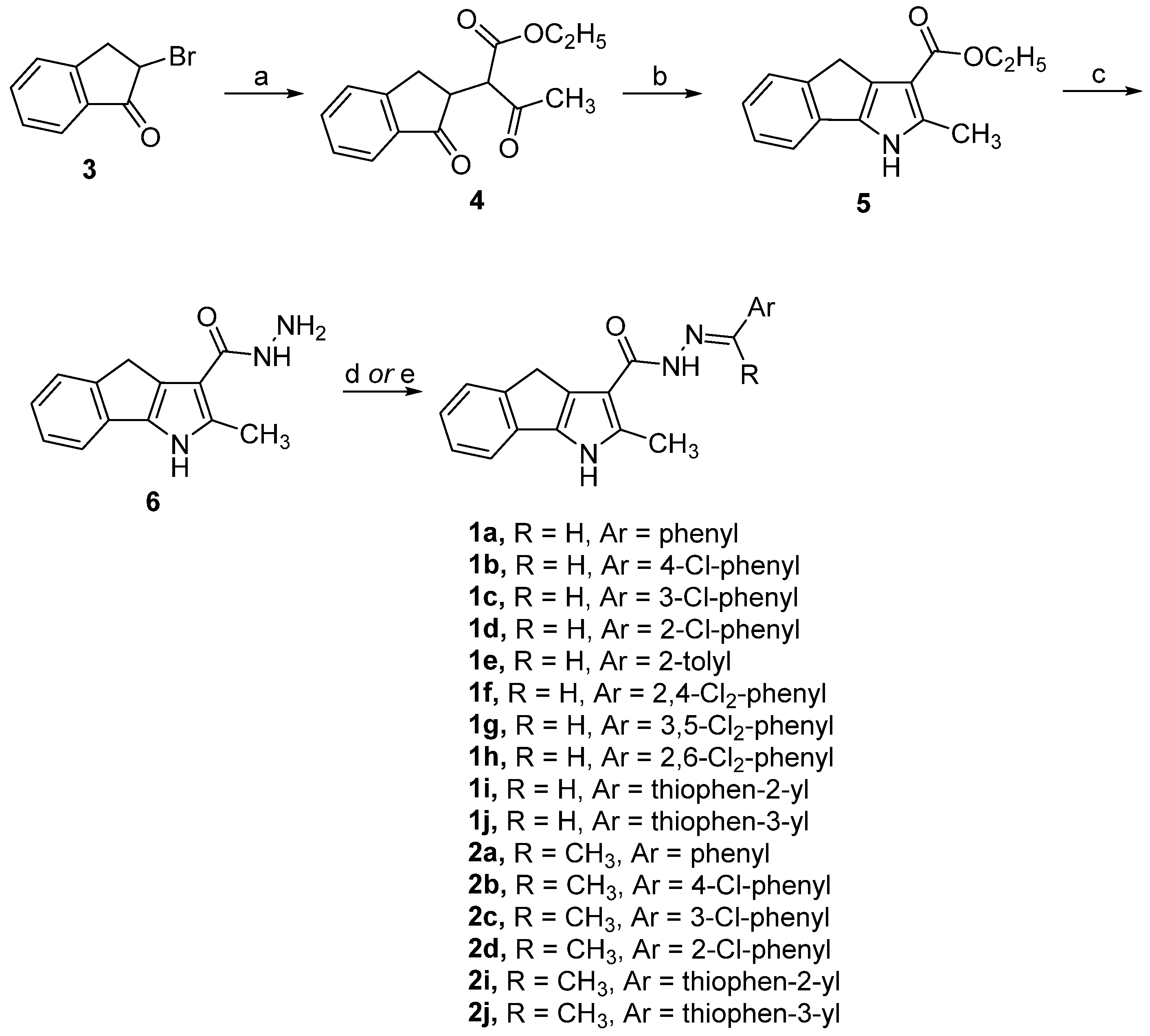
2.1. Synthesis of Carbohydrazides
2.2. Biology
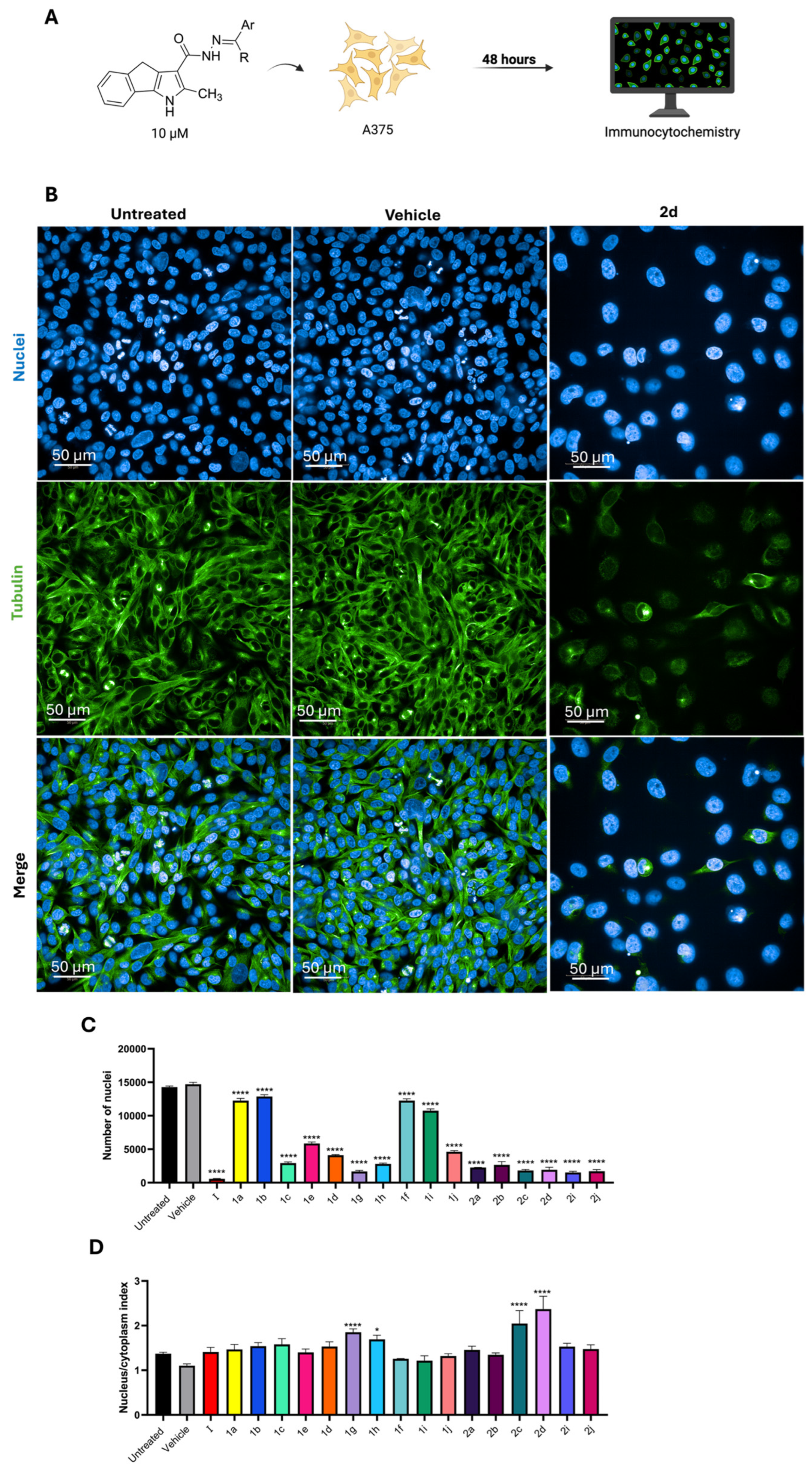
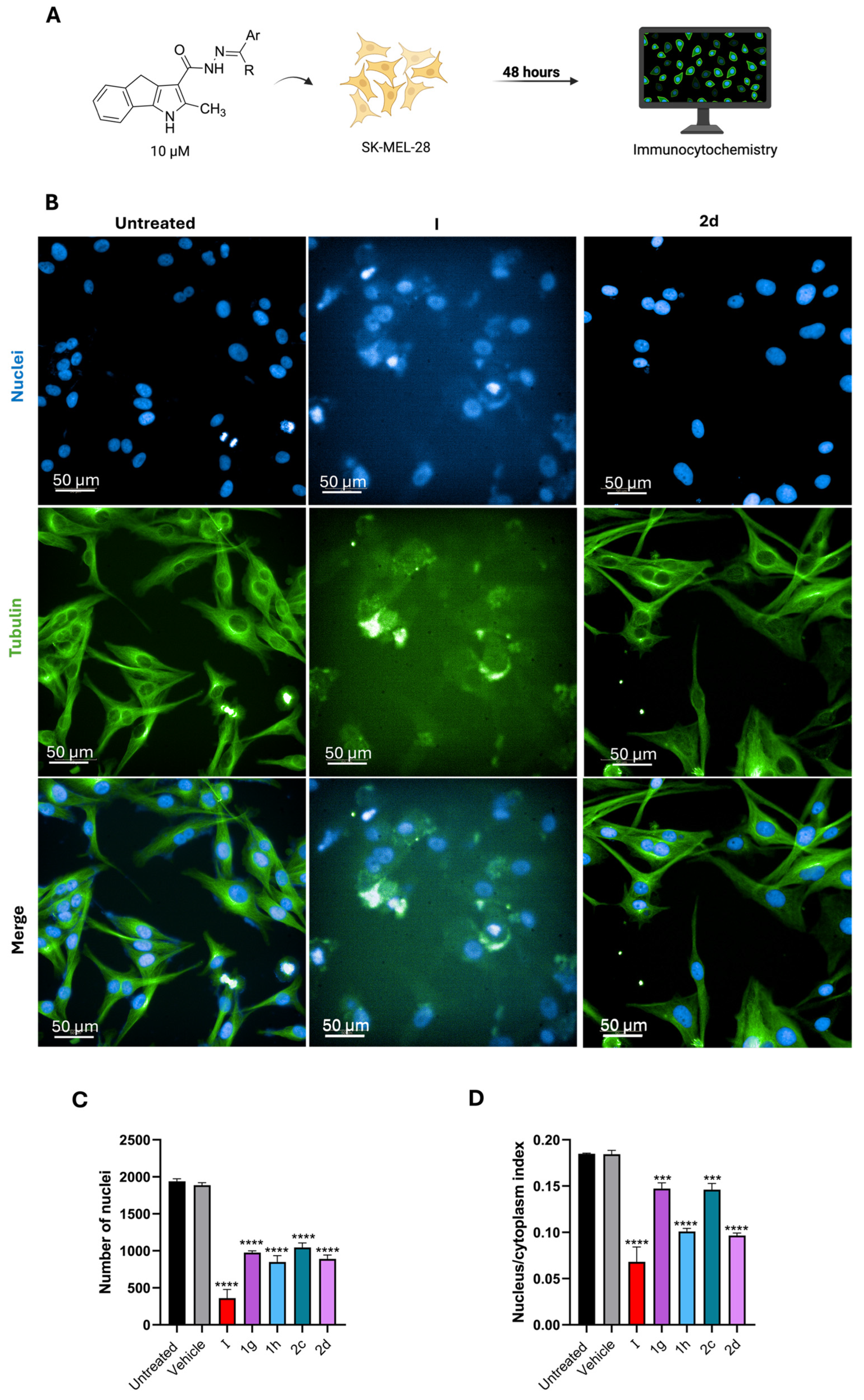
2.2.1. Immunocytochemistry
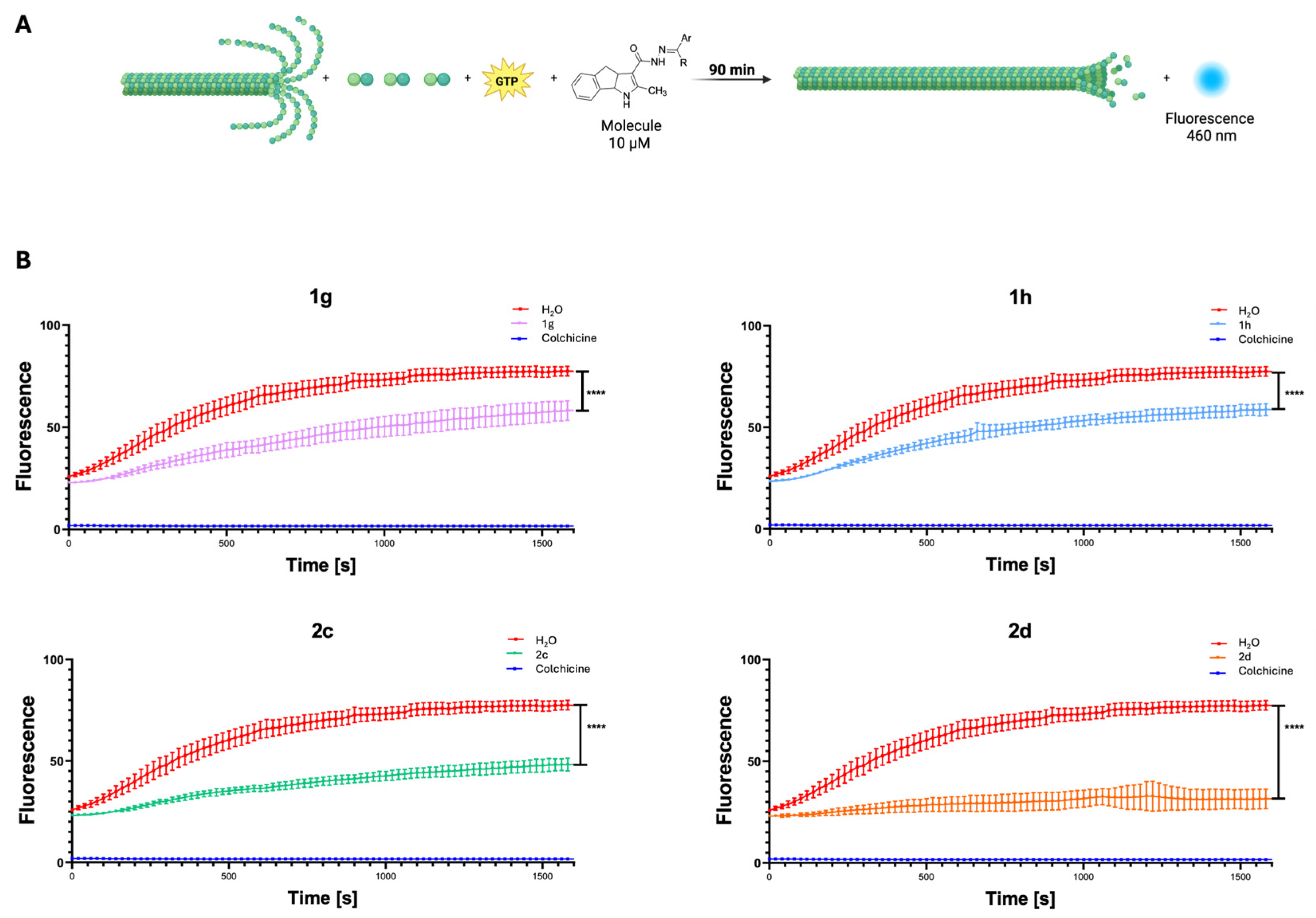
2.2.2. Tubulin Polymerization Assay
2.3. Computational Assay
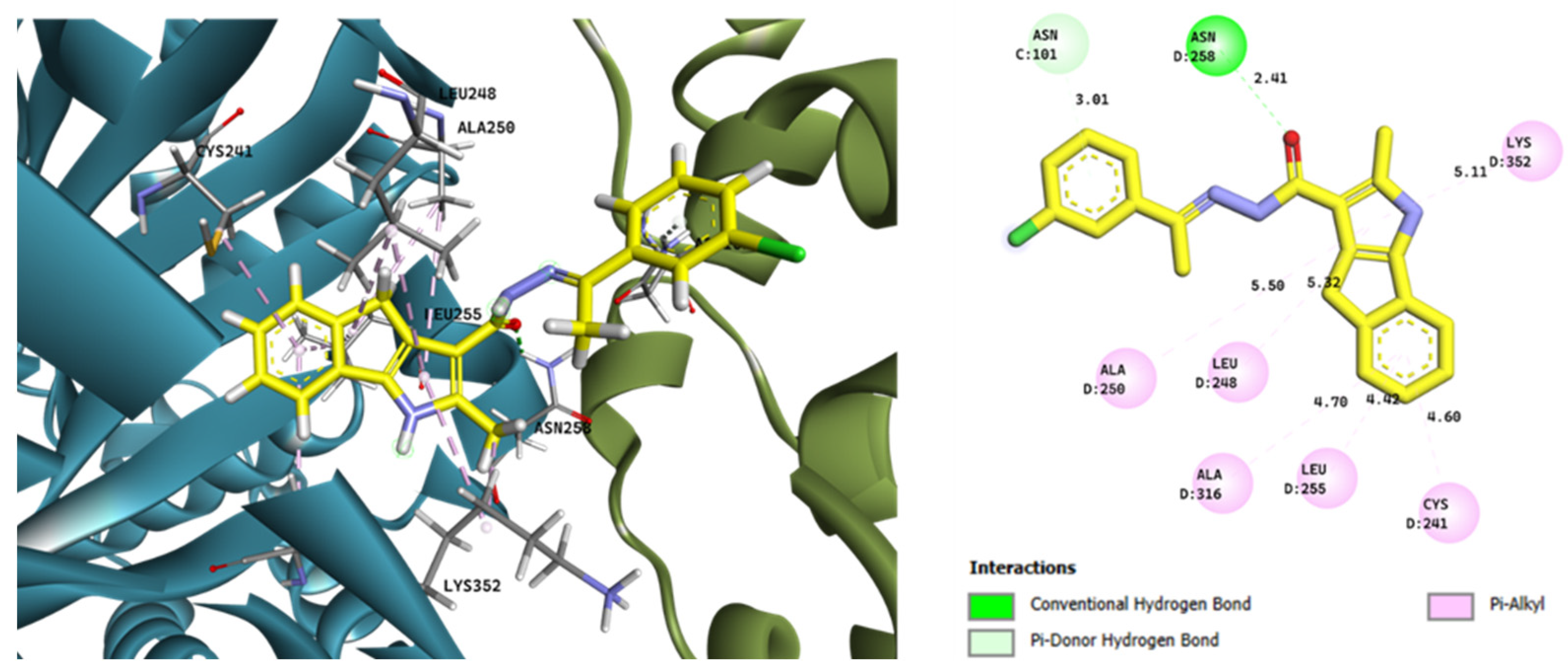
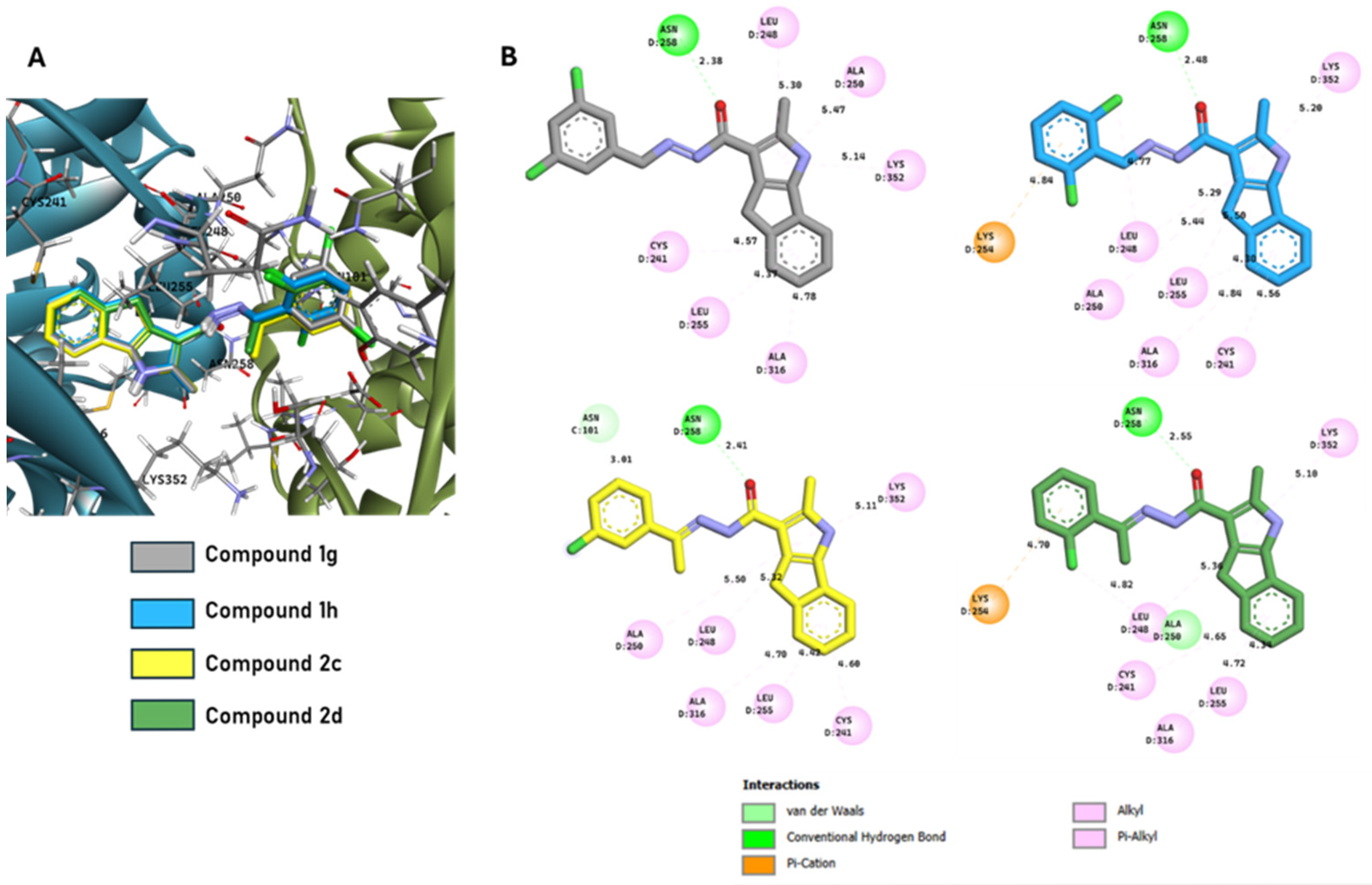
2.3.1. Computational Evaluation of Ligand–Tubulin Interactions
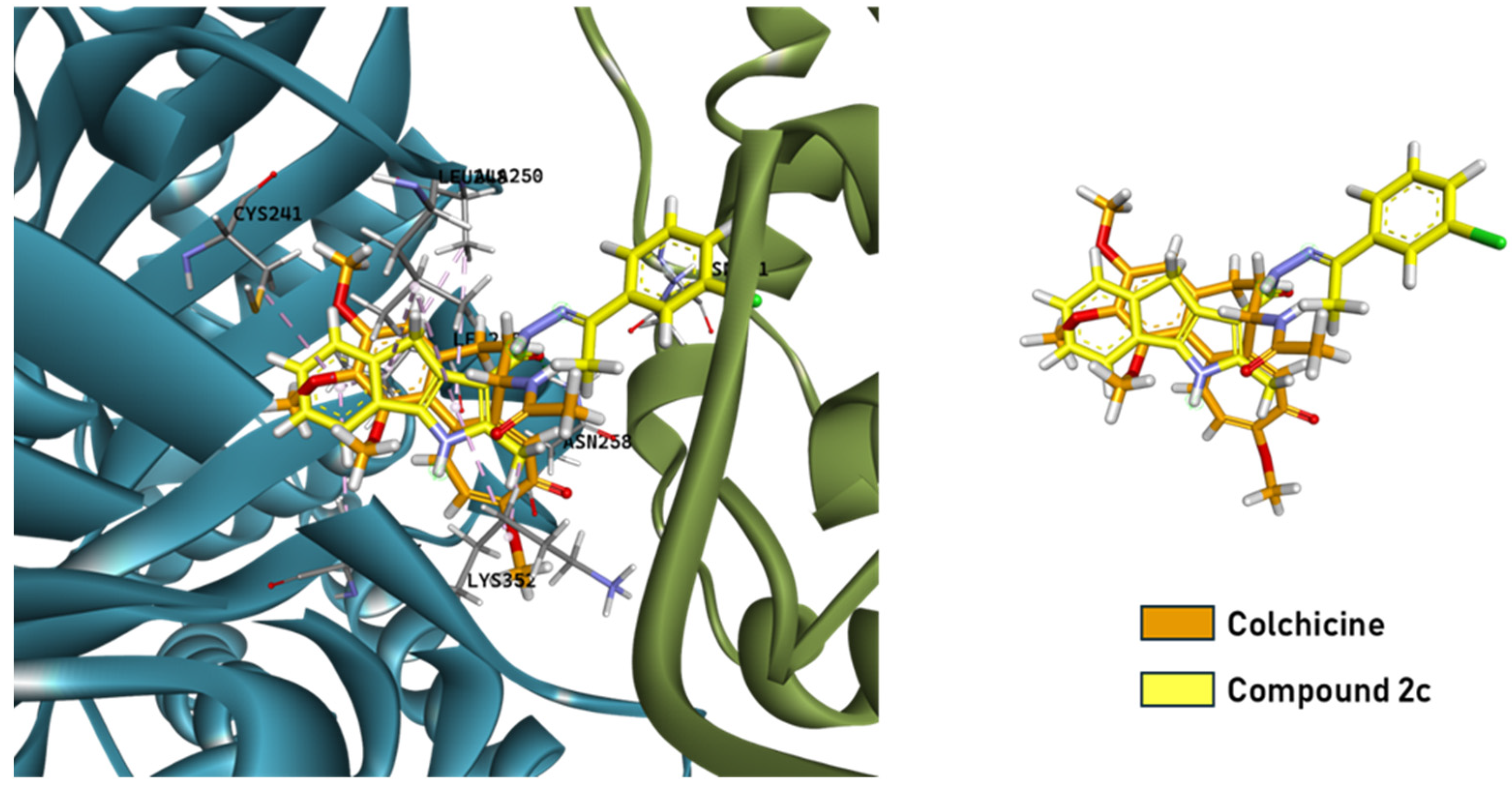
2.3.2. Binding Mode Analysis of Compound 2c
2.3.3. Structural Superimposition with Colchicine
2.3.4. Binding Patterns of the Most Active Ligands
2.3.5. Methodological Considerations
3. Methods and Materials
3.1. Chemistry
3.1.1. General Methods
3.1.2. General Procedure for the Synthesis of Carbohydrazides 1a–j and 2a–d,i,j
(E)-N′-(Benzylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1a)
(E)-N′-(4-Chlorobenzylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1b)
(E)-N′-(3-Chlorobenzylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1c)
(E)-N′-(2-Chlorobenzylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1d)
(E)-2-Methyl-N′-(2-methylbenzylidene)-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1e)
(E)-N′-(2,4-Dichlorobenzylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1f)
(E)-N′-(3,5-Dichlorobenzylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1g)
(E)-N′-(2,6-Dichlorobenzylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1h)
(E)-2-Methyl-N′-(thiophen-2-ylmethylene)-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1i)
(E)-2-Methyl-N′-(thiophen-3-ylmethylene)-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (1j)
(E)-2-Methyl-N′-(1-phenylethylidene)-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (2a)
(E)-N′-(1-(4-Chlorophenyl)ethylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (2b)
(E)-N′-(1-(3-Chlorophenyl)ethylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (2c)
(E)-N′-(1-(2-Chlorophenyl)ethylidene)-2-methyl-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (2d)
(E)-2-Methyl-N′-(1-(thiophen-2-yl)ethylidene)-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (2i)
(E)-2-Methyl-N′-(1-(thiophen-3-yl)ethylidene)-1,4-dihydroindeno[1,2-b]pyrrole-3-carbohydrazide (2j)
3.2. Biological Methods
3.2.1. A375 Cell Culture
3.2.2. Immunocytochemistry
3.2.3. Confocal Microscopy
3.2.4. Tubulin Polymerization Assay
3.3. Computational Methods
Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Crivelli, F.; Perfetti, E.; Sartorello, B.; Cantele, V.; Ostano, P.; Ilardi, F.; Di Massimo, D.S.; Clerico, M. Survey on symptoms in patients receiving chemotherapy in an Italian department of oncology: A comparison between period prevalences. Support. Care Cancer 2021, 29, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Bérubé, G. How to utilize academic research efforts in cancer drug discovery. Expert Opin. Drug Discov. 2019, 14, 331–334. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.; Elshamsy, A.M.; Ali, T.F.S.; Youssif, B.G.M.; Bräse, S.; Abdel-Aziz, M.; El-Koussi, N.A. Design and synthesis of new dihydropyrimidine derivatives with a cytotoxic effect as dual EGFR/VEGFR-2 inhibitors. ACS Omega 2024, 9, 34358–34369. [Google Scholar] [CrossRef]
- Prakash, S.; Tyagi, P.; Singh, P.; Rajkumar; Singh, A.P. Recent advancement in drug designing as small Molecules in targeted cancer therapy: Challenges and future directions. Curr. Cancer Drug Targets 2024, 25, 1364–1396. [Google Scholar] [CrossRef]
- Nieddu, V.; Pinna, G.; Marchesi, I.; Sanna, L.; Asproni, B.; Pinna, G.G.; Bagella, L.; Murineddu, G. Synthesis and antineoplastic evaluation of novel unsymmetrical 1,3,4-oxadiazoles. J. Med. Chem. 2016, 59, 10451–10469. [Google Scholar] [CrossRef] [PubMed]
- Lyu, W.; Sanna, L.; Bordoni, V.; Zeng, T.; Li, C.; Murineddu, G.; Pinna, G.A.; Kelvin, D.J.; Bagella, L. Target identification of a novel unsymmetrical 1,3,4-oxadiazole derivative with antiproliferative properties. J. Cell. Physiol. 2021, 236, 3789–3799. [Google Scholar] [CrossRef]
- Zoroddu, S.; Corona, P.; Sanna, L.; Borghi, F.; Bordoni, V.; Asproni, B.; Pinna, G.A.; Bagella, L.; Murineddu, G. Novel 1,3,4-oxadiazole chalcogen analogues: Synthesis and cytotoxic activity. Eur. J. Med. Chem. 2022, 238, 114440. [Google Scholar] [CrossRef]
- Kostova, I.; Saso, L. Advances in Research of Schiff-Base Metal Complexes as Potent Antioxidants. Curr. Med. Chem. 2013, 20, 4609–4632. [Google Scholar] [CrossRef]
- de Oliveira Carneiro Brum, J.; França, T.C.C.; LaPlante, S.R.; Villar, J.D.F. Synthesis and Biological Activity of Hydrazones and Derivatives: A Review. Mini Rev. Med. Chem. 2020, 20, 342–368. [Google Scholar] [CrossRef] [PubMed]
- Demurtas, M.; Baldisserotto, A.; Lampronti, I.; Moi, D.; Balboni, G.; Pacifico, S.; Vertuani, S.; Manfredini, S.; Onnis, V. Indole Derivatives as Multifunctional Drugs: Synthesis and Evaluation of Antioxidant, Photoprotective and Antiproliferative Activity of Indole Hydrazones. Bioorg. Chem. 2019, 85, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Han, M.İ.; Baysal, Ö.D.Y.; Başaran, G.Ş.; Sezer, G.; Telci, D.; Küçükgüzel, Ş.G. Design, Synthesis and Anticancer Activity Studies of Novel 4-Butylaminophenyl Hydrazide-Hydrazones as Apoptotic Inducers. Tetrahedron 2022, 115, 132797. [Google Scholar] [CrossRef]
- Tumosienè, I.; Stasevych, M.; Zvarych, V.; Jonuškiene, I.; Kantminienè, K.; Petrikatè, V. Novel 5-Oxopyrrolidine-3-carbohydrazides as Potent Protein Kinase Inhibitors: Synthesis, Anticancer Evaluation, and Molecular Modeling. Int. J. Mol. Sci. 2025, 26, 3162. [Google Scholar] [CrossRef]
- Natarelli, N.; Aleman, S.J.; MarK, I.M.; Tran, J.T.; Kwak, S.; Botto, E.; Aflatooni, S.; Diaz, M.J.; Lipner, S.R.; Shah, A. A Review of Current and Pipeline Drugs for Treatment of Melanoma. Pharmaceuticals 2024, 17, 214. [Google Scholar] [CrossRef]
- Palla, G.; Pelizzi, C.; Predieri, G.; Vignali, C. Conformational study on N-acylhydrazones of aromatic aldehydes by NMR spectroscopy. Gazz. Chim. Ital. 1982, 112, 339–341. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- BIOVIA Discovery Studio Visualizer, V25.1.0.24284; Dassault Systèmes: San Diego, CA, USA, 2025.
- Lu, Y.; Chen, J.; Xiao, M.; Li, W.; Miller, D.D. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm. Res. 2012, 29, 2943–2971. [Google Scholar] [CrossRef]
- Vilar, S.; Costanzi, S. Predicting Biological Activities through QSAR Analysis and Docking-Based Scoring. In Methods in Molecular Biology; Gohlke, H., Ed.; Humana Press: Totowa, NJ, USA, 2012; Volume 819, pp. 315–338. [Google Scholar] [CrossRef]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Li, J.; Fu, A.; Zhang, L. An Overview of Scoring Functions Used for Protein–Ligand Interactions in Molecular Docking. Interdiscip. Sci. 2019, 11, 320–328. [Google Scholar] [CrossRef]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.W.; Zhang, Y. DockRMSD: An Open-Source Tool for Atom Mapping and RMSD Calculation of Symmetric Molecules through Graph Isomorphism. J. Cheminform. 2019, 11, 40. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeerschd, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software News and Updates AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]

| Compound | Structure | Ar |
|---|---|---|
| 1a | 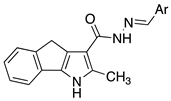 | phenyl |
| 1b | 4-Cl-phenyl | |
| 1c | 3-Cl-phenyl | |
| 1d | 2-Cl-phenyl | |
| 1e | 2-tolyl | |
| 1f | 2,4-Cl2-phenyl | |
| 1g | 3,5-Cl2-phenyl | |
| 1h | 2,6-Cl2-phenyl | |
| 1i | thiophen-2-yl | |
| 1j | thiophen-3-yl | |
| 2a | 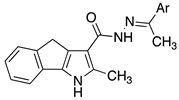 | phenyl |
| 2b | 4-Cl-phenyl | |
| 2c | 3-Cl-phenyl | |
| 2d | 2-Cl-phenyl | |
| 2i | thiophen-2-yl | |
| 2j | thiophen-3-yl |
| Compound | Best Predicted Pose Binding Affinity (kcal/mol) | Selected Pose Binding Affinity (kcal/mol) | Chosen Pose Rank |
|---|---|---|---|
| 1a | −9.4 | −9.1 | 4 |
| 1b | −9.6 | −9.3 | 2 |
| 1c | −9.8 | −9.4 | 4 |
| 1d | −9.8 | −9.1 | 4 |
| 1e | −9.7 | −9.0 | 5 |
| 1f | −9.8 | −9.4 | 2 |
| 1g | −9.3 | −8.6 | 3 |
| 1h | −9.6 | −9.3 | 3 |
| 1i | −8.9 | −8.2 | 5 |
| 1j | −9.0 | −7.7 | 2 |
| 2a | −9.6 | −9.4 | 2 |
| 2b | −9.6 | −9.6 | 1 |
| 2c | −10.0 | −9.4 | 3 |
| 2d | −9.8 | −9.7 | 2 |
| 2i | −8.8 | −8.2 | 4 |
| 2j | −9.1 | −9.1 | 1 |
| Colchicine | −10.7 | −10.7 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corona, P.; Lai, M.; Asproni, B.; Sciandrone, G.; Lupinu, I.; Ibba, R.; Piras, S.; Carta, A.; Murineddu, G. Synthesis of Novel Tricyclic N-Acylhydrazones as Tubulin Polymerization Inhibitors. Int. J. Mol. Sci. 2025, 26, 9212. https://doi.org/10.3390/ijms26189212
Corona P, Lai M, Asproni B, Sciandrone G, Lupinu I, Ibba R, Piras S, Carta A, Murineddu G. Synthesis of Novel Tricyclic N-Acylhydrazones as Tubulin Polymerization Inhibitors. International Journal of Molecular Sciences. 2025; 26(18):9212. https://doi.org/10.3390/ijms26189212
Chicago/Turabian StyleCorona, Paola, Michele Lai, Battistina Asproni, Giulia Sciandrone, Ilenia Lupinu, Roberta Ibba, Sandra Piras, Antonio Carta, and Gabriele Murineddu. 2025. "Synthesis of Novel Tricyclic N-Acylhydrazones as Tubulin Polymerization Inhibitors" International Journal of Molecular Sciences 26, no. 18: 9212. https://doi.org/10.3390/ijms26189212
APA StyleCorona, P., Lai, M., Asproni, B., Sciandrone, G., Lupinu, I., Ibba, R., Piras, S., Carta, A., & Murineddu, G. (2025). Synthesis of Novel Tricyclic N-Acylhydrazones as Tubulin Polymerization Inhibitors. International Journal of Molecular Sciences, 26(18), 9212. https://doi.org/10.3390/ijms26189212







