Nor1 and Mitophagy: An Insight into Sertoli Cell Function Regulating Spermatogenesis Using a Transgenic Rat Model
Abstract
1. Introduction
2. Results
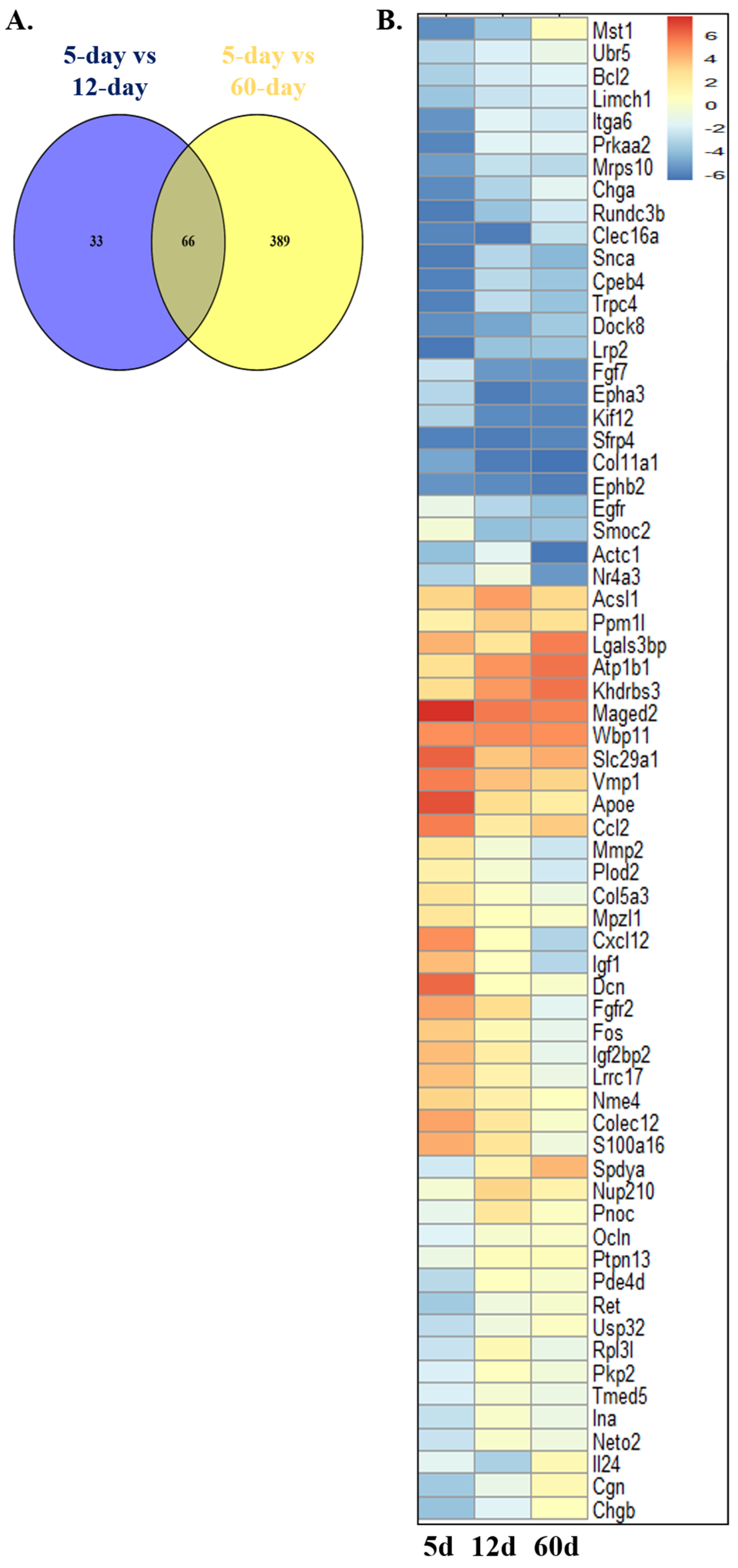
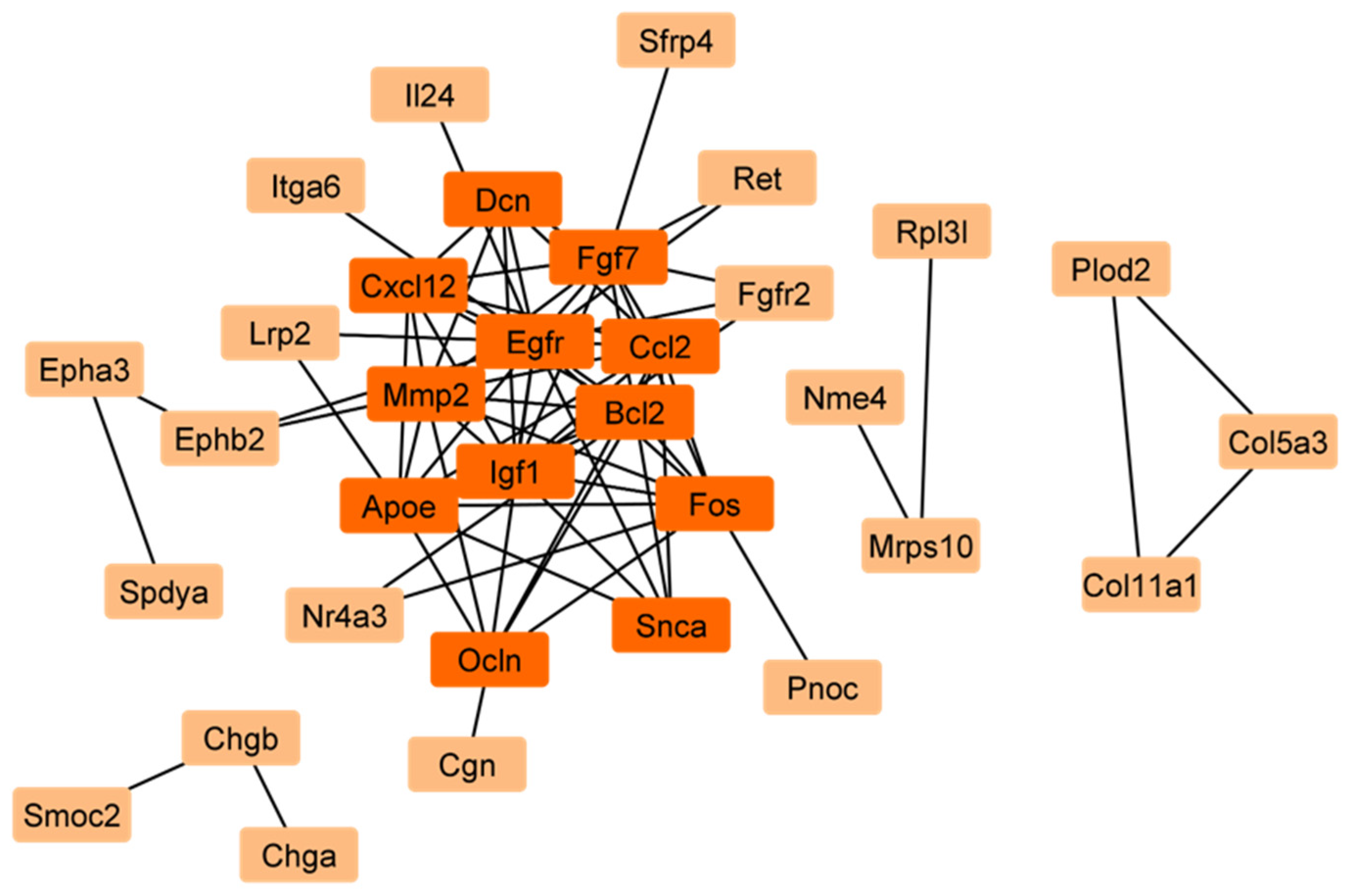
2.1. Identification of Mitophagy-Related Genes (MRGs) During the Development of Scs
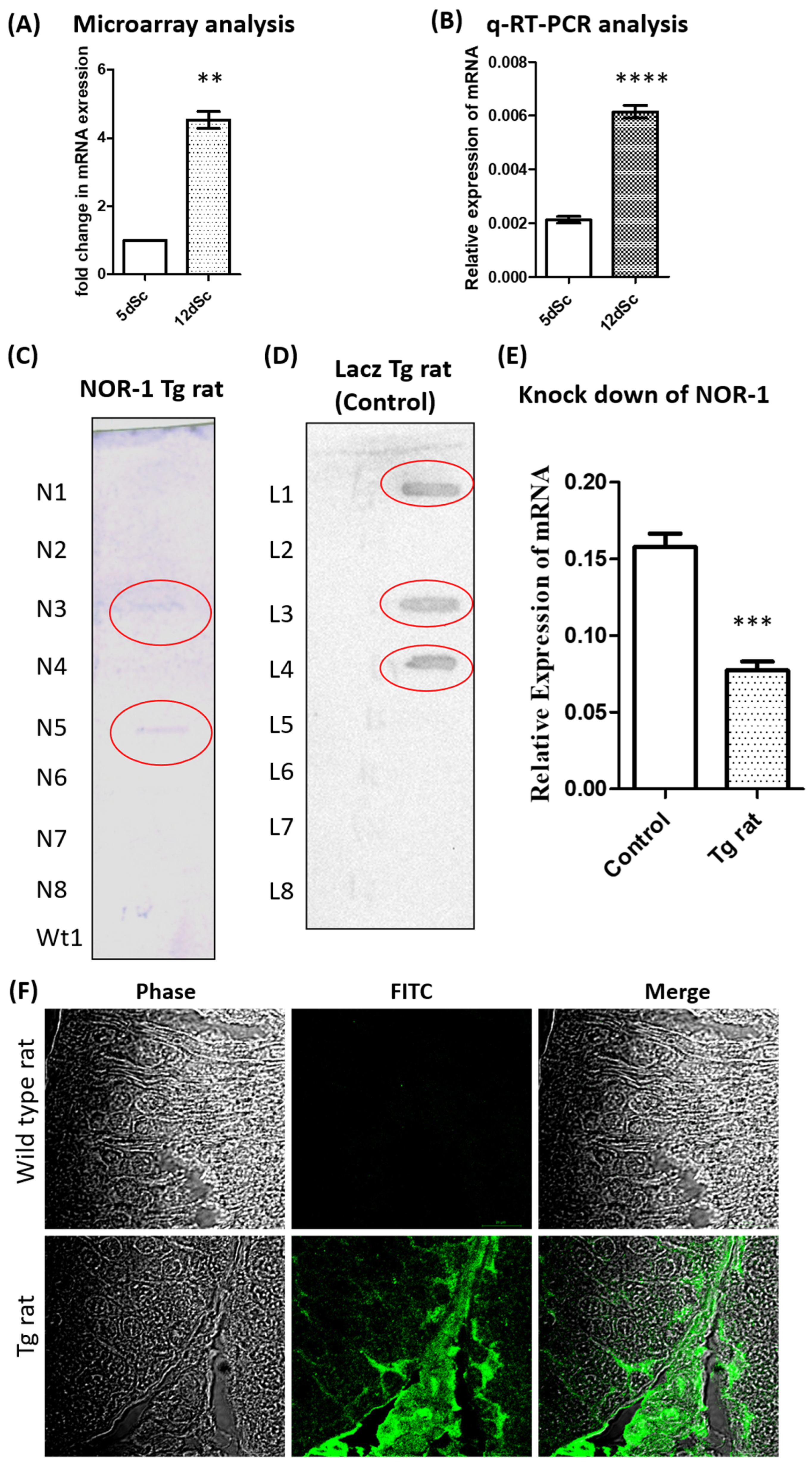
2.2. Validation of the Differential Expression of Nor1 in Scs
2.3. Generation of Transgenic Rat with Sc-Specific NOR1 Knockdown
2.4. Attenuation of Spermatogenesis in Testes of Sc-Specific Nor1 Knockdown Tg Rats
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Screening of Hub Genes for Sertoli Cell Development in Rats
4.3. Functional Enrichment Analysis
4.4. Differential Expression Analysis of Nor1 in 5-Day-Old and 12-Day-Old Rat Sc Cultures
4.5. Plasmids and Cloning
4.6. Generation of Transgenic (Tg) Rats with Reduced NOR1 in Scs
4.7. Validation of Knockdown of NOR1 by q-RT-PCR
4.8. Immunofluorescence Microscopy
4.9. Tissue Histology
4.10. Fertility Analyses of Nor1 Knockdown Rats
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Gc | Germ cell |
| Sc | Sertoli cell |
| Bcl2 | B-Cell Leukemia/Lymphoma 2 |
| Fos | FBJ murine osteosarcoma viral oncogene homolog |
| LC3 | Microtubule-associated protein 1A/1B light chain 3 |
| Mfn1 | mitofusin 1 |
| Mfn2 | mitofusin 2 |
| Opa1 | Optic Atrophy 1, mitochondrial dynamin-like GTPase |
| PTEN | phosphatase and tensin homolog |
| PINK1 | phosphatase and tensin homolog induced kinase I |
| UPS | ubiquitin proteasome system |
| mtDNA | mitochondrial DNA |
| POLG | polymerase gamma |
| Mff | mitochondrial fission factor |
| Atg7 | autophagy related 7 |
| GATA4 | GATA binding protein 4 |
| MRG | mitophagy-related genes |
| Tg rat | Sertoli-cell-specific Nor1 knockdown transgenic rat |
| Pgc1α | peroxisome proliferator-activated receptor gamma coactivator 1 α |
| Tfam | Transcription factor A, mitochondrial |
| Smad3 | SMAD family member 3 |
| DEGs | differentially expressed genes |
| MRDEGs | mitophagy-related differentially expressed genes |
| PPIN | protein–protein interaction network |
| Egfr | epidermal growth factor receptor |
| Ccl2 | C-C motif chemokine ligand 2 |
| Mmp2 | matrix metallopeptidase 2 |
| Igf1 | insulin-like growth factor 1 |
| Fgf7 | fibroblast growth factor 7 |
| Apoe | apolipoprotein E |
| Cxcl12 | C-X-C motif chemokine ligand 12 |
| Ocln | occludin |
| Dcn | decorin |
| Scna | synuclein alpha |
| GFP | green fluorescent protein |
| Mmp8 | Matrix metallo protease8 |
| ROS | reactive oxygen species |
| Amh | anti mullerian hormone |
| GEO | Gene Expression Omnibus |
| FDR | False discovery rate |
| MRGSCD | mitophagy-related genes in Sc development |
References
- Agarwal, A.; Baskaran, S.; Parekh, N.; Cho, C.L.; Henkel, R.; Vij, S.; Arafa, M.; Panner Selvam, M.K.; Shah, R. Male infertility. Lancet 2021, 397, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Potiris, A.; Moustakli, E.; Trismpioti, E.; Drakaki, E.; Mavrogianni, D.; Matsas, A.; Zikopoulos, A.; Sfakianakis, A.; Tsakiridis, I.; Dagklis, T.; et al. From Inflammation to Infertility: How Oxidative Stress and Infections Disrupt Male Reproductive Health. Metabolites 2025, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wang, Q. Exploring the Characters of Non-Coding RNAs in Spermatogenesis and Mal e Infertility. Int. J. Mol. Sci. 2025, 26, 1128. [Google Scholar] [CrossRef]
- Qu, N. Pharmacological Effects and Immune Mechanisms of Oriental Medicines in Restoring Male Infertility. Int. J. Mol. Sci. 2025, 26, 4642. [Google Scholar] [CrossRef]
- Sharpe, R.M. Sperm counts and fertility in men: A rocky road ahead. Science & Socie ty Series on Sex and Science. EMBO Rep. 2012, 13, 398–403. [Google Scholar]
- Vander Borght, M.; Wyns, C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018, 62, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Olesen, I.A.; Andersson, A.M.; Aksglaede, L.; Skakkebaek, N.E.; Rajpert-de Meyts, E.; Joergensen, N.; Juul, A. Clinical, genetic, biochemical, and testicular biopsy findings among 1,213 men evaluated for infertility. Fertil. Steril. 2017, 107, 74–82.e7. [Google Scholar] [CrossRef]
- Tournaye, H.; Krausz, C.; Oates, R.D. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol. 2017, 5, 544–553. [Google Scholar] [CrossRef]
- Zecevic, N.; Veselinovic, A.; Perovic, M.; Stojsavljevic, A. Association Between Zinc Levels and the Impact of Its Deficiency on Idiopathic Male Infertility: An Up-to-Date Review. Antioxidants 2025, 14, 165. [Google Scholar] [CrossRef]
- Bouvattier, C. Neonatal gonadotropin therapy in male congenital hypogonadotropic hypo gonadism. Nat. Rev. Endocrinol. 2011, 8, 172–182. [Google Scholar] [CrossRef]
- Irvine, D.S. Epidemiology and aetiology of male infertility. Hum. Reprod. 1998, 13 (Suppl. 1), 33–44. [Google Scholar] [CrossRef]
- Maroto, M.; Torvisco, S.N.; García-Merino, C.; Fernández-González, R.; Pericuesta, E. Mechanisms of Hormonal, Genetic, and Temperature Regulation of Germ Ce ll Proliferation, Differentiation, and Death During Spermatogenesis. Biomolecules 2025, 15, 500. [Google Scholar] [CrossRef]
- Santi, D.; Corona, G.; Salonia, A.; Ferlin, A. Current drawbacks and future perspectives in the diagnosis and treatme nt of male factor infertility, with a focus on FSH treatment: An exper t opinion. J. Endocrinol. Investig. 2025, 48, 1085–1100. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Chen, W.; Cheng, Z.; Wu, S.; He, J.; Han, L.; He, Z.; Qin, W. Novel Gene Regulation in Normal and Abnormal Spermatogenesis. Cells 2021, 10, 666. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998, 9, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.F.; Shen, Y.F.; Gao, D.W.; Lin, D.F.; Ma, W.Z.; Chang, D.G. Metabolic pathways and male fertility: Exploring the role of Sertoli c ells in energy homeostasis and spermatogenesis. Am. J. Physiol. Endocrinol. Metab. 2025, 329, E160–E178. [Google Scholar] [CrossRef]
- Shah, W.; Khan, R.; Shah, B.; Khan, A.; Dil, S.; Liu, W.; Wen, J.; Jiang, X. The Molecular Mechanism of Sex Hormones on Sertoli Cell Development an d Proliferation. Front. Endocrinol. 2021, 12, 648141. [Google Scholar] [CrossRef]
- Wang, L.; Bu, T.; Wu, X.; Gao, S.; Li, X.; Jesus, A.B.; Wong, C.K.C.; Chen, H.; Chung, N.P.Y.; Sun, F.; et al. Cell-Cell Interaction-Mediated Signaling in the Testis Induces Reprodu ctive Dysfunction-Lesson from the Toxicant/Pharmaceutical Models. Cells 2022, 11, 591. [Google Scholar] [CrossRef]
- Hai, Y. The roles and regulation of Sertoli cells in fate determinations of sp ermatogonial stem cells and spermatogenesis. Semin. Cell Dev. Biol. 2014, 29, 66–75. [Google Scholar] [CrossRef]
- Boguenet, M.; Bouet, P.E.; Spiers, A.; Reynier, P.; May-Panloup, P. Mitochondria: Their role in spermatozoa and in male infertility. Hum. Reprod. Update 2021, 27, 697–719. [Google Scholar] [CrossRef]
- Chang, Z.; Miao, L.; Wang, P. Mitochondrial Ribosome Regulation Drives Spermatogenesis and Male Fert ility. Biol. Cell 2025, 117, e12007. [Google Scholar] [CrossRef]
- Tesarik, J.; Mendoza-Tesarik, R. Mitochondria in Human Fertility and Infertility. Int. J. Mol. Sci. 2023, 24, 8950. [Google Scholar] [CrossRef]
- Costa, J.; Braga, P.C.; Rebelo, I.; Oliveira, P.F.; Alves, M.G. Mitochondria Quality Control and Male Fertility. Biology 2023, 12, 827. [Google Scholar] [CrossRef]
- Park, Y.J.; Pang, M.G. Mitochondrial Functionality in Male Fertility: From Spermatogenesis to Fertilization. Antioxidants 2021, 10, 98. [Google Scholar] [CrossRef]
- Wang, X.; Yin, L.; Wen, Y.; Yuan, S. Mitochondrial regulation during male germ cell development. Cell. Mol. Life Sci. 2022, 79, 91. [Google Scholar] [CrossRef]
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef]
- Mizushima, N.; Sugita, H.; Yoshimori, T.; Ohsumi, Y. A New Protein Conjugation System in Human the counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 1998, 273, 33889–33892. [Google Scholar] [CrossRef]
- Picca, A.; Faitg, J.; Auwerx, J.; Ferrucci, L.; D’Amico, D. Mitophagy in human health, ageing and disease. Nat. Metab. 2023, 5, 2047–2061. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Sacch aromyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Kuma, A.; Komatsu, M.; Mizushima, N. Autophagy-monitoring and autophagydeficient mice. Autophagy 2017, 13, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- McWilliams, T.G.; Prescott, A.R.; Allen, G.F.G.; Tamjar, J.; Munson, M.J.; Thomson, C.; Muqit, M.M.K.; Ganley, I.G. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 2016, 214, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Yun, J.; Liu, J.; Malide, D.; Liu, C.; Rovira, I.I.; Holmström, K.M.; Fergusson, M.M.; Yoo, Y.H.; Combs, C.A.; et al. Measuring in vivo Mitophagy. Mol. Cell 2015, 60, 685–696. [Google Scholar] [CrossRef]
- Chan, D.C. Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 235–259. [Google Scholar] [CrossRef]
- Tábara, L.C.; Segawa, M.; Prudent, J. Molecular mechanisms of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2025, 26, 123–146. [Google Scholar] [CrossRef]
- Zaman, M.; Shutt, T.E. The Role of Impaired Mitochondrial Dynamics in MFN2-Mediated Pathology. Front. Cell Dev. Biol. 2022, 10, 858286. [Google Scholar] [CrossRef]
- Zhao, S.; Heng, N.; Wang, H.; Wang, H.; Zhang, H.; Gong, J.; Hu, Z.; Zhu, H. Mitofusins: From mitochondria to fertility. Cell. Mol. Life Sci. 2022, 79, 370. [Google Scholar] [CrossRef]
- Chan, N.C.; Chan, D.C. Parkin uses the UPS to ship off dysfunctional mitochondria. Autophagy 2011, 7, 771–772. [Google Scholar] [CrossRef]
- Rakovic, A.; Ziegler, J.; Mårtensson, C.U.; Prasuhn, J.; Shurkewitsch, K.; König, P.; Paulson, H.L.; Klein, C. PINK1-dependent mitophagy is driven by the UPS and can occur independe ntly of LC3 conversion. Cell Death Differ. 2019, 26, 1428–1441. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Urbé, S. Diverse routes to mitophagy governed by ubiquitylation and mitochondri al import. Trends Cell Biol. 2025, 35, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Youle, R.J. The role of PINK1-Parkin in mitochondrial quality control. Nat. Cell Biol. 2024, 26, 1639–1651. [Google Scholar] [CrossRef]
- Sekine, S.; Youle, R.J. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol. 2018, 16, 2. [Google Scholar] [CrossRef]
- Baklouti-Gargouri, S.; Ghorbel, M.; Ben Mahmoud, A.; Mkaouar-Rebai, E.; Cherif, M.; Chakroun, N.; Sellami, A.; Fakhfakh, F.; Ammar-Keskes, L. Identification of a novel m.9588G > a missense mutation in the mitocho ndrial COIII gene in asthenozoospermic Tunisian infertile men. J. Assist. Reprod. Genet. 2014, 31, 595–600. [Google Scholar] [CrossRef]
- Carra, E.; Sangiorgi, D.; Gattuccio, F.; Rinaldi, A.M. Male infertility and mitochondrial DNA. Biochem. Biophys. Res. Commun. 2004, 322, 333–339. [Google Scholar] [CrossRef]
- Demain, L.A.; Conway, G.S.; Newman, W.G. Genetics of mitochondrial dysfunction and infertility. Clin. Genet. 2017, 91, 199–207. [Google Scholar] [CrossRef]
- Zhang, X.; Xiong, Y.; Tan, Y.; Wang, L.; Li, R.; Zhang, Y.; Huang, H. Melatonin rescues impaired penetration ability of human spermatozoa in duced by mitochondrial dysfunction. Reproduction 2019, 158, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Irigoyen, P.; Pintos-Polasky, P.; Rosa-Villagrán, L.; Skowronek, M.; Cassina, A.; Sapiro, R. Mitochondrial metabolism determines the functional status of human spe rm and correlates with semen parameters. Front. Cell Dev. Biol. 2022, 10, 926684. [Google Scholar] [CrossRef] [PubMed]
- Luoma, P.; Melberg, A.; Rinne, J.O.; Kaukonen, J.A.; Nupponen, N.N.; Chalmers, R.M.; Oldfors, A.; Rautakorpi, I.; Peltonen, L.; Majamaa, K.; et al. Parkinsonism, premature menopause, and mitochondrial DNA polymerase γ mutations: Clinical and molecular genetic study. Lancet 2004, 364, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Kujoth, G.C.; Hiona, A.; Pugh, T.D.; Someya, S.; Panzer, K.; Wohlgemuth, S.E.; Hofer, T.; Seo, A.Y.; Sullivan, R.; Jobling, W.A.; et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 2005, 309, 481–484. [Google Scholar] [CrossRef]
- Trifunovic, A.; Wredenberg, A.; Falkenberg, M.; Spelbrink, J.N.; Rovio, A.T.; Bruder, C.E.; Bohlooly, Y.M.; Gidlof, S.; Oldfors, A.; Wibom, R.; et al. Premature ageing in mice expressing defective mitochondrial DNA polyme rase. Nature 2004, 429, 417–423. [Google Scholar] [CrossRef]
- Varuzhanyan, G.; Rojansky, R.; Sweredoski, M.J.; Graham, R.L.; Hess, S.; Ladinsky, M.S.; Chan, D.C. Mitochondrial fusion is required for spermatogonial differentiation an d meiosis. eLife 2019, 8, 51601. [Google Scholar] [CrossRef]
- Chen, H.; Ren, S.; Clish, C.; Jain, M.; Mootha, V.; McCaffery, J.M.; Chan, D.C. Titration of mitochondrial fusion rescues Mff-deficient cardiomyopathy. J. Cell Biol. 2015, 211, 795–805. [Google Scholar] [CrossRef]
- Shang, Y.; Wang, H.; Jia, P.; Zhao, H.; Liu, C.; Liu, W.; Song, Z.; Xu, Z.; Yang, L.; Wang, Y. Autophagy regulates spermatid differentiation via degradation of PDLIM 1. Autophagy 2016, 12, 1575–1592. [Google Scholar] [CrossRef]
- Wang, H.; Wan, H.; Li, X.; Liu, W.; Chen, Q.; Wang, Y.; Yang, L.; Tang, H.; Zhang, X.; Duan, E. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014, 24, 852. [Google Scholar] [CrossRef]
- Dietert, S.E. Fine structure of the formation and fate of the residual bodies of mou se spermatozoa with evidence for the participation of lysosomes. J. Morphol. 1966, 120, 317–346. [Google Scholar] [CrossRef]
- Eid, N.; Ito, Y.; Horibe, A.; Hamaoka, H.; Kondo, Y. A Method for In Vivo Induction and Ultrastructural Detection of Mitoph agy in Sertoli Cells. Methods Mol. Biol. 2018, 1748, 103–112. [Google Scholar]
- Beckervordersandforth, R.; Ebert, B.; Schäffner, I.; Moss, J.; Fiebig, C.; Shin, J.; Moore, D.L.; Ghosh, L.; Trinchero, M.F.; Stockburger, C.; et al. Role of mitochondrial metabolism in the control of early lineage progr ession and aging phenotypes in adult hippocampal neurogenesis. Neuron 2017, 93, 560–573.e6. [Google Scholar] [CrossRef]
- Eid, N.; Ito, Y.; Otsuki, Y. Enhanced mitophagy in sertoli cells of ethanol-treated rats: Morpholog ical evidence and clinical relevance. J. Mol. Histol. 2011, 43, 71–80. [Google Scholar] [CrossRef]
- Yéfimova, M.; Messaddeq, N.; Harnois, T.; Meunier, A.; Clarhaut, J.; Noblanc, A.; Benzakour, O. A chimerical phagocytosis model reveals the recruitment by sertoli cel ls of autophagy for the degradation of ingested illegitimate substrate s. Autophagy 2013, 9, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, T.; Nakamura, S.; Yamano, Y.; Endo, T.; Yanagawa, K.; Tokumura, A.; Yoshimori, T. Rubicon prevents autophagic degradation of gata4 to promote sertoli ce ll function. PLoS Genet. 2021, 17, 1009688. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, I. A switch in Sertoli cell responsiveness to FSH may be responsible for robust onset of germ cell differentiation during prepubartal testicula r maturation in rats. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E886–E898. [Google Scholar] [CrossRef]
- Gautam, M.; Bhattacharya, I.; Rai, U.; Majumdar, S.S. Hormone induced differential transcriptome analysis of Sertoli cells d uring postnatal maturation of rat testes. PLoS ONE 2018, 13, e0191201. [Google Scholar] [CrossRef]
- Close, A.F.; Dadheech, N.; Villela, B.S.; Rouillard, C.; Buteau, J. The orphan nuclear receptor Nor1/Nr4a3 is a negative regulator of β-ce ll mass. J. Biol. Chem. 2019, 294, 4889–4897. [Google Scholar] [CrossRef]
- Close, A.F.; Dadheech, N.; Lemieux, H.; Wang, Q.; Buteau, J. Disruption of Beta-Cell Mitochondrial Networks by the Orphan Nuclear R eceptor Nor1/Nr4a3. Cells 2020, 9, 168. [Google Scholar] [CrossRef]
- Paez, H.G.; Ferrandi, P.J.; Pitzer, C.R.; Mohamed, J.S.; Alway, S.E. Loss of NOR1 represses muscle metabolism through mTORC1-mediated signa ling and mitochondrial gene expression in C2C12 myotubes. FASEB J. 2023, 37, e23050. [Google Scholar] [CrossRef] [PubMed]
- DeYoung, R.A.; Baker, J.C.; Cado, D.; Winoto, A. The orphan steroid receptor Nur77 family member Nor1 is essential for early mouse embryogenesis. J. Biol. Chem. 2003, 278, 47104–47109. [Google Scholar] [CrossRef]
- Ponnio, T.; Burton, Q.; Pereira, F.A.; Wu, D.K.; Conneely, O.M. The nuclear receptor Nor1 is essential for proliferation of the semici rcular canals of the mouse inner ear. Mol. Cell. Biol. 2002, 22, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.; Ganguli, N.; Sen Sharma, S.; Majumdar, S.S. Sertoli cell specific decline in NOR1 leads to germ cell apoptosis and reduced fertility. J. Cell. Biochem. 2018, 119, 6514–6526. [Google Scholar] [CrossRef]
- Sheng, Z.; Zhang, R.; Ji, Z.; Liu, Z.; Zhou, Y. Identification of mitophagy-related key genes and their correlation wi th immune cell infiltration in acute myocardial infarction via bioinfo rmatics analysis. Front. Cardiovasc. Med. 2025, 11, 1501608. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Wu, Y.; Zhao, D.; Zhang, H.; Lin, J.; Wang, Y. Six mitophagy-related hub genes as peripheral blood biomarkers of Alzh eimer’s disease and their immune cell infiltration correlation. Front. Neurosci. 2023, 17, 1125281. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Peng, F.; Weng, B.; Tang, X.; Chen, Y.; Yang, A.; Chen, B.; Ran, M. miR-222 Suppresses Immature Porcine Sertoli Cell Growth by Targeting t he GRB10 Gene Through Inactivating the PI3K/AKT Signaling Pathway. Front. Genet. 2020, 11, 581593. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wu, H.; He, D.; Hu, X.; Li, Y. Downregulation of BCL11A by siRNA induces apoptosis in B lymphoma cell lines. Biomed. Rep. 2013, 1, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Russell, L.D.; Chiarini-Garcia, H.; Korsmeyer, S.J.; Knudson, C.M. Bax-dependent spermatogonia apoptosis is required for testicular devel opment and spermatogenesis. Biol. Reprod. 2002, 66, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Steger, K.; Wilhelm, J.; Konrad, L.; Stalf, T.; Greb, R.; Diemer, T.; Kliesch, S.; Bergmann, M.; Weidner, W. Both protamine-1 to protamine-2 mRNA ratio and Bcl2 mRNA content in te sticular spermatids and ejaculated spermatozoa discriminate between fe rtile and infertile men. Hum. Reprod. 2008, 23, 11–16. [Google Scholar] [CrossRef]
- Hollville, E.; Carroll, R.G.; Cullen, S.P.; Martin, S.J. Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol. Cell 2014, 55, 451–466. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Pan, C.; Li, D.; Han, X. Microcystin-leucine-arginine causes blood-testis barrier disruption an d degradation of occludin mediated by matrix metalloproteinase-8. Cell. Mol. Life Sci. 2018, 75, 1117–1132. [Google Scholar] [CrossRef]
- Lv, D.; Ji, Y.; Zhang, Q.; Shi, Z.; Chen, T.; Zhang, C.; Wang, X.; Ren, T.; Gao, Z.; Zhong, C. Mailuoshutong pill for varicocele-associated male infertility-Phytoche mical characterisation and multitarget mechanism. Front. Pharmacol. 2022, 13, 961011. [Google Scholar] [CrossRef]
- Nakamura, B.N.; Lawson, G.; Chan, J.Y.; Banuelos, J.; Cortés, M.M.; Hoang, Y.D.; Ortiz, L.; Rau, B.A.; Luderer, U. Knockout of the transcription factor NRF2 disrupts spermatogenesis in an age-dependent manner. Free Radic. Biol. Med. 2010, 49, 1368–1379. [Google Scholar] [CrossRef]
- Kyrgiafini, M.A.; Sarafidou, T.; Mamuris, Z. The Role of Long Noncoding RNAs on Male Infertility: A Systematic Review and In Silico Analysis. Biology 2022, 11, 1510. [Google Scholar] [CrossRef]
- Che, T.F.; Lin, C.W.; Wu, Y.Y.; Chen, Y.J.; Han, C.L.; Chang, Y.L.; Wu, C.T.; Hsiao, T.H.; Hong, T.M.; Yang, P.C. Mitochondrial translocation of EGFR regulates mitochondria dynamics an d promotes metastasis in NSCLC. Oncotarget 2015, 6, 37349–37366. [Google Scholar] [CrossRef]
- Schneider, M.R.; Gratao, A.A.; Dahlhoff, M.; Boersma, A.; Angelis, M.; Hoang-Vu, C.; Wolf, E.; Klonisch, T. EGFR ligands exert diverging effects on male reproductive organs. Exp. Mol. Pathol. 2010, 88, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.W.; Kwan, R.W.; Mak, P.H.; Mak, K.K.; Sham, M.H.; Chan, S.Y. Overexpression of epidermal growth factor induced hypospermatogenesis in transgenic mice. J. Biol. Chem. 2000, 275, 18297–18301. [Google Scholar] [CrossRef]
- Murphy, M.P.; Hartley, R.C. Mitochondria as a therapeutic target for common pathologies. Nat. Rev. Drug Discov. 2018, 17, 865–886. [Google Scholar] [CrossRef]
- O’Connor, T.; Borsig, L.; Heikenwalder, M. CCL2-CCR2 Signaling in Disease Pathogenesis. Endocr. Metab. Immune Disord. Drug Targets 2015, 15, 105–118. [Google Scholar] [CrossRef]
- Luciano-Mateo, F.; Cabré, N.; Fernández-Arroyo, S.; Baiges-Gayà, G.; Hernández-Aguilera, A.; Rodríguez-Tomàs, E.; Joven, J. Chemokine c–c motif ligand 2 overexpression drives tissue-specific met abolic responses in the liver and muscle of mice. Sci. Rep. 2020, 10, 11954. [Google Scholar] [CrossRef]
- Bassiouni, W.; Valencia, R.; Mahmud, Z.; Seubert, J.M.; Schulz, R. Matrix metalloproteinase-2 proteolyzes mitofusin-2 and impairs mitocho ndrial function during myocardial ischemia-reperfusion injury. Basic Res. Cardiol. 2023, 118, 29. [Google Scholar] [CrossRef]
- Longin, J.; Le Magueresse-Battistoni, B. Evidence that MMP-2 and TIMP-2 are at play in the FSH-induced changes in Sertoli cells. Mol. Cell. Endocrinol. 2002, 189, 25–35. [Google Scholar] [CrossRef]
- Hou, X.; Li, Z.; Higashi, Y.; Delafontaine, P.; Sukhanov, S. Insulin-Like Growth Factor I Prevents Cellular Aging via Activation of Mitophagy. J. Aging Res. 2020, 2020, 4939310. [Google Scholar] [CrossRef]
- Riis, S.; Murray, J.B.; O’Connor, R. IGF-1 Signalling Regulates Mitochondria Dynamics and Turnover through a Conserved GSK-3β-Nrf2-BNIP3 Pathway. Cells 2020, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Yakar, S.; Liu, J.L.; Fernandez, A.M.; Wu, Y.; Schally, A.V.; Frystyk, J.; Chernausek, S.D.; Mejia, W.; Le Roith, D. Liver-specific igf-1 gene deletion leads to muscle insulin insensitivi ty. Diabetes 2001, 50, 1110–1118. [Google Scholar] [CrossRef]
- Giallongo, C.; Dulcamare, I.; Tibullo, D.; Del Fabro, V.; Vicario, N.; Parrinello, N.; Romano, A.; Scandura, G.; Lazzarino, G.; Conticello, C.; et al. CXCL12/CXCR4 axis supports mitochondrial trafficking in tumor myeloma microenvironment. Oncogenesis 2022, 11, 6. [Google Scholar] [CrossRef]
- Hu, E.; Mueller, E.; Oliviero, S.; Papaioannou, V.E.; Johnson, R.; Spiegelman, B.M. Targeted disruption of the c-fos gene demonstrates c-fos-dependent and -independent pathways for gene expression stimulated by growth factor s or oncogenes. EMBO J. 1994, 13, 3094–3103. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, X.; Niu, C.; Yang, Y.; Huang, Z.; Xie, J. Fibroblast growth factor 7 protects osteoblasts against oxidative dama ge through targeting mitochondria. FASEB J. 2024, 38, e23524. [Google Scholar] [CrossRef]
- Neill, T.; Torres, A.; Buraschi, S.; Owens, R.T.; Hoek, J.B.; Baffa, R.; Iozzo, R.V. Decorin induces mitophagy in breast carcinoma cells via peroxisome pro liferator-activated receptor γ coactivator-1α (PGC-1α) and mitostatin. J. Biol. Chem. 2014, 289, 4952–4968. [Google Scholar] [CrossRef] [PubMed]
- Wynne, M.E.; Ogunbona, O.; Lane, A.R.; Gokhale, A.; Zlatic, S.A.; Xu, C.; Wen, Z.; Duong, D.M.; Rayaprolu, S.; Ivanova, A.; et al. APOE expression and secretion are modulated by mitochondrial dysfuncti on. eLife 2023, 12, e85779. [Google Scholar] [CrossRef]
- Saitou, M.; Furuse, M.; Sasaki, H.; Schulzke, J.D.; Fromm, M.; Takano, H.; Noda, T.; Tsukita, S. Complex phenotype of mice lacking occludin, a component of tight junct ion strands. Mol. Biol. Cell 2000, 11, 4131–4142. [Google Scholar] [CrossRef]
- Beeman, N.E.; Baumgartner, H.K.; Webb, P.G.; Schaack, J.B.; Neville, M.C. Disruption of occludin function in polarized epithelial cells activate s the extrinsic pathway of apoptosis leading to cell extrusion without loss of transepithelial resistance. BMC Cell Biol. 2009, 10, 85. [Google Scholar] [CrossRef] [PubMed]
- Pathak, D.; Berthet, A.; Bendor, J.T.; Yu, K.; Sellnow, R.C.; Orr, A.L.; Nguyen, M.K.; Edwards, R.H.; Manfredsson, F.P.; Nakamura, K. Loss of alpha-Synuclein Does Not Affect Mitochondrial Bioenergetics in Rodent Neurons. eNeuro 2017, 4, ENEURO.0216-16.2017. [Google Scholar] [CrossRef]
- Choubey, V.; Safiulina, D.; Vaarmann, A.; Cagalinec, M.; Wareski, P.; Kuum, M.; Zharkovsky, A.; Kaasik, A. Mutant A53T alpha-synuclein induces neuronal death by increasing mitoc hondrial autophagy. J. Biol. Chem. 2011, 286, 10814–10824. [Google Scholar] [CrossRef]
- Pradhan, B.S.; Majumdar, S.S. An Efficient Method for Generation of Transgenic Rats Avoiding Embryo Manipulation. Mol. Ther. Nucleic Acids 2016, 5, e293. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, S.; Shetty, J.; Hou, L.; Delcher, A.; Zhu, B.; Osoegawa, K.; Jong, P.; Nierman, W.C.; Strausberg, R.L.; Fraser, C.M. Human, mouse, and rat genome large-scale rearrangements: Stability ver sus speciation. Genome Res. 2004, 14, 1851–1860. [Google Scholar] [CrossRef]
- Ahmed, E.A.; de Rooij, D.G. Staging of mouse seminiferous tubule cross-sections. Methods Mol. Biol. 2009, 558, 263–277. [Google Scholar]
- Clouthier, D.E.; Avarbock, M.R.; Maika, S.D.; Hammer, R.E.; Brinster, R.L. Rat spermatogenesis in mouse testis. Nature 1996, 381, 418–421. [Google Scholar] [CrossRef]
- El-Domyati, M.; Al-Din, A.; Barakat, M.; El-Fakahany, H.; Xu, J.; Sakkas, D. Deoxyribonucleic acid repair and apoptosis in testicular germ cells of aging fertile men: The role of the poly(adenosine diphosphate-ribosyl)ation pathway. Fertil. Steril. 2009, 91, 2221–2229. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.K.; Wayne, C.M.; Meistrich, M.L.; Wilkinson, M.F. Pem homeobox gene promoter sequences that direct transcription in a Se rtoli cell-specific, stage-specific, and androgen-dependent manner in the testis in vivo. Mol. Endocrinol. 2003, 17, 223–233. [Google Scholar] [CrossRef][Green Version]
- Münsterberg, A.; Lovell-Badge, R. Expression of the mouse anti-müllerian hormone gene suggests a role in both male and female sexual differentiation. Development 1991, 113, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Giuili, G.; Shen, W.H.; Ingraham, H.A. The nuclear receptor SF-1 mediates sexually dimorphic expression of Mu llerian Inhibiting Substance, in vivo. Development 1997, 124, 1799–1807. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and m icroarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology informatio n. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecula r interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Pradhan, B.S.; Bhattacharya, I.; Sarkar, R.; Majumdar, S.S. Downregulation of Sostdc1 in Testicular Sertoli Cells is Prerequisite for Onset of Robust Spermatogenesis at Puberty. Sci. Rep. 2019, 9, 11458. [Google Scholar] [CrossRef]
- Pradhan, B.S.; Bhattacharya, I.; Sarkar, R.; Majumdar, S.S. Pubertal down-regulation of Tetraspanin 8 in testicular Sertoli cells is crucial for male fertility. Mol. Hum. Reprod. 2020, 26, 760–772. [Google Scholar] [CrossRef]
- Basu, S.; Arya, S.P.; Usmani, A.; Pradhan, B.S.; Sarkar, R.K.; Ganguli, N.; Shukla, M.; Mandal, K.; Singh, S.; Sarda, K.; et al. Defective Wnt3 expression by testicular Sertoli cells compromise male fertility. Cell Tissue Res. 2018, 371, 351–363. [Google Scholar] [CrossRef]
- Mandal, K.; Sarkar, R.K.; Sen Sharma, S.; Jain, A.; Majumdar, S.S. Sertoli cell specific knockdown of RAR-related orphan receptor (ROR) a lpha at puberty reduces sperm count in rats. Gene 2018, 641, 18–24. [Google Scholar] [CrossRef]
- Sarkar, H.; Batta, S.R.; Wadhwa, N.; Majumdar, S.S.; Pradhan, B.S. Generation of a Transgenic Mouse Model for Investigating Mitochondria in Sperm. Cells 2025, 14, 296. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pradhan, B.S.; Das, D.; Sarkar, H.; Bhattacharya, I.; Wadhwa, N.; Majumdar, S.S. Nor1 and Mitophagy: An Insight into Sertoli Cell Function Regulating Spermatogenesis Using a Transgenic Rat Model. Int. J. Mol. Sci. 2025, 26, 9209. https://doi.org/10.3390/ijms26189209
Pradhan BS, Das D, Sarkar H, Bhattacharya I, Wadhwa N, Majumdar SS. Nor1 and Mitophagy: An Insight into Sertoli Cell Function Regulating Spermatogenesis Using a Transgenic Rat Model. International Journal of Molecular Sciences. 2025; 26(18):9209. https://doi.org/10.3390/ijms26189209
Chicago/Turabian StylePradhan, Bhola Shankar, Deepyaman Das, Hironmoy Sarkar, Indrashis Bhattacharya, Neerja Wadhwa, and Subeer S. Majumdar. 2025. "Nor1 and Mitophagy: An Insight into Sertoli Cell Function Regulating Spermatogenesis Using a Transgenic Rat Model" International Journal of Molecular Sciences 26, no. 18: 9209. https://doi.org/10.3390/ijms26189209
APA StylePradhan, B. S., Das, D., Sarkar, H., Bhattacharya, I., Wadhwa, N., & Majumdar, S. S. (2025). Nor1 and Mitophagy: An Insight into Sertoli Cell Function Regulating Spermatogenesis Using a Transgenic Rat Model. International Journal of Molecular Sciences, 26(18), 9209. https://doi.org/10.3390/ijms26189209





