Unraveling the Roles of Epigenetic Regulators During the Embryonic Development of Rhipicephalus microplus
Abstract
1. Introduction
2. Results
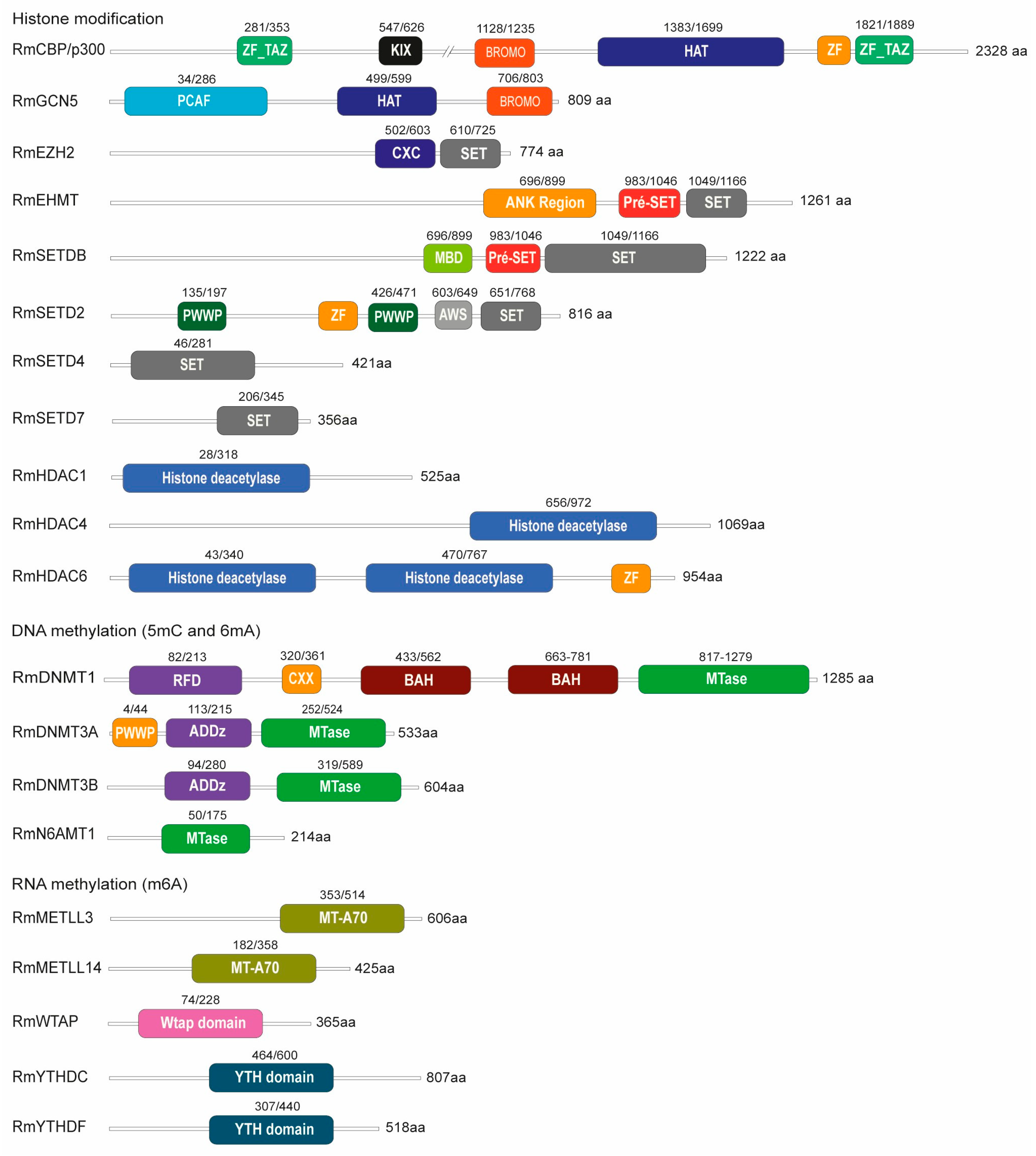
2.1. The Epigenetic Machinery Is Encoded Within the R. microplus Genome
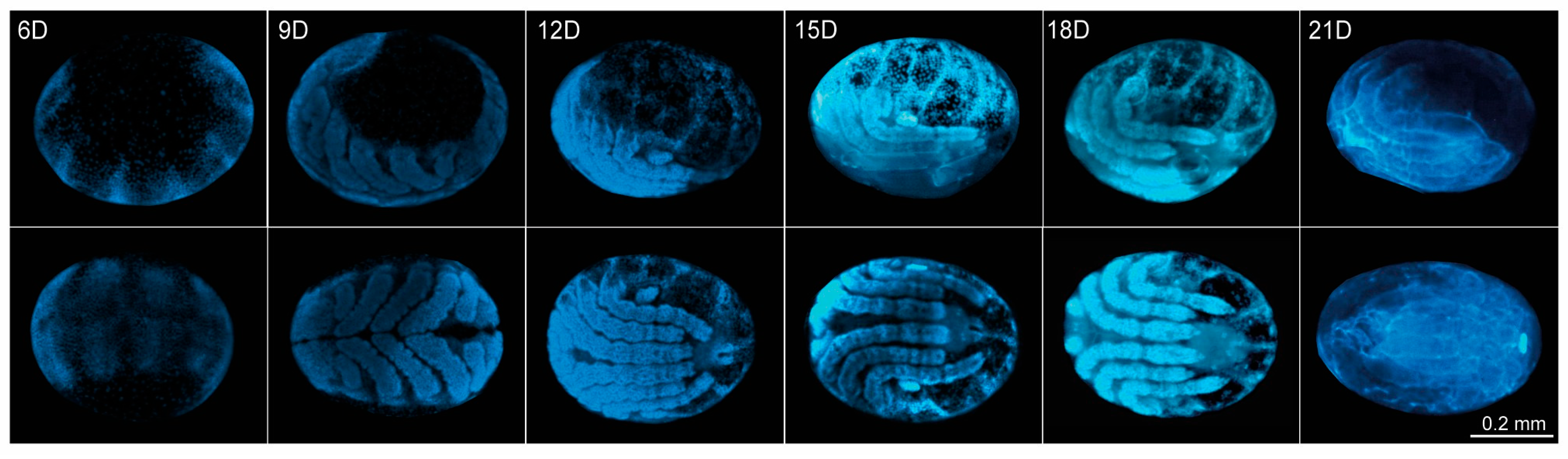
2.2. Embryonic Development of R. microplus
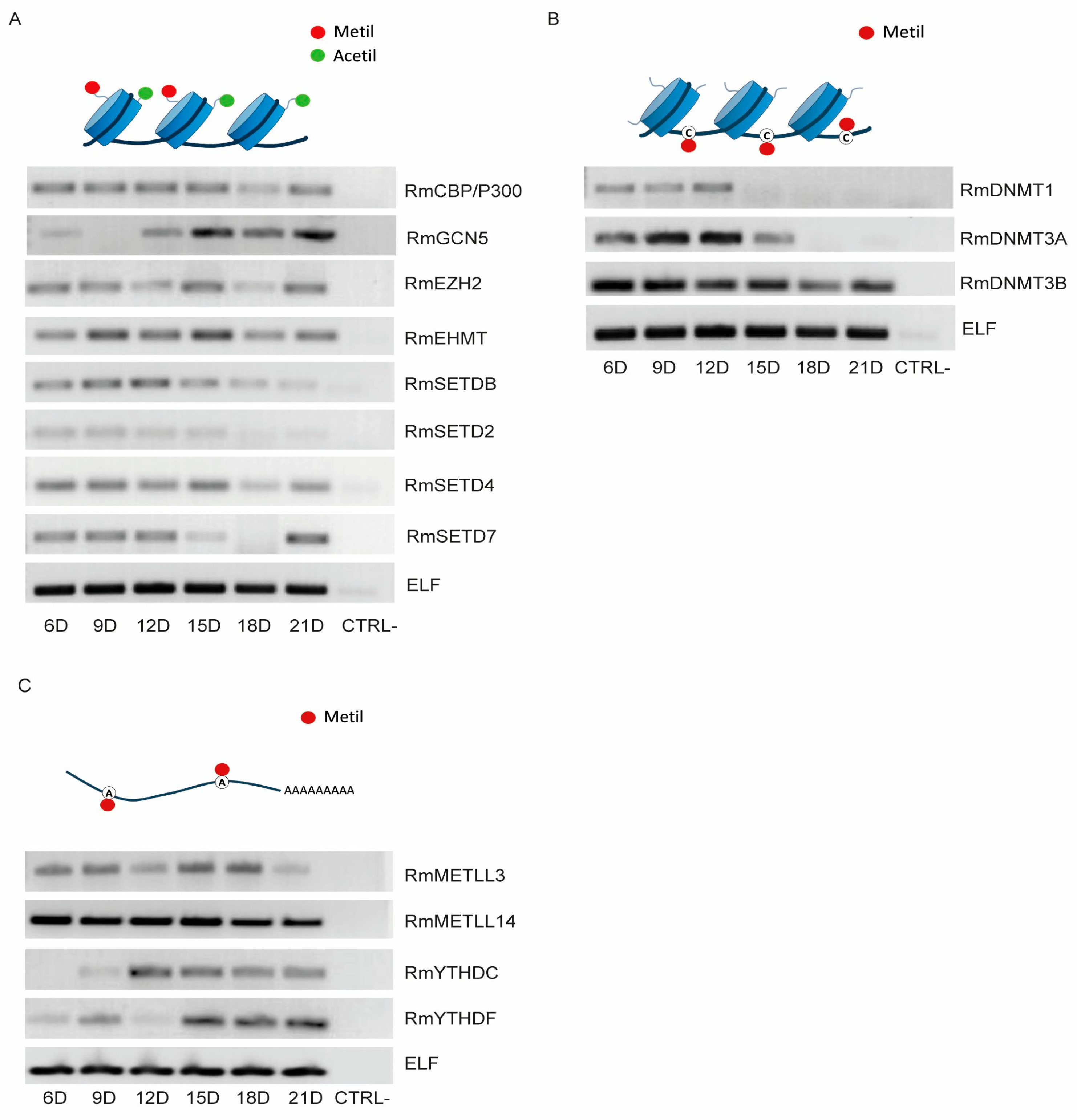
2.3. Transcriptional Profile of Epigenetic Proteins Throughout R. microplus Embryonic Development
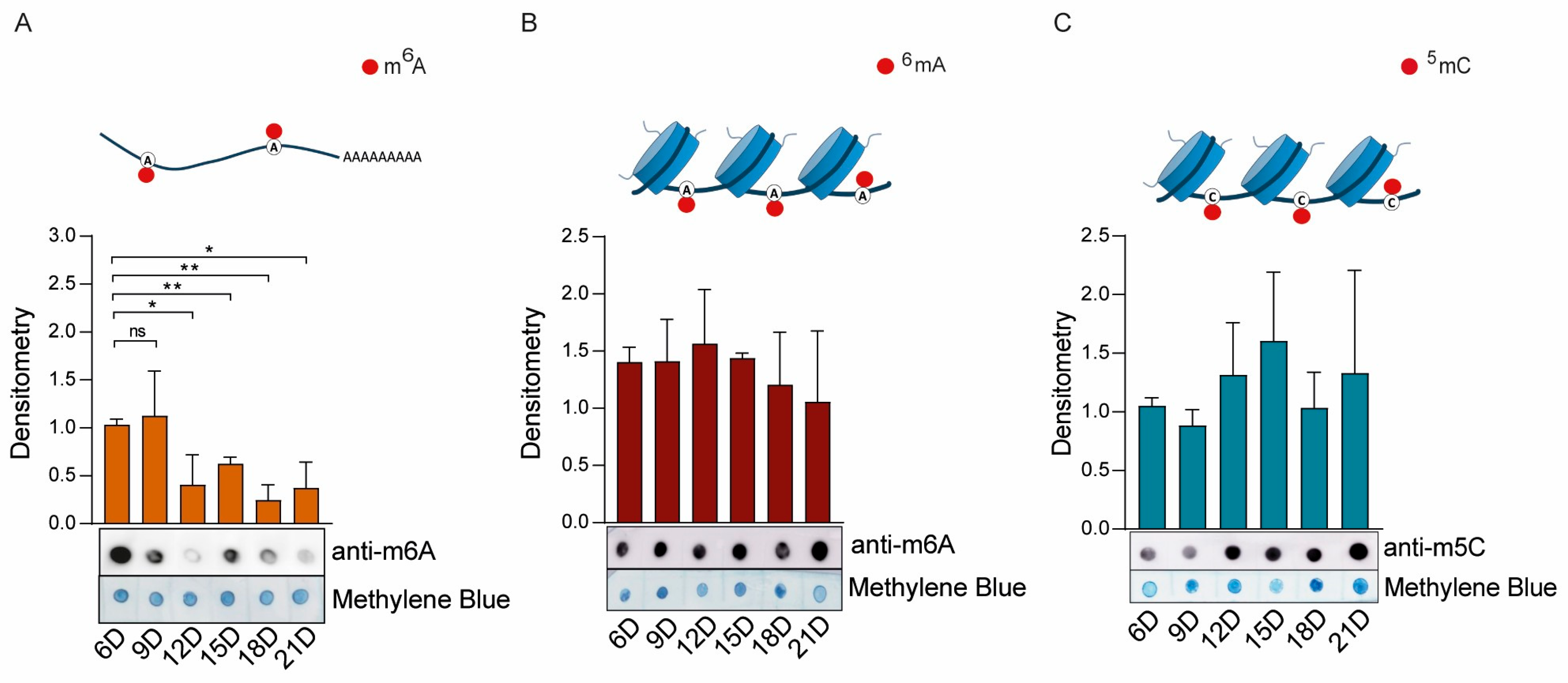
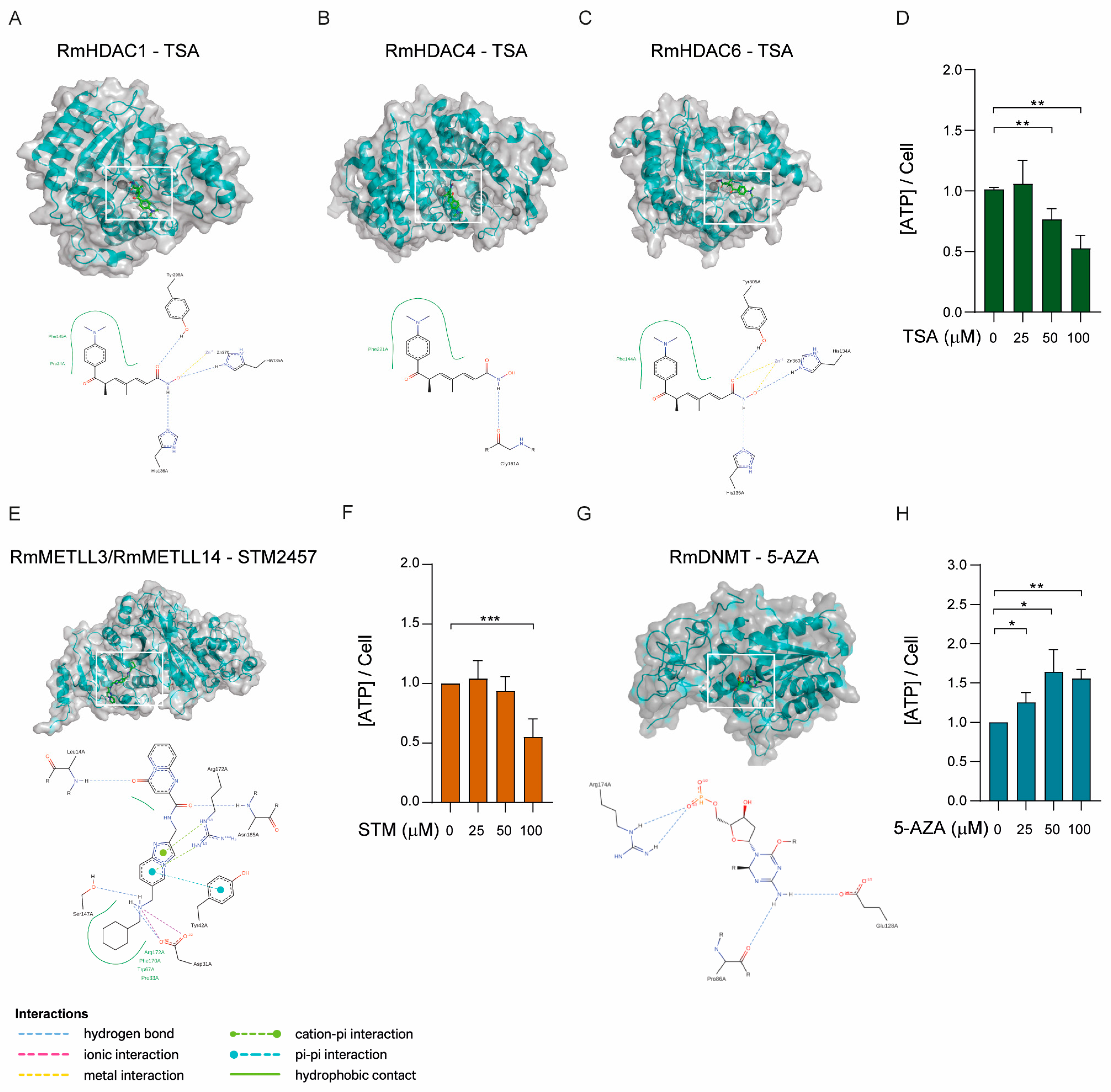
2.4. Activity of the Epigenetic Enzymes Throughout R. microplus Embryonic Development
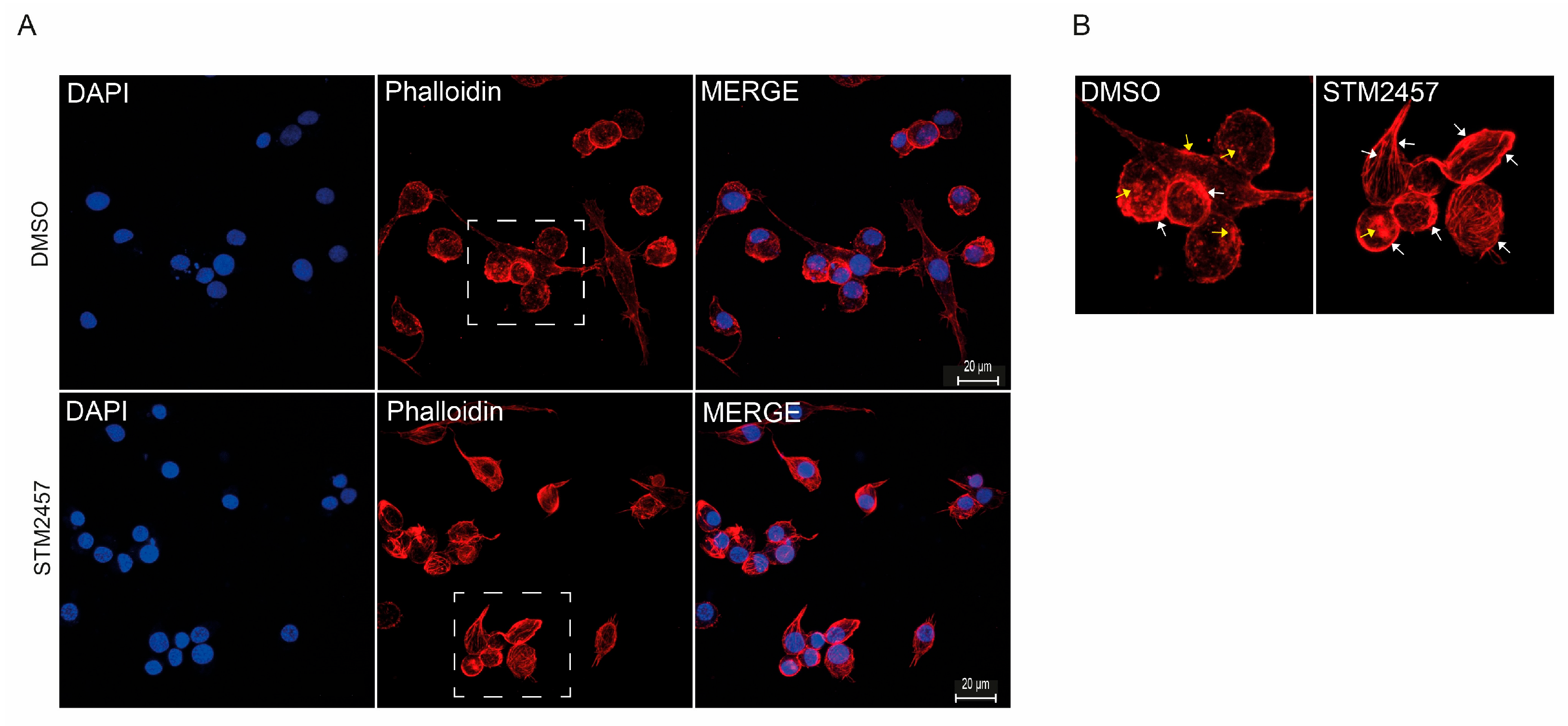
2.5. Biological Effects of Epigenetic Inhibitors on BME Cells
2.6. m5C Methylation of the Nuclear Genome and Electron Transport Chain Function in BME Cells
3. Discussion
4. Materials and Methods
4.1. Tick Maintenance
4.2. Genome-Wide Identification of Epigenetic Enzymes in R. microplus
4.3. Phylogenetic Analysis
4.4. Molecular Docking
4.5. Nucleic Acid Isolation and RT-PCR
4.6. Dot Blot
4.7. Western Blot
4.8. Statistical Analysis
4.9. Cell Culture and Cell Viability Assay
4.10. Fluorescence Microscopy
4.11. Respirometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jongejan, F.; Uilenberg, G. The global importance of ticks. Parasitology 2004, 129, S3–S14. [Google Scholar] [CrossRef]
- Martins, R.; Ruiz, N.; Fonseca, R.N.D.; Vaz Junior, I.D.S.; Logullo, C. The dynamics of energy metabolism in the tick embryo. Rev. Bras. Parasitol. Vet. 2018, 27, 259–266. [Google Scholar] [CrossRef]
- Jonsson, N.N. The productivity effects of cattle tick (Boophilus microplus) infestation on cattle, with particular reference to Bos indicus cattle and their crosses. Vet. Parasitol. 2006, 137, 1–10. [Google Scholar] [CrossRef]
- Seixas, A.; Oliveira, P.; Termignoni, C.; Logullo, C.; Masuda, A.; da Silva Vaz, I. Rhipicephalus (Boophilus) microplus embryo proteins as target for tick vaccine. Vet. Immunol. Immunopathol. 2012, 148, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Bishop, L.J.; Stutzer, C.; Maritz-Olivier, C. More than Three Decades of Bm86: What We Know and Where to Go. Pathogens 2023, 12, 1071. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, J.; Almazán, C.; Canales, M.; de la Lastra, J.M.P.; Kocan, K.M.; Willadsen, P. A ten-year review of commercial vaccine performance for control of tick infestations on cattle. Anim. Health Res. Rev. 2007, 8, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Suraweera, A.; O’bYrne, K.J.; Richard, D.J. Epigenetic drugs in cancer therapy. Cancer Metast. Rev. 2025, 44, 37. [Google Scholar] [CrossRef]
- Watzlowik, M.T.; Silberhorn, E.; Das, S.; Singhal, R.; Venugopal, K.; Holzinger, S.; Stokes, B.; Schadt, E.; Sollelis, L.; Bonnell, V.A.; et al. Plasmodium blood stage development requires the chromatin remodeller Snf2L. Nature 2025, 639, 1069–1075. [Google Scholar] [CrossRef]
- Hossain, M.U.; Ferdous, N.; Reza, M.N.; Ahammad, I.; Tiernan, Z.; Wang, Y.; O’hAnlon, F.; Wu, Z.; Sarker, S.; Mohiuddin, A.K.M.; et al. Pathogen-driven gene expression patterns lead to a novel approach to the identification of common therapeutic targets. Sci. Rep. 2022, 12, 21070. [Google Scholar] [CrossRef]
- Walters, H.A.; Temesvari, L.A. Target acquired: Transcriptional regulators as drug targets for protozoan parasites. Int. J. Parasitol. 2021, 51, 599–611. [Google Scholar] [CrossRef]
- Tian, Z.; Wang, K.; Guo, S.; Wang, X.-P. The PBAP chromatin remodeling complex mediates summer diapause via H3K4me3-driven juvenile hormone regulation in Colaphellus bowringi. Proc. Natl. Acad. Sci. USA 2025, 122, e2422328122. [Google Scholar] [CrossRef]
- Yuichiro, S.; Stephanie, A.; Brian, T.; Paula, G. Molecular mechanisms underlying the evolution of a color polyphenism by genetic accommodation in the tobacco hornworm, Manduca sexta. Proc. Natl. Acad. Sci. USA 2025, 122, e2425004122. [Google Scholar] [CrossRef]
- Alvarado, S.; Rajakumar, R.; Abouheif, E.; Szyf, M. Epigenetic variation in the Egfr gene generates quantitative variation in a complex trait in ants. Nat. Commun. 2015, 6, 6513. [Google Scholar] [CrossRef]
- Ayushi, G.; Suresh, N. Epigenetic processes in insect adaptation to environmental stress. Curr. Opin. Insect Sci. 2025, 67, 101294. [Google Scholar] [CrossRef]
- Piunti, A.; Shilatifard, A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cell Biol. 2021, 22, 326–345, Erratum in Nat. Rev. Mol. Cell Biol. 2022, 23, 444. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Xie, H. Genome-wide analysis of bivalent histone modifications during Drosophila embryogenesis. Genesis 2022, 60, e23502. [Google Scholar] [CrossRef]
- Atinbayeva, N.; Valent, I.; Zenk, F.; Loeser, E.; Rauer, M.; Herur, S.; Quarato, P.; Pyrowolakis, G.; Gomez-Auli, A.; Mittler, G.; et al. Inheritance of H3K9 methylation regulates genome architecture in Drosophila early embryos. EMBO J. 2024, 43, 2685–2714. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Wang, S. Epigenetic Regulation in Insect-Microbe Interactions. Annu. Rev. Entomol. 2025, 28, 31. [Google Scholar] [CrossRef]
- Amarante, A.D.; da Silva, I.C.; Carneiro, V.C.; Vicentino, A.R.; Pinto, M.D.; Higa, L.M.; Moharana, K.C.; Talyuli, O.A.; Venancio, T.M.; de Oliveira, P.L.; et al. Zika virus infection drives epigenetic modulation of immunity by the histone acetyltransferase CBP of Aedes aegypti. PLoS Negl. Trop. Dis. 2022, 16, e0010559. [Google Scholar] [CrossRef] [PubMed]
- Gomes, F.M.; Tyner, M.D.W.; Barletta, A.B.F.; Saha, B.; Yenkoidiok-Douti, L. Double peroxidase and histone acetyltransferase AgTip60 maintain innate immune memory in primed mosquitoes. Proc. Natl. Acad. Sci. USA 2021, 118, e2114242118. [Google Scholar] [CrossRef]
- Raddatz, G.; Guzzardo, P.M.; Olova, N.; Fantappié, M.R.; Rampp, M.; Schaefer, M.; Reik, W.; Hannon, G.J.; Lyko, F. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc. Natl. Acad. Sci. USA 2013, 110, 8627–8631. [Google Scholar] [CrossRef]
- Kotsarenko, K.; Vechtova, P.; Hammerova, Z.; Langova, N.; Malinovska, L.; Wimmerova, M.; Sterba, J.; Grubhoffer, L. Newly identified DNA methyltransferases of Ixodes ricinus ticks. Ticks Tick Borne Dis. 2019, 11, 101348. [Google Scholar] [CrossRef]
- Pozo, M.I.; Hunt, B.J.; Van Kemenade, G.; Guerra-Sanz, J.M.; Wäckers, F.; Mallon, E.B.; Jacquemyn, H. The effect of DNA methylation on bumblebee colony development. BMC Genom. 2021, 22, 73. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.C.; Dohmen, E.; George, S.; Sillam-Dussès, D.; Séité, S.; Vasseur-Cognet, M. Complex regulatory role of DNA methylation in caste- and age-specific expression of a termite. Open Biol. 2022, 12, 220047. [Google Scholar] [CrossRef] [PubMed]
- Duncan, E.J.; Cunningham, C.B.; Dearden, P.K. Phenotypic Plasticity: What Has DNA Methylation Got to Do with It? Insects 2022, 13, 110. [Google Scholar] [CrossRef] [PubMed]
- Seal, R.L.; Denny, P.; Bruford, E.A.; Gribkova, A.K.; Landsman, D.; Marzluff, W.F.; McAndrews, M.; Panchenko, A.R.; Shaytan, A.K.; Talbert, P.B. A standardized nomenclature for mammalian histone genes. Epigenet. Chromatin 2022, 15, 34. [Google Scholar] [CrossRef]
- Dai, Z.; Ramesh, V.; Locasale, J.W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 2020, 21, 737–753, Erratum in Nat. Rev. Genet. 2020, 21, 782. [Google Scholar] [CrossRef]
- Mannervik, M. Beyond histones: The elusive substrates of chromatin regulators. Genes Dev. 2024, 38, 357–359. [Google Scholar] [CrossRef]
- Wojciechowski, M.; Lowe, R.; Maleszka, J.; Conn, D.; Maleszka, R.; Hurd, P.J. Phenotypically distinct female castes in honey bees are defined by alternative chromatin states during larval development. Genome Res. 2018, 28, 1532–1542. [Google Scholar] [CrossRef]
- Choppin, M.; Feldmeyer, B.; Foitzik, S. Histone acetylation regulates the expression of genes involved in worker reproduction in the ant Temnothorax rugatulus. BMC Genom. 2021, 22, 871. [Google Scholar] [CrossRef]
- Yuan, D.; Zhang, X.; Yang, Y.; Wei, L.; Li, H.; Zhao, T.; Guo, M.; Li, Z.; Huang, Z.; Wang, M.; et al. Schlank orchestrates insect developmental transition by switching H3K27 acetylation to trimethylation in the prothoracic gland. Proc. Natl. Acad. Sci. USA 2024, 121, e2401861121. [Google Scholar] [CrossRef] [PubMed]
- Glastad, K.M.; Hunt, B.G.; Goodisman, M.A.D. Epigenetics in insects: Genome regulation and the generation of phenotypic diversity. Annu. Rev. Entomol. 2019, 64, 185–203. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Chen, Y.; Liu, Y.; Zhuang, L.; Chen, W.; Zeng, B.; Liao, X.; Guo, G.; Wang, Y.; Wang, X. m6A methylation promotes white-to-beige fat transition by facilitating Hif1a translation. EMBO Rep. 2021, 22, e52348. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Roignant, J.-Y.; Soller, M. m6A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017, 33, 380–390. [Google Scholar] [CrossRef]
- Wu, R.; Jiang, D.; Wang, Y.; Wang, X. N6-Methyladenosine (m6A) Methylation in mRNA with A Dynamic and Reversible Epigenetic Modification. Mol. Biotechnol. 2016, 58, 450–459. [Google Scholar] [CrossRef]
- Gui, Y.; Yuan, S. Epigenetic regulations in mammalian spermatogenesis: RNA-m6A modification and beyond. Cell. Mol. Life Sci. 2021, 78, 4893–4905. [Google Scholar] [CrossRef]
- Xiang, Y.; Laurent, B.; Hsu, C.-H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m6 A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576, Erratum in Nature, 2017, 552, 430. [Google Scholar] [CrossRef]
- Zhong, X.; Yu, J.; Frazier, K.; Weng, X.; Li, Y.; Cham, C.M.; Dolan, K.; Zhu, X.; Hubert, N.; Tao, Y.; et al. Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m6A mRNA Methylation. Cell Rep. 2018, 25, 1816–1828. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Flavell, R.A.; Li, H.B. RNA m6A modification and its function in diseases. Front. Med. 2018, 12, 481–489. [Google Scholar] [CrossRef]
- Tang, C.; Hu, W. Epigenetic modifications during embryonic development: Gene reprogramming and regulatory networks. J. Reprod. Immunol. 2024, 165, 104311. [Google Scholar] [CrossRef] [PubMed]
- Guynes, K.; Sarre, L.A.; Carrillo-Baltodano, A.M.; Davies, B.E.; Xu, L.; Liang, Y.; Martín-Zamora, F.M.; Hurd, P.J.; de Mendoza, A.; Martín-Durán, J.M. Annelid methylomes reveal ancestral developmental and aging-associated epigenetic erosion across Bilateria. Genome Biol. 2024, 25, 204. [Google Scholar] [CrossRef]
- Li, B.; Hu, P.; Huang, Z.H.; Yang, J.Y.; Wang, J.; Xie, X.Z.; Wang, Y.H.; Li, C.C.; Xu, J.P. RNA methyltransferase BmMettl3 and BmMettl14 in silkworm (Bombyx mori) and the regulation of silkworm embryonic development. Arch. Insect Biochem. Physiol. 2023, 113, e22005. [Google Scholar] [CrossRef]
- Drewell, R.A.; Bush, E.C.; Remnant, E.J.; Wong, G.T.; Beeler, S.M.; Stringham, J.L.; Lim, J.; Oldroyd, B.P. The dynamic DNA methylation cycle from egg to sperm in the honey bee Apis mellifera. Development 2014, 141, 2702–2711. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Alberdi, P.; Ayllón, N.; Valdés, J.J.; Pierce, R.; Villar, M.; de la Fuente, J. Anaplasma phagocytophilum increases the levels of histone modifying enzymes to inhibit cell apoptosis and facilitate pathogen infection in the tick vector Ixodes scapularis. Epigenetics 2016, 11, 303–319. [Google Scholar] [CrossRef]
- De, S.; Kitsou, C.; Sonenshine, D.E.; Pedra, J.H.F.; Fikrig, E.; Kassis, J.A.; Pal, U. Epigenetic Regulation of Tick Biology and Vectorial Capacity. Trends Genet. 2021, 37, 8–11. [Google Scholar] [CrossRef]
- Parizi, L.F.; Pohl, P.C.; Masuda, A.; Vaz Ida, S., Jr. New approaches toward anti-Rhipicephalus (Boophilus) microplus tick vaccine. Rev. Bras. Parasitol. Vet. 2009, 18, 1–7. [Google Scholar] [CrossRef]
- Palli, S.R. Epigenetic regulation of post-embryonic development. Curr. Opin. Insect Sci. 2021, 43, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Christman, J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 2002, 21, 5483–5495. [Google Scholar] [CrossRef] [PubMed]
- Pasini, D.; Malatesta, M.; Jung, H.R.; Walfridsson, J.; Willer, A.; Olsson, L.; Skotte, J.; Wutz, A.; Porse, B.; Jensen, O.N.; et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010, 38, 4958–4969. [Google Scholar] [CrossRef] [PubMed]
- Tie, F.; Banerjee, R.; Conrad, P.A.; Scacheri, P.C.; Harte, P.J. Histone Demethylase UTX and Chromatin Remodeler BRM Bind Directly to CBP and Modulate Acetylation of Histone H3 Lysine 27. Mol. Cell Biol. 2012, 32, 2323–2334. [Google Scholar] [CrossRef] [PubMed]
- Tie, F.; Banerjee, R.; Stratton, C.A.; Prasad-Sinha, J.; Stepanik, V.; Zlobin, A.; Diaz, M.O.; Scacheri, P.C.; Harte, P.J. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009, 136, 3131–3141. [Google Scholar] [CrossRef]
- Albert, J.; Renata, Z.J. DNA Methyltransferases—Role and Function, 2nd ed.; Springer: Cham, Switzerland, 2022; Volume 1389. [Google Scholar]
- Sun, T.; Xu, Y.; Xiang, Y.; Ou, J.; Soderblom, E.J.; Diao, Y. Crosstalk between RNA m6A and DNA methylation regulates transposable element chromatin activation and cell fate in human pluripotent stem cells. Nat. Genet. 2023, 55, 1324–1335. [Google Scholar] [CrossRef]
- Zhao, B.S.; Wang, X.; Beadell, A.V.; Lu, Z.; Shi, H.; Kuuspalu, A.; Ho, R.K.; He, C. M6 A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 2017, 542, 475–478. [Google Scholar] [CrossRef]
- Santos, V.T.; Ribeiro, L.; Fraga, A.; de Barros, C.M.; Campos, E.; Moraes, J.; Fontenele, M.R.; Araujo, H.M.; Feitosa, N.M.; Logullo, C.; et al. The embryogenesis of the Tick Rhipicephalus (Boophilus) microplus: The establishment of a new chelicerate model system. Genesis 2013, 51, 803–818. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef]
- Mogal, A.; Abdulkadir, S.A. Effects of Histone Deacetylase Inhibitor (HDACi); Trichostatin-A (TSA) on the expression of housekeeping genes. Mol. Cell. Probes 2006, 20, 81–86. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033–3042. [Google Scholar] [CrossRef]
- Li, X.Y.; Harrison, M.M.; Villalta, J.E.; Kaplan, T.; Eisen, M.B. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 2014, 3, 3737. [Google Scholar] [CrossRef]
- He, S.; Zhang, G.; Wang, J.; Gao, Y.; Sun, R.; Cao, Z.; Chen, Z.; Zheng, X.; Yuan, J.; Luo, Y.; et al. 6mA-DNA-binding factor Jumu controls maternal-to-zygotic transition upstream of Zelda. Nat. Commun. 2019, 10, 2219. [Google Scholar] [CrossRef]
- Wang, M.; Xiao, Y.; Li, Y.; Wang, X.; Qi, S.; Wang, Y.; Zhao, L.; Wang, K.; Peng, W.; Luo, G.-Z.; et al. RNA m6A Modification Functions in Larval Development and Caste Differentiation in Honeybee (Apis mellifera). Cell Rep. 2021, 34, 108580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Xu, Y.; Wang, X.; Zhu, Y.; Wang, L.; Zhang, W.; Wang, Y.; Gao, Y.; Wu, X.; Cheng, Y.; et al. Dynamic FMR1 granule phase switch instructed by m6A modification contributes to maternal RNA decay. Nat. Commun. 2022, 13, 859. [Google Scholar] [CrossRef] [PubMed]
- Broix, L.; Roy, R.; Shohayeb, B.; Oomoto, I.; Umeshima, H.; Wang, D.O. m6A RNA methylation-mediated control of global APC expression is required for local translation of β-actin and axon development. Cell Rep. 2025, 44, 115727. [Google Scholar] [CrossRef]
- Jiao, Y.; Palli, S.R. N6-adenosine (m6A) mRNA methylation is required for Tribolium castaneum development and reproduction. Insect Biochem. Mol. Biol. 2023, 159, 103985. [Google Scholar] [CrossRef]
- Ramalho-Santos, J.; Varum, S.; Amaral, S.; Mota, P.C.; Sousa, A.P.; Amaral, A. Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 2009, 15, 553–572. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Zhang, C.; Tao, L.; Hao, J.; Tan, K.; Miao, K.; Yu, Y.; Sui, L.; Wu, Z.; Tian, J.; et al. High-resolution profiles of gene expression and DNA methylation highlight mitochondrial modifications during early embryonic development. J. Reprod. Dev. 2017, 63, 247–261. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Rosignoli, S.; Pacelli, M.; Manganiello, F.; Paiardini, A. An outlook on structural biology after AlphaFold: Tools, limits and perspectives. FEBS Open Bio. 2024, 15, 202–222. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Rosignoli, S.; Paiardini, A. DockingPie: A consensus docking plugin for PyMOL. Bioinformatics 2022, 38, 4233–4234. [Google Scholar] [CrossRef]
- Vieira, I.H.P.; Botelho, E.B.; de Souza Gomes, T.J.; Kist, R.; Caceres, R.A.; Zanchi, F.B. Visual dynamics: A WEB application for molecular dynamics simulation using GROMACS. BMC Bioinform. 2023, 24, 107. [Google Scholar] [CrossRef] [PubMed]
- Nijhof, A.M.; Balk, J.A.; Postigo, M.; Jongejan, F. Selection of reference genes for quantitative RT-PCR studies in Rhipicephalus (Boophilus) microplus and Rhipicephalus appendiculatus ticks and determination of the expression profile of Bm86. BMC Mol. Biol. 2009, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Esteves, E.; Lara, F.A.; Lorenzini, D.M.; Costa, G.H.; Fukuzawa, A.H.; Pressinotti, L.N.; Silva, J.R.; Ferro, J.A.; Kurtti, T.J.; Munderloh, U.G.; et al. Cellular and molecular characterization of an embryonic cell line (BME26) from the tick Rhipicephalus (Boophilus) microplus. Insect Biochem. Mol. Biol. 2008, 38, 568–580. [Google Scholar] [CrossRef]
- Zonderland, J.; Wieringa, P.; Moroni, L. A quantitative method to analyse F-actin distribution in cells. MethodsX 2019, 6, 2562–2569, Erratum in MethodsX 2023, 10, 102139. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amarante, A.M.; de Oliveira, D.M.; de Souza, M.P.N.C.; Fonseca-Oliveira, M.; Galina, A.; Rosignoli, S.; Arcanjo, A.F.; Moraes, B.; Paiardini, A.; Rotili, D.; et al. Unraveling the Roles of Epigenetic Regulators During the Embryonic Development of Rhipicephalus microplus. Int. J. Mol. Sci. 2025, 26, 9171. https://doi.org/10.3390/ijms26189171
Amarante AM, de Oliveira DM, de Souza MPNC, Fonseca-Oliveira M, Galina A, Rosignoli S, Arcanjo AF, Moraes B, Paiardini A, Rotili D, et al. Unraveling the Roles of Epigenetic Regulators During the Embryonic Development of Rhipicephalus microplus. International Journal of Molecular Sciences. 2025; 26(18):9171. https://doi.org/10.3390/ijms26189171
Chicago/Turabian StyleAmarante, Anderson Mendonça, Daniel Martins de Oliveira, Marcos Paulo Nicolich Camargo de Souza, Manoel Fonseca-Oliveira, Antonio Galina, Serena Rosignoli, Angélica Fernandes Arcanjo, Bruno Moraes, Alessandro Paiardini, Dante Rotili, and et al. 2025. "Unraveling the Roles of Epigenetic Regulators During the Embryonic Development of Rhipicephalus microplus" International Journal of Molecular Sciences 26, no. 18: 9171. https://doi.org/10.3390/ijms26189171
APA StyleAmarante, A. M., de Oliveira, D. M., de Souza, M. P. N. C., Fonseca-Oliveira, M., Galina, A., Rosignoli, S., Arcanjo, A. F., Moraes, B., Paiardini, A., Rotili, D., de Paula Li Yasumura, J. D., Henaut-Jacobs, S., Venancio, T. M., Uhl, M., Nunes-da-Fonseca, R., Parizi, L. F., da Silva Vaz Junior, I., dos Santos Mermelstein, C., Rios, T., ... Fantappié, M. R. (2025). Unraveling the Roles of Epigenetic Regulators During the Embryonic Development of Rhipicephalus microplus. International Journal of Molecular Sciences, 26(18), 9171. https://doi.org/10.3390/ijms26189171









