Proteomics Integrated with Transcriptomics of Clubroot Resistant and Susceptible Brassica napus in Response to Plasmodiophora brassicae Infection
Abstract
1. Introduction
2. Results
2.1. P. brassicae Inoculation and Disease Symptom Assessment
2.2. Time-Course Proteomic Profiling of CR- and CS-NIL Roots Samples of B. napus
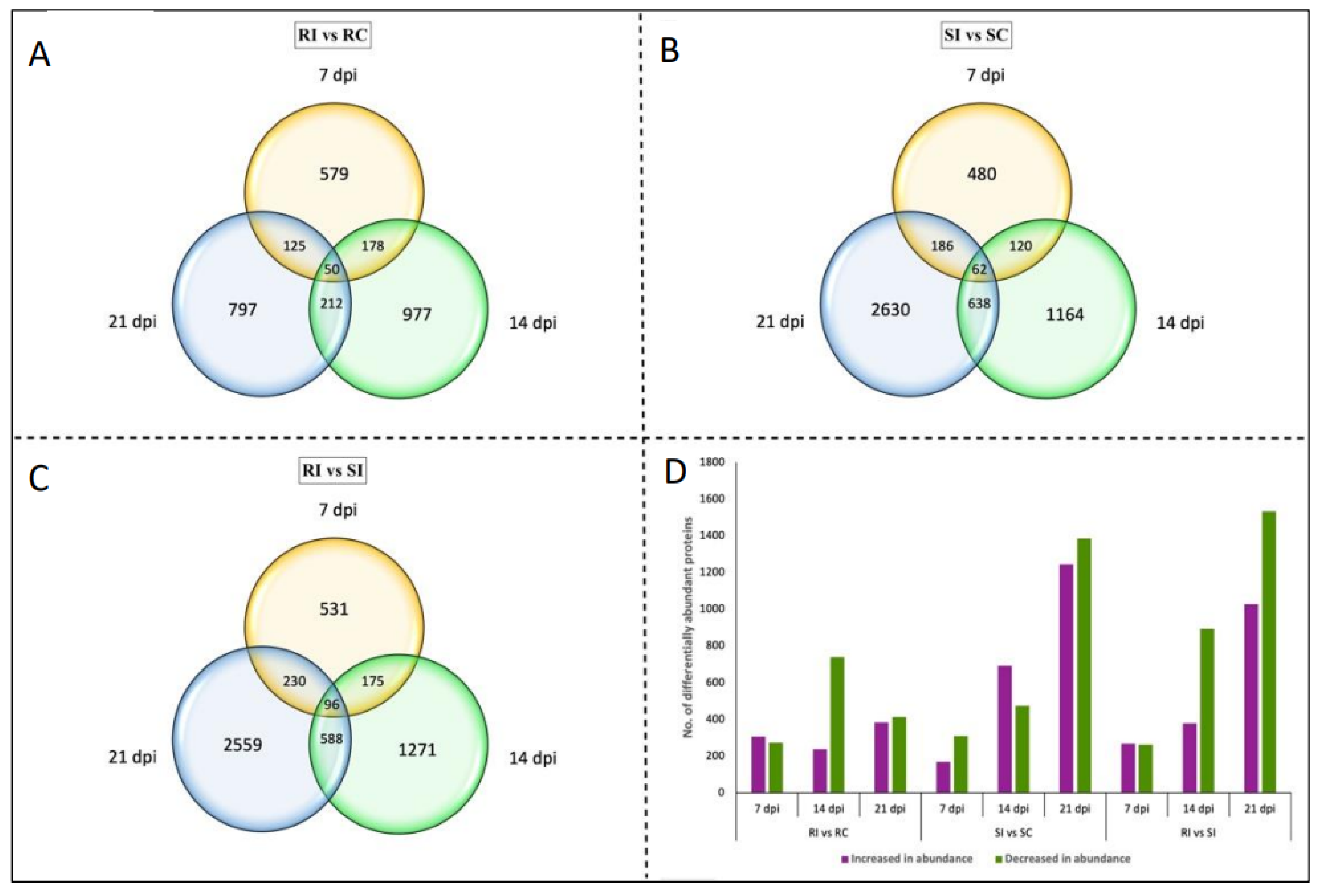
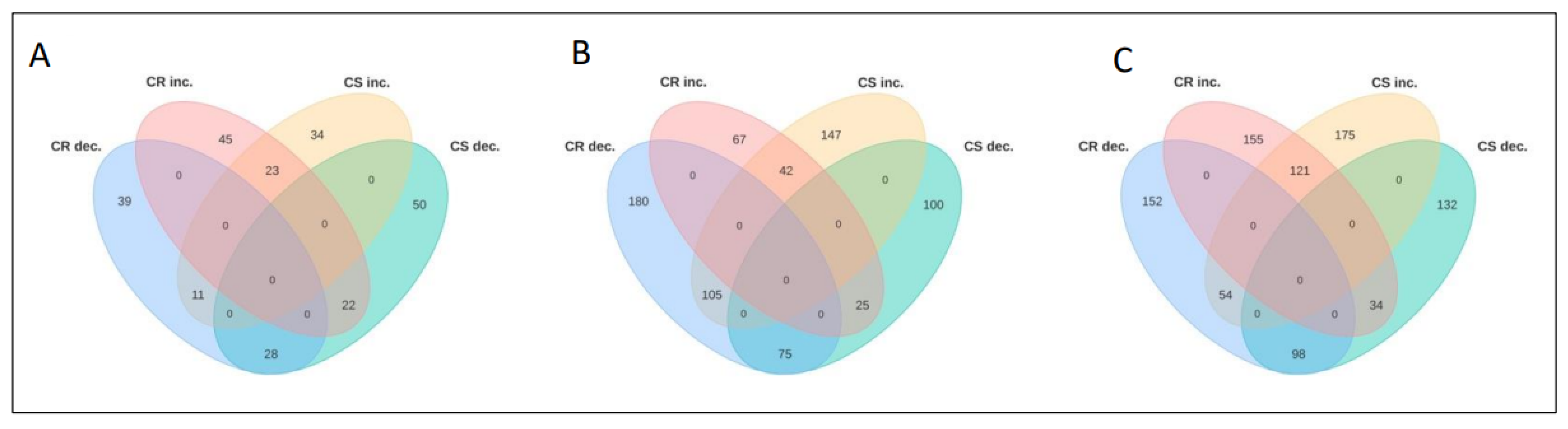
2.3. Temporal Changes in the CR- and CS-NIL Root Proteomes in Response to Pathogen Infection (RI vs. RC and SI vs. SC)
2.3.1. 7 Days Post-Inoculation
2.3.2. 14 Days Post-Inoculation
2.3.3. 21 Days Post-Inoculation
2.4. Temporal Changes in the CR- and CS-NILs Root Proteomes upon Infection by P. brassicae (RI vs. SI)
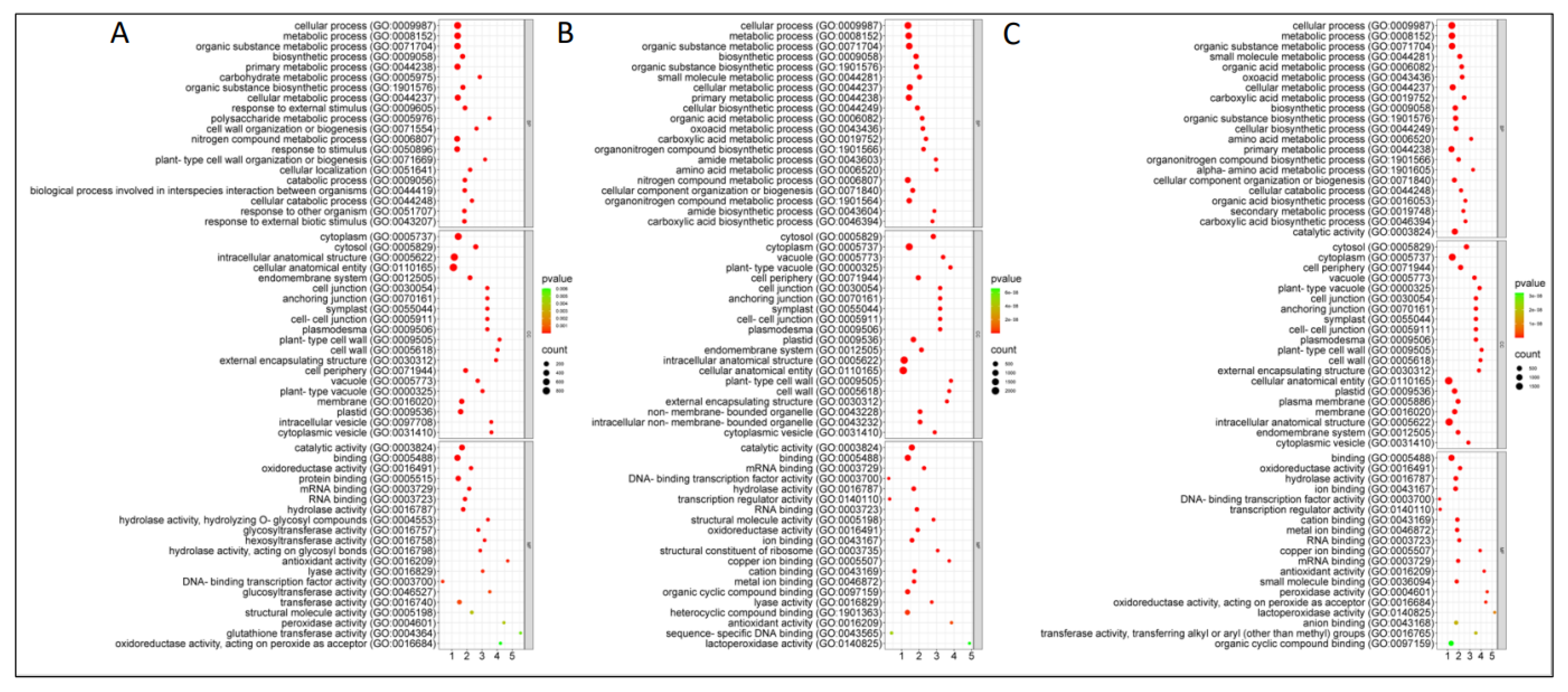
2.5. GO Enrichment of Differentially Abundant Proteins (DAPs)
2.6. Identification of Putative DAPs Involved in Clubroot Resistance
2.6.1. Phytohormone-Mediated Signaling
2.6.2. Calcium-Mediated Signaling
2.6.3. Reactive Oxygen Species-Mediated Signaling
2.6.4. Glucosinolates
2.6.5. Resistance Proteins
2.6.6. General Stress-Related Proteins
2.6.7. Lipid Metabolism
2.6.8. Cell Wall Modifications
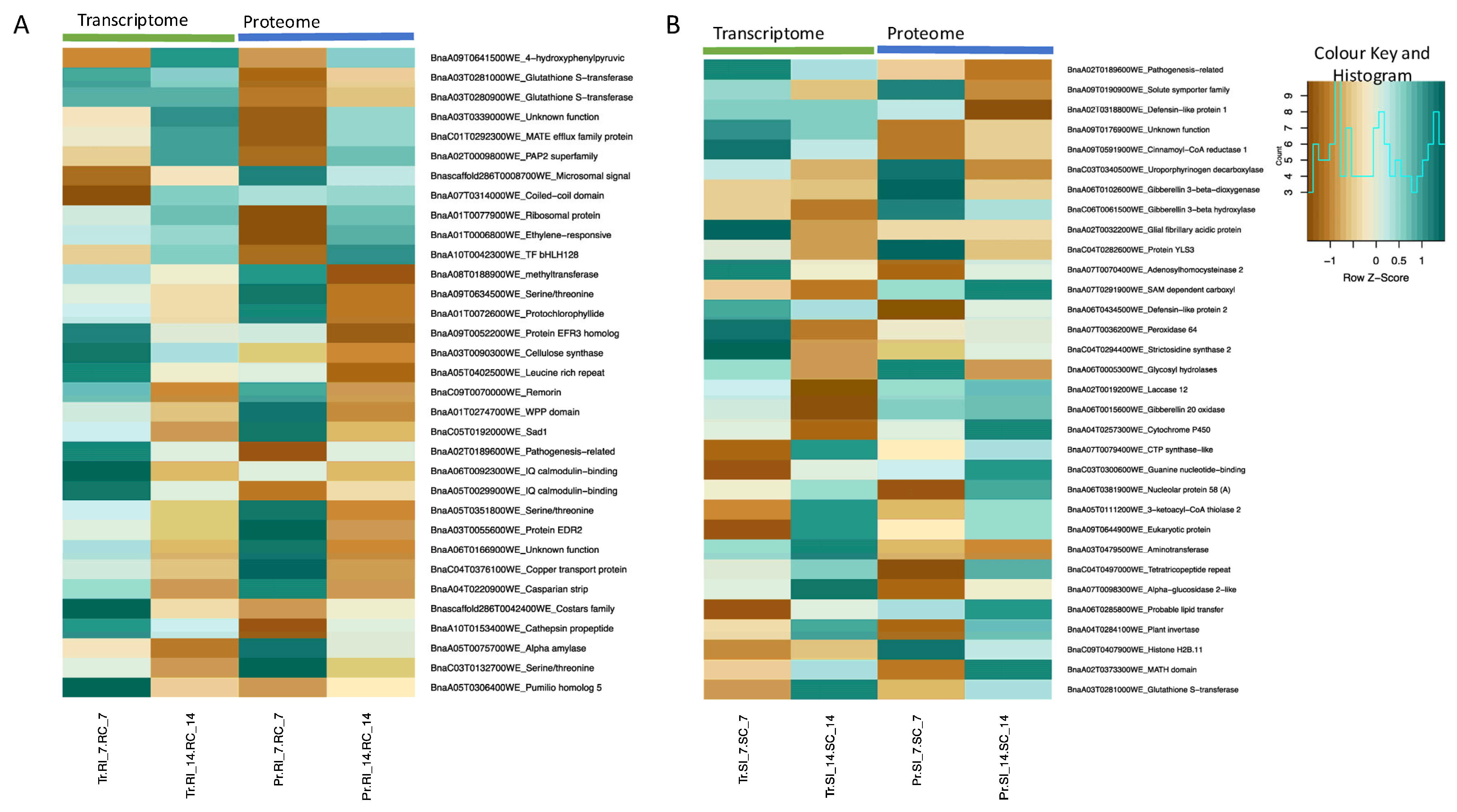
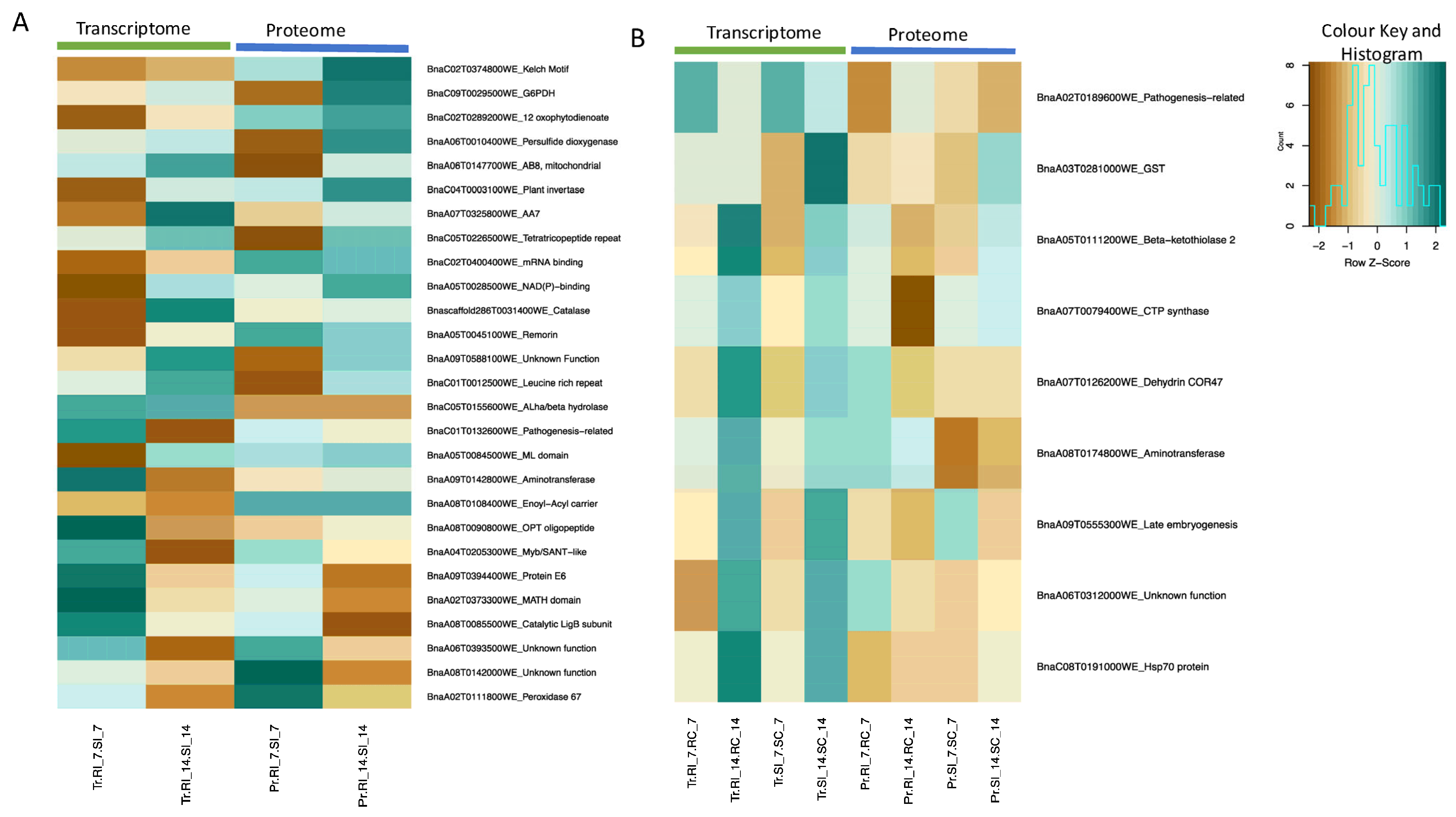
2.7. Multi-Omics Integration: Transcriptomics and Proteomics
3. Discussion
3.1. Perception of Infection Signals
3.2. Phytohormone-Mediated Signaling Implications
3.3. Resistance Proteins (R-Proteins)
3.4. Glucosinolate Metabolism
3.5. Multi-Omics Analysis Reveals ROS Signaling and Cell Wall Reinforcement as a Likely Strategy for P. brassicae Defense
4. Study Limitations
5. Conclusions
6. Materials and Methods
6.1. Plant Materials
6.2. Preparation of the Inoculum and Inoculation Technique
6.3. Sample Collection for Protein Extraction
6.4. Protein Extraction for Proteome Analysis
6.5. Nanoflow LC-MS/MS Analysis
6.6. Functional Annotation and Enrichment of Differentially Abundant Proteins
6.7. Integration of Transcriptomics with Proteomics
6.8. Correlation Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Buczacki, S.T. Plasmodiophora. An inter-relationship between biological and practical problems. In Zoosporic Plant Pathogens: A Modern Perspective; Buczacki, S.T., Ed.; Academic Press Inc.: London, UK, 1983; pp. 161–191. [Google Scholar]
- Statista. Worldwide Oilseed Production by Type 2021/22. Available online: https://www.statista.com/statistics/267271/worldwide-oilseed-production-since-2008/ (accessed on 27 March 2023).
- Canola Council of Canada. Industry Overview. 2021. Available online: https://www.canolacouncil.org/about-canola/industry/ (accessed on 27 March 2023).
- Dixon, G.R. The Occurrence and Economic Impact of Plasmodiophora brassicae and Clubroot Disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Deora, A.; Gossen, B.D.; McDonald, M.R. Infection and development of Plasmodiophora brassicae in resistant and susceptible canola cultivars. Can. J. Plant Sci. 2012, 34, 239–247. [Google Scholar]
- Kageyama, K.; Asano, T. Life Cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 203. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Prinsen, E.; Rolfe, S.A.; Scholes, J.D. Metabolism and plant hormone action during clubroot disease. J. Plant Growth Regul. 2009, 28, 229–244. [Google Scholar] [CrossRef]
- Donald, C.; Porter, I. Integrated control of clubroot. J. Plant Growth Regul. 2009, 28, 289–303. [Google Scholar] [CrossRef]
- Hollman, K.B.; Hwang, S.F.; Manolii, V.P.; Strelkov, S.E. Pathotypes of Plasmodiophora brassicae collected from clubroot resistant canola (Brassica napus L.) cultivars in western Canada in 2017–2018. Can. J. Plant Pathol. 2021, 43, 622–630. [Google Scholar] [CrossRef]
- Chen, J.; Pang, W.; Chen, B.; Zhang, C.; Piao, Z. Transcriptome analysis of Brassica rapa near-isogenic lines carrying clubroot-resistant and -susceptible alleles in response to Plasmodiophora brassicae during early infection. Front. Plant Sci. 2016, 6, 1183. [Google Scholar] [CrossRef]
- Chu, M.; Song, T.; Falk, K.C.; Zhang, X.; Liu, X.; Chang, A.; Lahlali, R.; McGregor, L.; Gossen, B.D.; Yu, F.; et al. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genom. 2014, 15, 1166. [Google Scholar] [CrossRef]
- Fu, P.; Piao, Y.; Zhan, Z.; Zhao, Y.; Pang, W.; Li, X.; Piao, Z. Transcriptome profile of Brassica rapa L. reveals the involvement of jasmonic acid, ethylene, and brassinosteroid signaling pathways in clubroot resistance. Agronomy 2019, 9, 589. [Google Scholar] [CrossRef]
- Gludovacz, T.V.; Deora, A.; McDonald, M.R.; Gossen, B.D. Cortical colonization by Plasmodiophora brassicae in susceptible and resistant cabbage cultivars. Eur. J. Plant Pathol. 2014, 140, 859–862. [Google Scholar] [CrossRef]
- Ji, R.; Gao, S.; Bi, Q.; Wang, Y.; Lv, M.; Ge, W.; Feng, H. The salicylic acid signaling pathway plays an important role in the resistant process of Brassica rapa L. ssp. pekinensis to Plasmodiophora brassicae Woronin. J. Plant Growth Regul. 2021, 40, 405–422. [Google Scholar] [CrossRef]
- Jia, H.; Wei, X.; Yang, Y.; Yuan, Y.; Wei, F.; Zhao, Y.; Yang, S.; Yao, Q.; Wang, Z.; Tian, B.; et al. Root RNA-seq analysis reveals a distinct transcriptome landscape between clubroot-susceptible and clubroot-resistant Chinese cabbage lines after Plasmodiophora brassicae infection. Plant Soil 2017, 421, 93–105. [Google Scholar] [CrossRef]
- Lan, M.; Li, G.; Hu, J.; Yang, H.; Zhang, L.; Xu, X.; Liu, J.; He, J.; Sun, R. iTRAQ-based quantitative analysis reveals proteomic changes in Chinese cabbage (Brassica rapa L.) in response to Plasmodiophora brassicae infection. Sci. Rep. 2019, 9, 12058, Erratum in Sci. Rep. 2020, 10, 1849. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, Y.; Zhao, Y.; Xie, Z.; Hossain, M.R.; Yang, S.; Shi, G.; Lv, Y.; Wang, Z.; Tian, B.; et al. Root transcriptome and metabolome profiling reveal key phytohormone-related genes and pathways involved clubroot resistance in Brassica rapa L. Front. Plant Sci. 2021, 12, 759623. [Google Scholar]
- Yuan, Y.; Qin, L.; Su, H.; Yang, S.; Wei, X.; Wang, Z.; Zhao, Y.; Li, L.; Liu, H.; Tian, B.; et al. Transcriptome and coexpression network analyses reveal hub genes in Chinese cabbage (Brassica rapa L. ssp. pekinensis) during different stages of Plasmodiophora brassicae infection. Front. Plant Sci. 2021, 12, 650252. [Google Scholar] [CrossRef]
- Adhikary, D.; Mehta, D.; Uhrig, R.G.; Rahman, H.; Kav, N.N.V. A proteome-level investigation into Plasmodiophora brassicae resistance in Brassica napus canola. Front. Plant Sci. 2022, 13, 860393, Correction in Front. Plant Sci. 2025, 16, 1597953. [Google Scholar] [CrossRef]
- Cao, T.; Srivastava, S.; Rahman, M.H.; Kav, N.N.V.; Hotte, N.; Deyholos, M.K.; Strelkov, S.E. Proteome-level changes in the roots of Brassica napus as a result of Plasmodiophora brassicae infection. Plant Sci. 2008, 174, 97–115. [Google Scholar] [CrossRef]
- Ji, R.; Wang, Y.; Wang, X.; Liu, Y.; Shen, X.; Feng, H. Proteomic analysis of the interaction between Plasmodiophora brassicae and Chinese cabbage (Brassica rapa L. ssp. pekinensis) at the initial infection stage. Sci. Hortic. 2018, 233, 386–393. [Google Scholar] [CrossRef]
- Moon, J.; Kim, S.; Choi, G.; Kwon, S.Y.; Cho, H.S.; Kim, H.S.; Moon, J.S.; Park, J.M. Comparative proteomic analysis of host responses to Plasmodiophora brassicae infection in susceptible and resistant Brassica oleracea. Plant Biotechnol. Rep. 2020, 14, 263–274. [Google Scholar] [CrossRef]
- Song, T.; Chu, M.; Lahlali, R.; Yu, F.; Peng, G. Shotgun label-free proteomic analysis of clubroot (Plasmodiophora brassicae) resistance conferred by the gene Rcr1 in Brassica rapa. Front. Plant Sci. 2016, 7, 1013. [Google Scholar] [CrossRef]
- Hasan, J.; Megha, S.; Rahman, H. Clubroot in Brassica: Recent advances in genomics, breeding, and disease management. Genome 2021, 64, 735–760. [Google Scholar] [CrossRef]
- Zhang, L.; Du, L.; Poovaiah, B.W. Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 2014, 9, e973818. [Google Scholar] [CrossRef]
- Su, T.; Yu, S.; Wang, W.; Li, P.; Zhang, F.; Yu, Y.; Zhang, D.; Zhao, X. iTRAQ analysis of protein profile during the secondary stage of infection of Plasmodiophora brassicae in Chinese cabbage (Brassica rapa subsp. pekinensis). J. Plant Pathol. 2018, 100, 533–542. [Google Scholar] [CrossRef]
- Adhikary, D.; Mehta, D.; Kisiala, A.; Basu, U.; Uhrig, R.G.; Emery, R.N.; Rahman, H.; Kav, N.N.V. Proteome- and metabolome-level changes during early stages of clubroot infection in Brassica napus canola. Mol. Omics 2024, 20, 265–282. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsen, M.; Honoré, B. Transcriptomics and Proteomics: Integration? In eLS; John Wiley & Sons, Ltd.: Chichester, UK; American Cancer Society: Atlanta, GA, USA, 2018; pp. 1–7. [Google Scholar]
- Kaur, K.; Basu, U.; Kav, N.N.V.; Rahman, H. Transcriptome analysis of Brassica napus near-isogenic lines carrying clubroot resistance of turnip (B. rapa var. rapifera). Genome 2025, 68, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ferdausi, A.; Megha, S.; Liu, Y.; Rahman, H. Metabolomic and transcript level changes reveal the role of polyphenols and flavonols in response to Plasmodiophora brassicae infection in Brassica napus. J. Plant Pathol. 2023, 105, 1449–1464. [Google Scholar] [CrossRef]
- Wagner, G.; Laperche, A.; Lariagon, C.; Marnet, N.; Renault, D.; Guitton, Y.; Bouchereau, A.; Delourme, R.; Manzanares-Dauleux, M.J.; Gravot, A. Resolution of quantitative resistance to clubroot into QTL-specific metabolic modules. J. Exp. Bot. 2019, 70, 5375–5390. [Google Scholar] [CrossRef]
- Wang, Z.; Megha, S.; Kebebe, S.; Kav, N.N.V.; Rahman, H. Genetic and molecular analysis reveals that two major loci and their interaction confer clubroot resistance in canola introgressed from rutabaga. Plant Genome 2022, 15, e20241. [Google Scholar] [CrossRef]
- Takele Assefa, A.; Vandesompele, J.; Thas, O. On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments. BMC Genom. 2020, 21, 312, Correction in BMC Genom. 2020, 21, 384. [Google Scholar] [CrossRef]
- Biswas, S.; Agrawal, Y.N.; Mucyn, T.S.; Dangl, J.L.; Jones, C.D. Biological Averaging in RNA-Seq. arXiv 2013, arXiv:1309.0670. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signaling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Liu, P.L.; Du, L.; Huang, Y.; Gao, S.M.; Yu, M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 2017, 17, 47. [Google Scholar] [CrossRef]
- Campos, W.F.; Dressano, K.; Ceciliato, P.H.O.; Guerrero-Abad, J.C.; Silva, A.L.; Fiori, C.S.; Do Canto, A.M.; Bergonci, T.; Claus, L.A.N.; Silva-Filho, M.C.; et al. Arabidopsis thaliana rapid alkalinization factor 1-mediated root growth inhibition is dependent on calmodulin-like protein 38. J. Biol. Chem. 2018, 293, 2159–2171. [Google Scholar] [CrossRef] [PubMed]
- Köster, P.; DeFalco, T.A.; Zipfel, C. Ca2+ signals in plant immunity. EMBO J. 2022, 41, e110741. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef]
- Zhao, C.; Zayed, O.; Yu, Z.; Zhu, J.K. Leucine-rich repeat extension proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Wang, Q.; Kaspi, R.; Parrella, M.P.; Abel, S. Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant. J. 2005, 43, 79–96. [Google Scholar] [CrossRef]
- Göhre, V.; Spallek, T.; Häweker, H.; Mersmann, S.; Mentzel, T.; Boller, T.; Torres, M.D.; Mansfield, J.W.; Robatzek, S. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 2008, 18, 1824–1832. [Google Scholar] [CrossRef]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef]
- Trujillo, M.; Shirasu, K. Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 2010, 13, 402–408. [Google Scholar] [CrossRef]
- Abel, S.; Ballas, N.; Wong, L.M.; Theologis, A. DNA elements responsive to auxin. Bioessays 1996, 18, 647–654. [Google Scholar] [CrossRef]
- Harper, R.M.; Stowe-Evans, E.L.; Luesse, D.R.; Muto, H.; Tatematsu, K.; Watahiki, M.K.; Yamamoto, K.; Liscum, E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 2000, 12, 757–770. [Google Scholar] [CrossRef]
- Jahn, L.; Mucha, S.; Bergmann, S.; Horn, C.; Staswick, P.; Steffens, B.; Siemens, J.; Ludwig-Müller, J. The Clubroot pathogen (Plasmodiophora brassicae) influences auxin signaling to regulate auxin homeostasis in Arabidopsis. Plants 2013, 2, 726–749. [Google Scholar] [CrossRef] [PubMed]
- LeClere, S.; Tellez, R.; Rampey, R.A.; Matsuda, S.P.; Bartel, B. Characterization of a family of IAA- amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002, 277, 20446–20452. [Google Scholar] [CrossRef] [PubMed]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Robin, A.H.K.; Saha, G.; Laila, R.; Park, J.I.; Kim, H.T.; Nou, I.S. Expression and role of biosynthetic, transporter, receptor, and responsive genes for auxin signaling during clubroot disease development. Int. J. Mol. Sci. 2020, 21, 5554. [Google Scholar] [CrossRef]
- Zhou, J.M.; Trifa, Y.; Silva, H.; Pontier, D.; Lam, E.; Shah, J.; Klessig, D.F. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 2000, 13, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tessaro, M.J.; Lassner, M.; Li, X. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 2003, 15, 2647–2653. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, Y.; Huang, Q.; Yin, G.; Pennerman, K.K.; Yu, J.; Liu, Z.; Li, D.; Guo, A. Analysis of key genes of jasmonic acid mediated signal pathway for defense against insect damages by comparative transcriptome sequencing. Sci. Rep. 2015, 5, 16500. [Google Scholar] [CrossRef]
- Xu, L.; Yang, H.; Ren, L.; Chen, W.; Liu, L.; Liu, F.; Zeng, L.; Yan, R.; Chen, K.; Fang, X. Jasmonic acid-mediated aliphatic glucosinolate metabolism is involved in clubroot disease development in Brassica napus L. Front. Plant Sci. 2018, 9, 750. [Google Scholar] [PubMed]
- Müller, M.; Munné-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Vorwerk, S.; Schiff, C.; Santamaria, M.; Koh, S.; Nishimura, M.; Vogel, J.; Somerville, C.; Somerville, S. EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol. 2007, 7, 35. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, J.; An, L.; Doerge, R.W.; Chen, Z.J.; Grau, C.R.; Meng, J.; Osborn, T.C. Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta 2007, 227, 13–24. [Google Scholar] [CrossRef]
- Choi, H.W.; Lee, B.G.; Kim, N.H.; Park, Y.; Lim, C.W.; Song, H.K.; Hwang, B.K. A role for a menthone reductase in resistance against microbial pathogens in plants. Plant Physiol. 2008, 148, 383–401. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Hasegawa, A.; Taninaka, A.; Mizutani, M.; Sugimoto, Y. NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J. Biol. Chem. 2011, 286, 6999–7009. [Google Scholar] [CrossRef] [PubMed]
- Selote, D.; Robin, G.P.; Kachroo, A. GmRIN4 protein family members function nonredundantly in soybean race-specific resistance against Pseudomonas syringae. New Phytol. 2013, 197, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Wilton, M.; Subramaniam, R.; Elmore, J.; Felsensteiner, C.; Coaker, G.; Desveaux, D. The type III effector HopF2(Pto) targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. USA 2010, 107, 2349–2354. [Google Scholar] [CrossRef] [PubMed]
- Summanwar, A.; Basu, U.; Kav, N.N.V.; Rahman, H. Identification of lncRNAs in response to infection by Plasmodiophora brassicae in Brassica napus and development of lncRNA-based SSR markers. Genome 2020, 64, 547–566. [Google Scholar] [CrossRef]
- Brader, G.; Mikkelsen, M.D.; Halkier, B.A.; Tapio Palva, E. Altering glucosinolate profiles modulates disease resistance in plants. Plant J. 2006, 46, 758–767. [Google Scholar] [CrossRef]
- Kissen, R.; Bones, A.M. Nitrile-specifier proteins involved in glucosinolate hydrolysis in Arabidopsis thaliana. J. Biol. Chem. 2009, 284, 12057–12070. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2–oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Qin, L.; Karunakaran, C.; Wei, Y.; Peng, G. Lignin accumulation in cell wall plays a role in clubroot resistance. Front. Plant Sci. 2024, 15, 1401265. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Baek, K.H. Protective roles of cytosolic and plastidal proteasomes on abiotic stress and pathogen invasion. Plants 2020, 9, 832. [Google Scholar] [CrossRef]
- Zhang, X.; Gou, M.; Liu, C.J. Arabidopsis Kelch repeat f-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 2013, 25, 4994–5010. [Google Scholar] [CrossRef]
- Govender, N.T.; Mahmood, M.; Seman, I.A.; Wong, M.Y. The phenylpropanoid pathway and lignin in defense against Ganoderma boninense colonized root tissues in oil palm (Elaeis guineensis Jacq.). Front. Plant Sci. 2017, 8, 1395. [Google Scholar] [CrossRef]
- Wakao, S.; Andre, C.; Benning, C. Functional analyses of cytosolic glucose-6-phosphate dehydrogenases and their contribution to seed oil accumulation in Arabidopsis. Plant Physiol. 2008, 146, 277–288. [Google Scholar] [CrossRef]
- Laloi, C.; Mestres-Ortega, D.; Marco, Y.; Meyer, Y.; Reichheld, J.P. The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 2004, 134, 1006–1016. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef]
- Liu, J.; Fu, C.; Li, G.; Khan, M.N.; Wu, H. ROS homeostasis and plant salt tolerance: Plant nanobiotechnology updates. Sustainability 2021, 13, 3552. [Google Scholar] [CrossRef]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Breusegem, F.V.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef]
- Ono, M.; Isono, K.; Sakata, Y.; Taji, T. CATALASE2 plays a crucial role in long-term heat tolerance of Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2021, 534, 747–751. [Google Scholar] [CrossRef]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef] [PubMed]
- Palma, J.M.; Mateos, R.M.; López-Jaramillo, J.; Rodríguez-Ruiz, M.; González-Gordo, S.; Lechuga-Sancho, A.M.; Corpas, F.J. Plant catalases as NO and H2S targets. Redox Biol. 2020, 34, 101525. [Google Scholar] [CrossRef] [PubMed]
- Skamnioti, P.; Henderson, C.; Zhang, Z.; Robinson, Z.; Gurr, S.J. A novel role for catalase B in the maintenance of fungal cell-wall integrity during host invasion in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 2007, 20, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13, Correction in Genome Biol. 2016, 17, 181. [Google Scholar] [CrossRef]
- Lamarre, S.; Frasse, P.; Zouine, M.; Labourdette, D.; Sainderichin, E.; Hu, G.; Le Berre-Anton, V.; Bouzayen, M.; Maza, E. Optimization of an RNA-Seq differential gene expression analysis depending on biological replicate number and library size. Front. Plant Sci. 2018, 9, 108. [Google Scholar] [CrossRef]
- Kaur, K.; Liu, Y.; Rahman, H. Introgression of resistance to multiple pathotypes of Plasmodiophora brassicae from turnip (Brassica rapa ssp. rapifera) into spring B. napus canola. Agron. J. 2022, 12, 1225. [Google Scholar] [CrossRef]
- Summanwar, A.; Basu, U.; Rahman, H.; Kav, N. Identification of lncRNAs responsive to infection by Plasmodiophora brassicae in clubroot-susceptible and-resistant Brassica napus lines carrying resistance introgressed from rutabaga. Mol. Plant Microbe Interact. 2019, 32, 1360–1377. [Google Scholar] [CrossRef]
- Leutert, M.; Rodríguez-Mias, R.A.; Fukuda, N.K.; Villén, J. R2-P2 rapid-robotic phosphoproteomics enables multidimensional cell signaling studies. Mol. Syst. Biol. 2019, 15, e9021. [Google Scholar] [CrossRef]
- Uhrig, R.G.; Schläpfer, P.; Roschitzki, B.; Hirsch-Hoffmann, M.; Gruissem, W. Diurnal changes in concerted plant protein phosphorylation and acetylation in Arabidopsis organs and seedlings. Plant J. 2019, 99, 176–194. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Scandola, S.; Uhrig, R.G. BoxCar and Library-free data-independent acquisition substantially improve the depth, range, and completeness of label-free quantitative proteomics. Ann. Chem. 2022, 94, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Song, J.M.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R.; et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. 4. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, 412–419. [Google Scholar] [CrossRef]
- Dennis, G.D.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Goecks, J.; Nekrutenko, A.; Taylor, J. Galaxy Team Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, K.; Adhikary, D.; Kav, N.N.V.; Scandola, S.; Uhrig, R.G.; Rahman, H. Proteomics Integrated with Transcriptomics of Clubroot Resistant and Susceptible Brassica napus in Response to Plasmodiophora brassicae Infection. Int. J. Mol. Sci. 2025, 26, 9157. https://doi.org/10.3390/ijms26189157
Kaur K, Adhikary D, Kav NNV, Scandola S, Uhrig RG, Rahman H. Proteomics Integrated with Transcriptomics of Clubroot Resistant and Susceptible Brassica napus in Response to Plasmodiophora brassicae Infection. International Journal of Molecular Sciences. 2025; 26(18):9157. https://doi.org/10.3390/ijms26189157
Chicago/Turabian StyleKaur, Kawalpreet, Dinesh Adhikary, Nat N. V. Kav, Sabine Scandola, R. Glen Uhrig, and Habibur Rahman. 2025. "Proteomics Integrated with Transcriptomics of Clubroot Resistant and Susceptible Brassica napus in Response to Plasmodiophora brassicae Infection" International Journal of Molecular Sciences 26, no. 18: 9157. https://doi.org/10.3390/ijms26189157
APA StyleKaur, K., Adhikary, D., Kav, N. N. V., Scandola, S., Uhrig, R. G., & Rahman, H. (2025). Proteomics Integrated with Transcriptomics of Clubroot Resistant and Susceptible Brassica napus in Response to Plasmodiophora brassicae Infection. International Journal of Molecular Sciences, 26(18), 9157. https://doi.org/10.3390/ijms26189157





