Effect of Reoxygenation on Radioresistance of Chronically Hypoxic A549 Non-Small Cell Lung Cancer (NSCLC) Cells Following X-Ray and Carbon Ion Exposure
Abstract
1. Introduction
2. Results
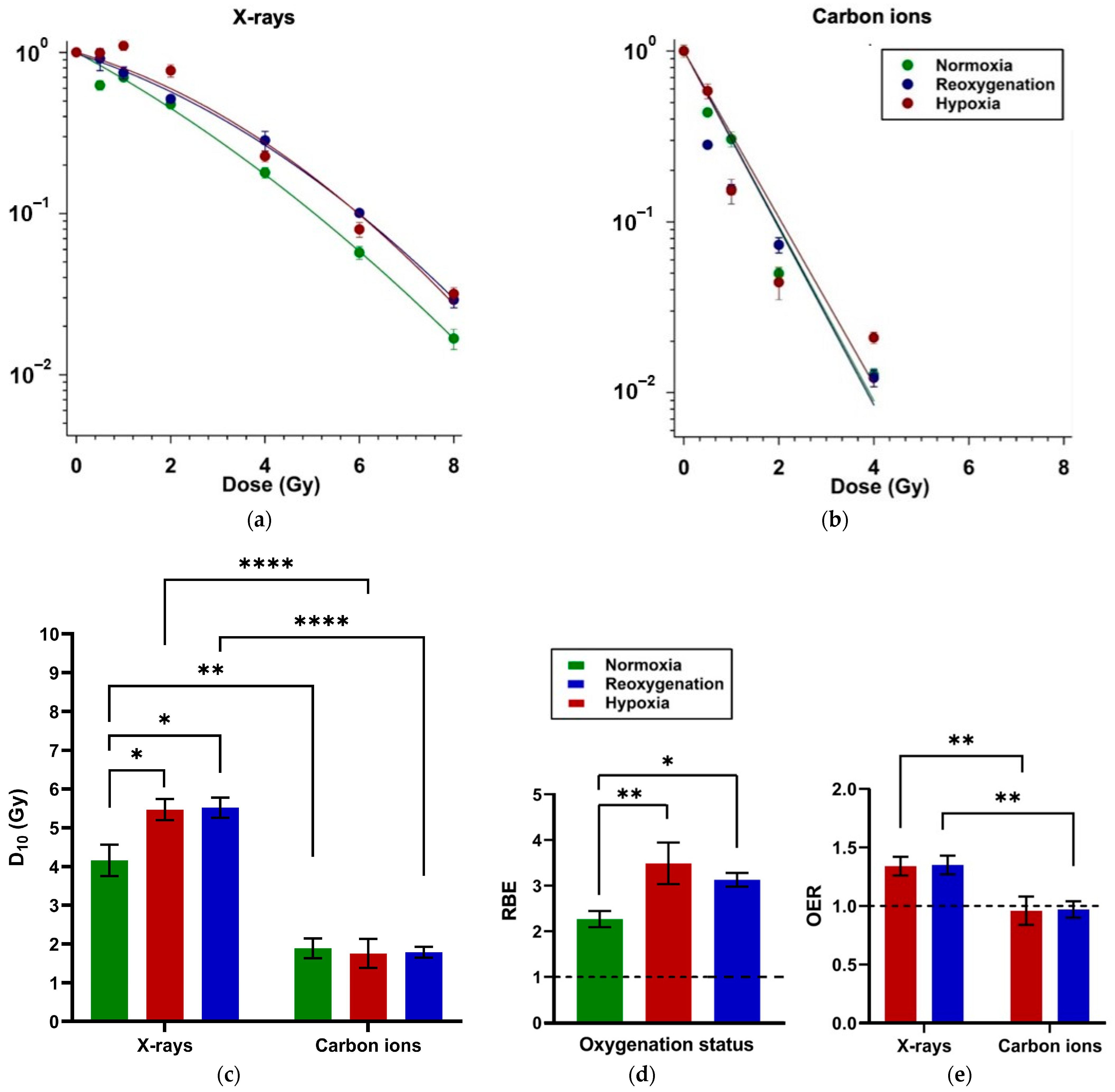
2.1. Minimal Impact of Reoxygenation on Clonogenic Survival of A549 Cells After X-Ray and Carbon Ion Exposure
2.2. Distinct G1 and G2 Phase Radiation Responses in Reoxygenated Compared to Continuously Hypoxic A549 Cells
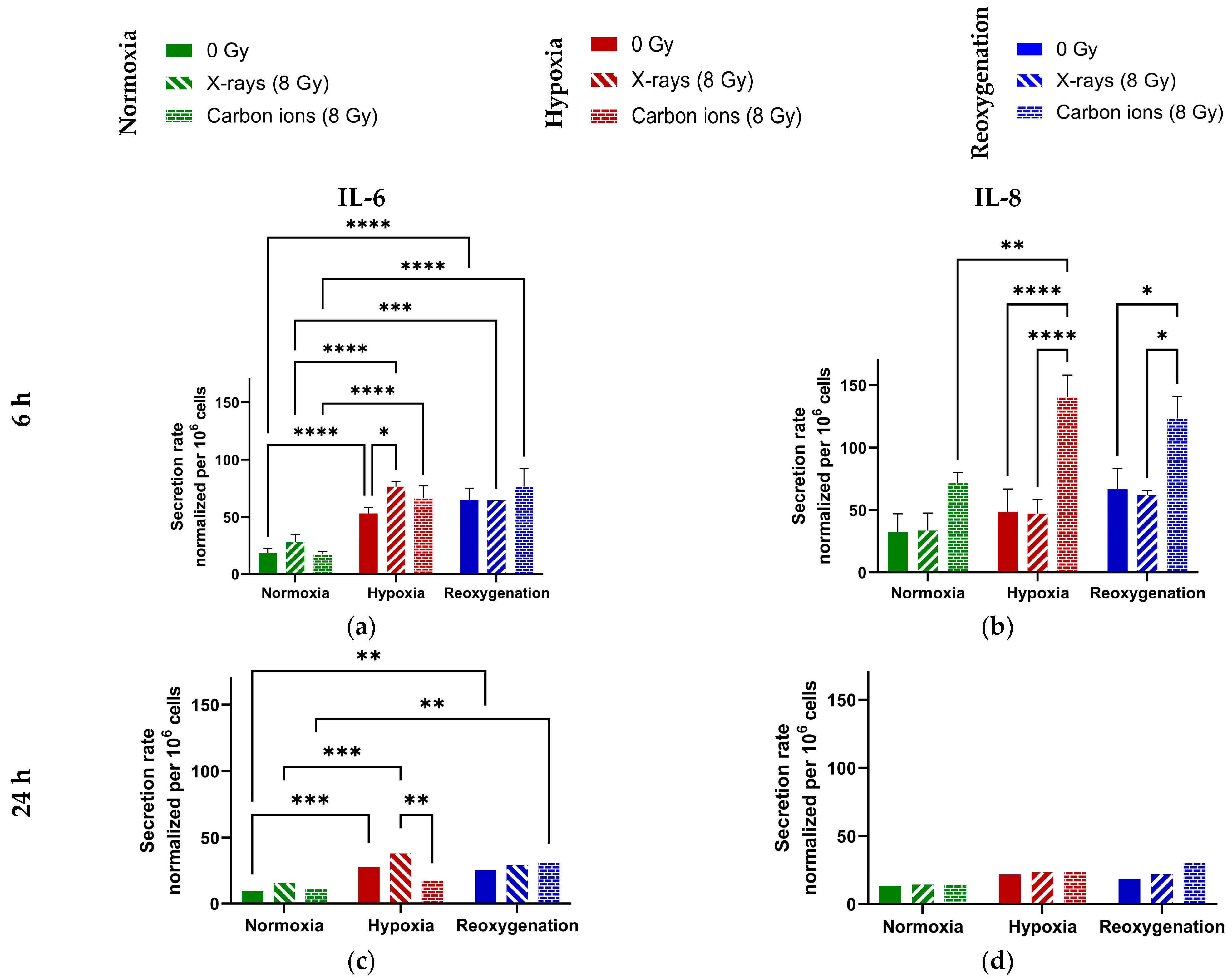
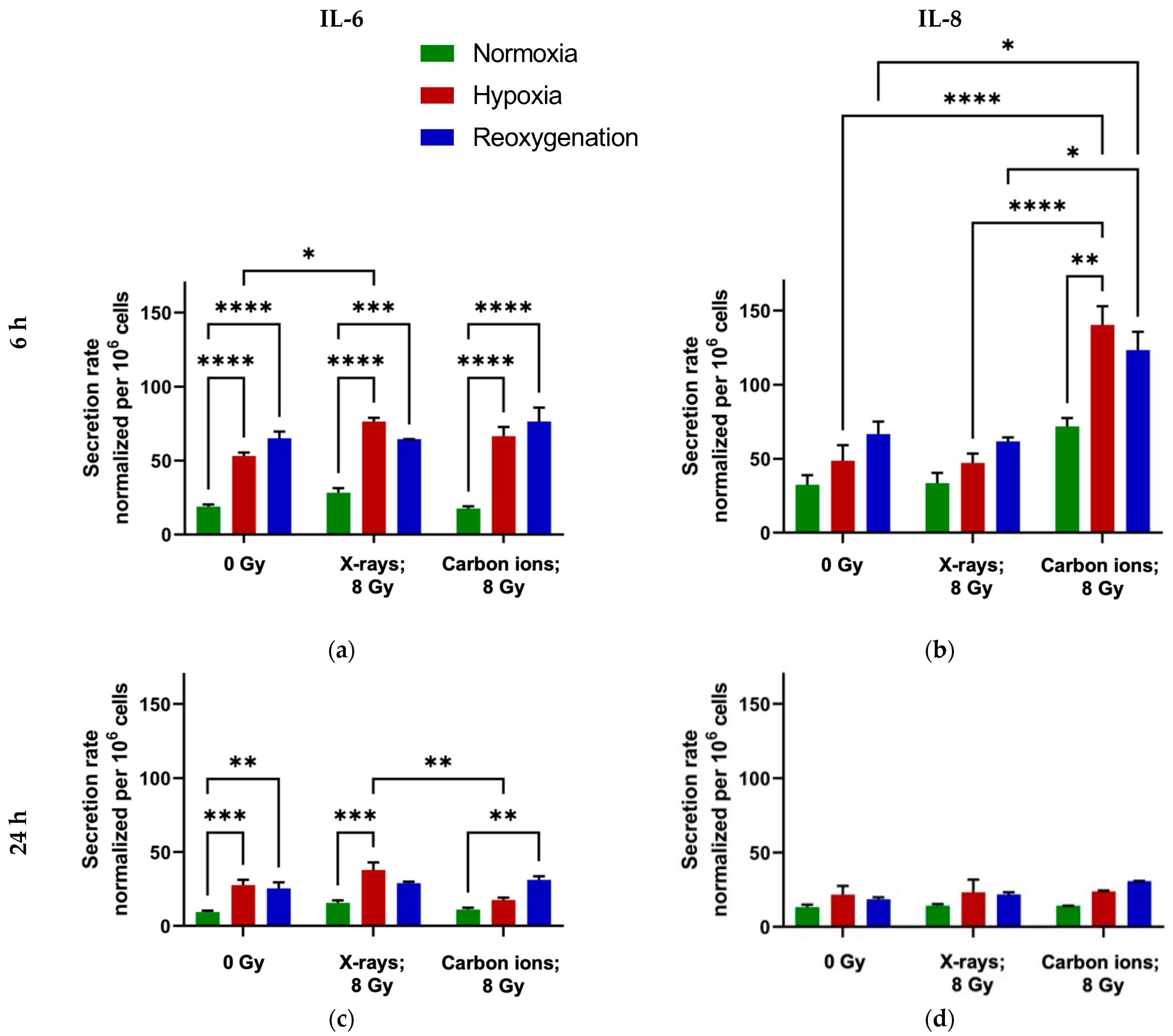
2.3. Minimal Modulation of IL-6 and IL-8 Secretion by Reoxygenation in A549 Cells
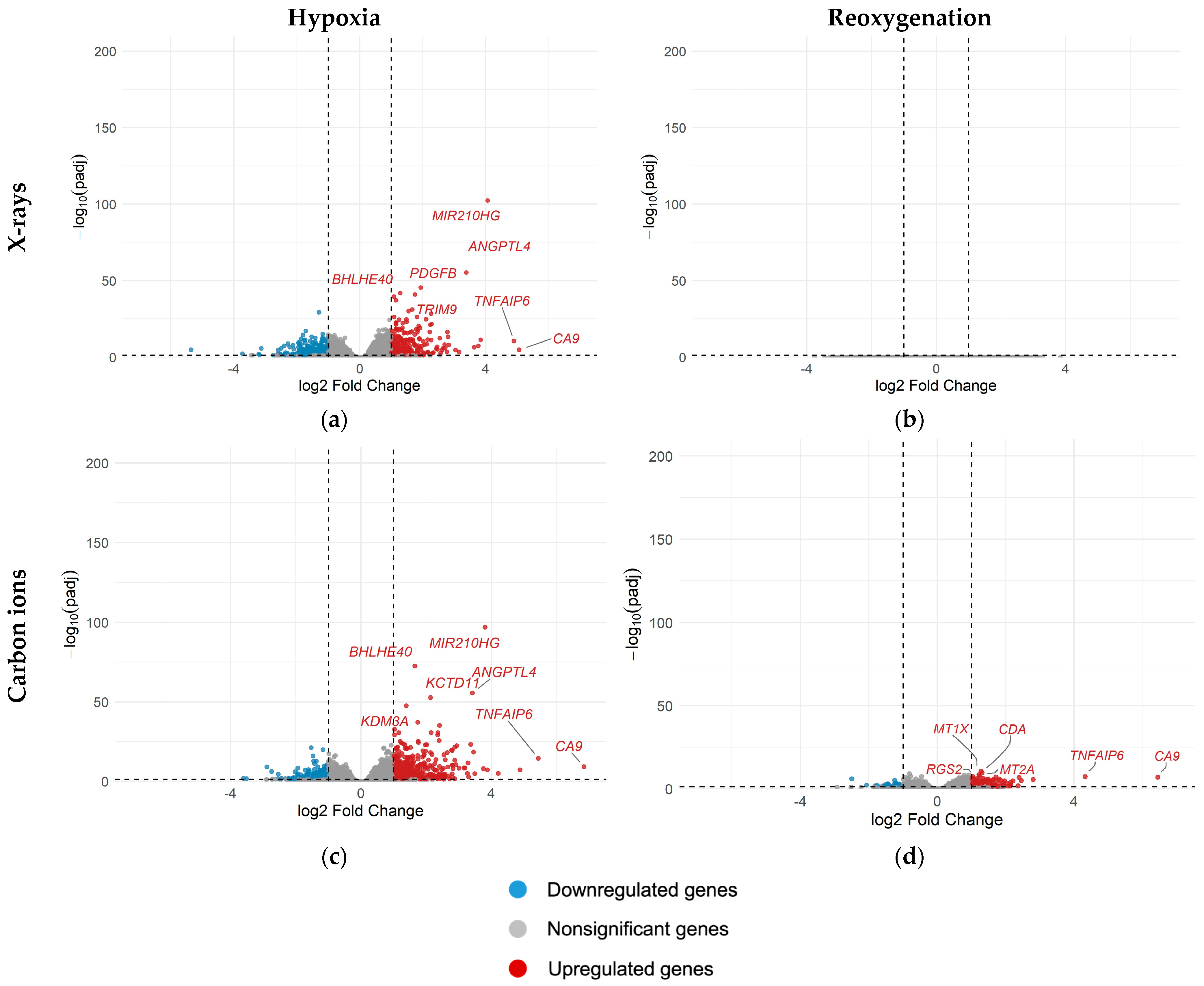
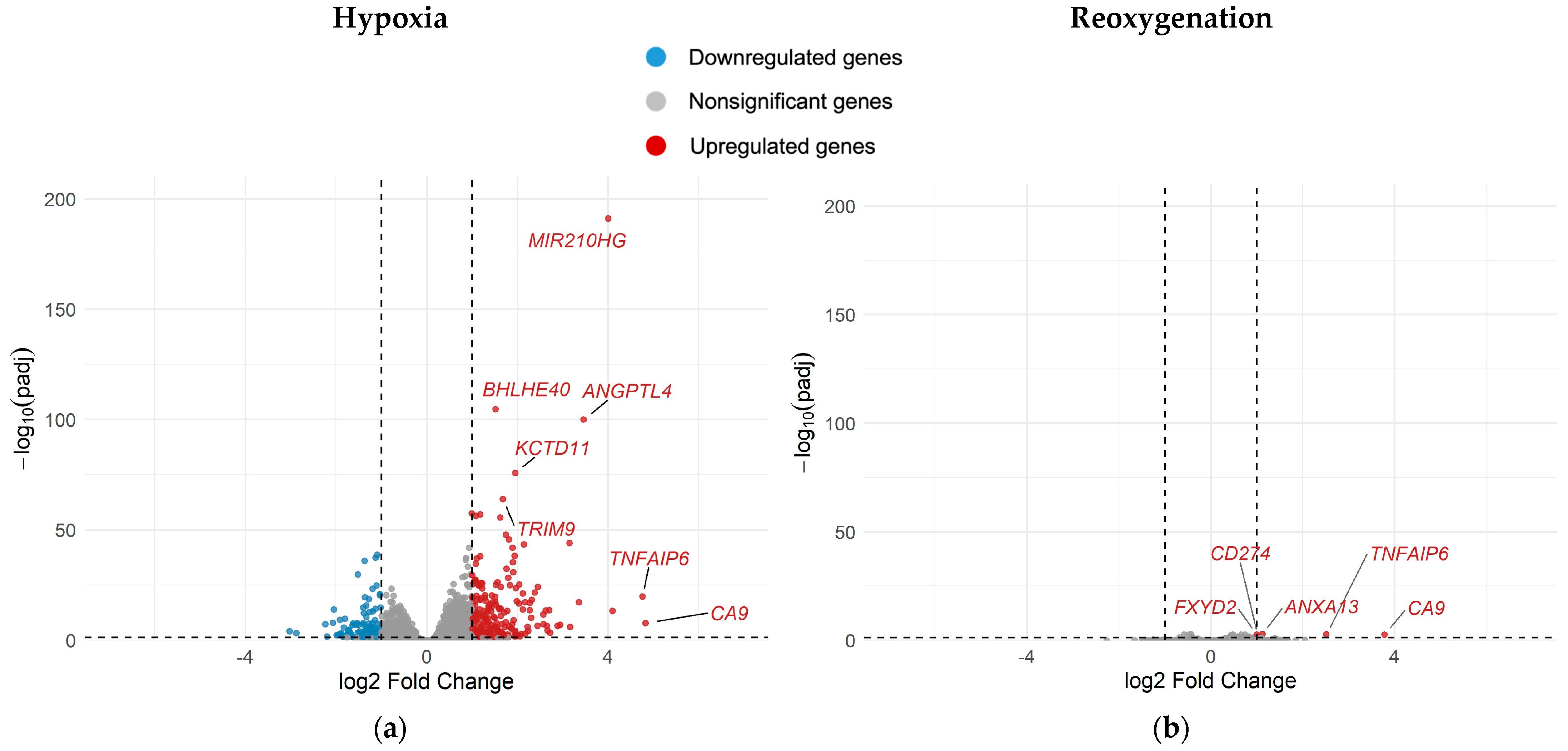
2.4. Differential Gene Expression in A549 Cells Under Different Oxygenation Conditions with and Without Irradiation
- Continuous hypoxia without irradiation (H0) vs. normoxia without irradiation (N0)
- Reoxygenation without irradiation (R0) vs. normoxia without irradiation (N0)
- Continuous hypoxia after irradiation with 8 Gy (H8) vs. normoxia after irradiation with 8 Gy (N8)
- Reoxygenation after irradiation with 8 Gy (R8) vs. normoxia after irradiation with 8 Gy (N8)
- Normoxia after irradiation with 8 Gy (N8) vs. normoxia without irradiation (N0)
- Hypoxia after irradiation with 8 Gy (H8) vs. hypoxia without irradiation (H0)
- Reoxygenation after irradiation with 8 Gy (R8) vs. reoxygenation without irradiation (R0)
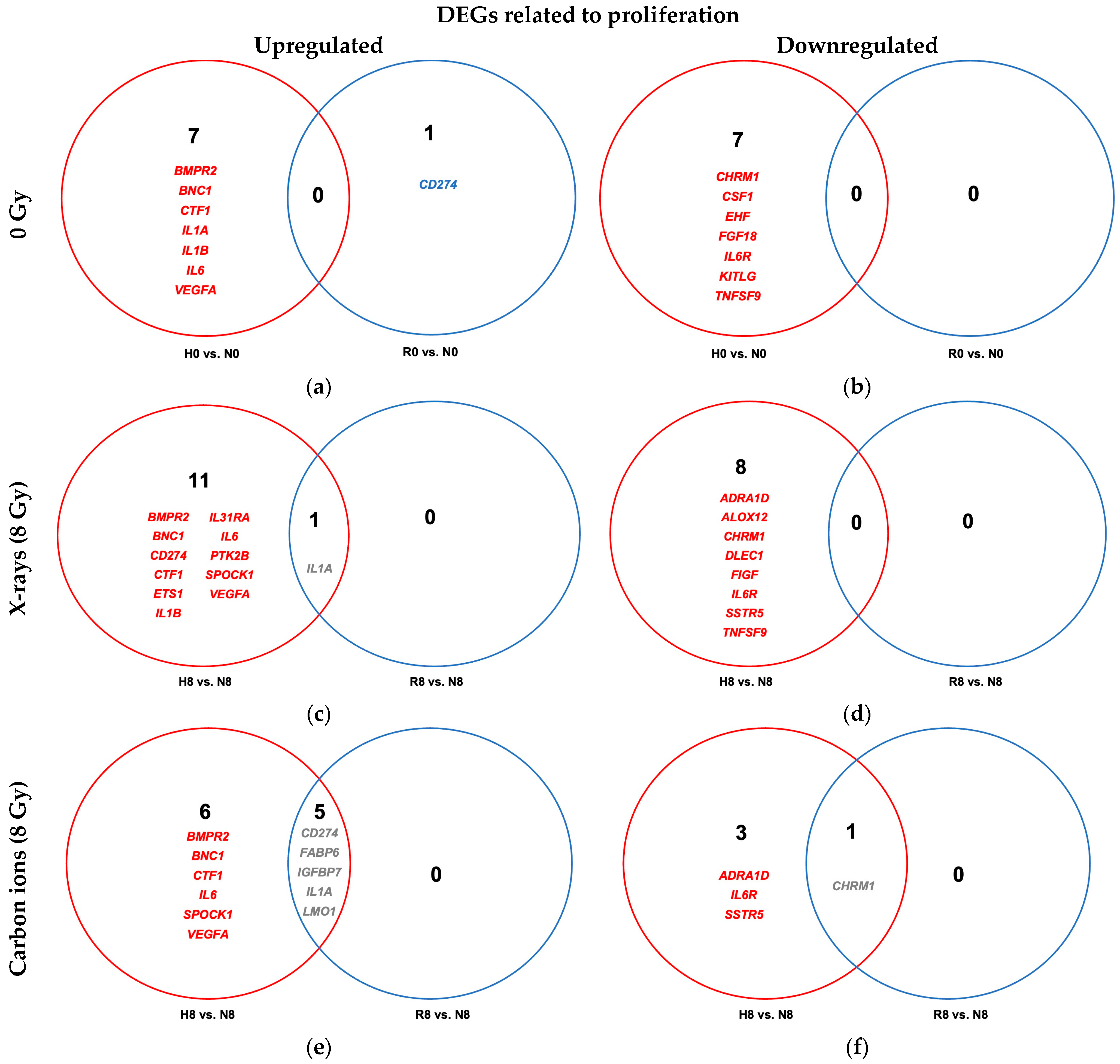
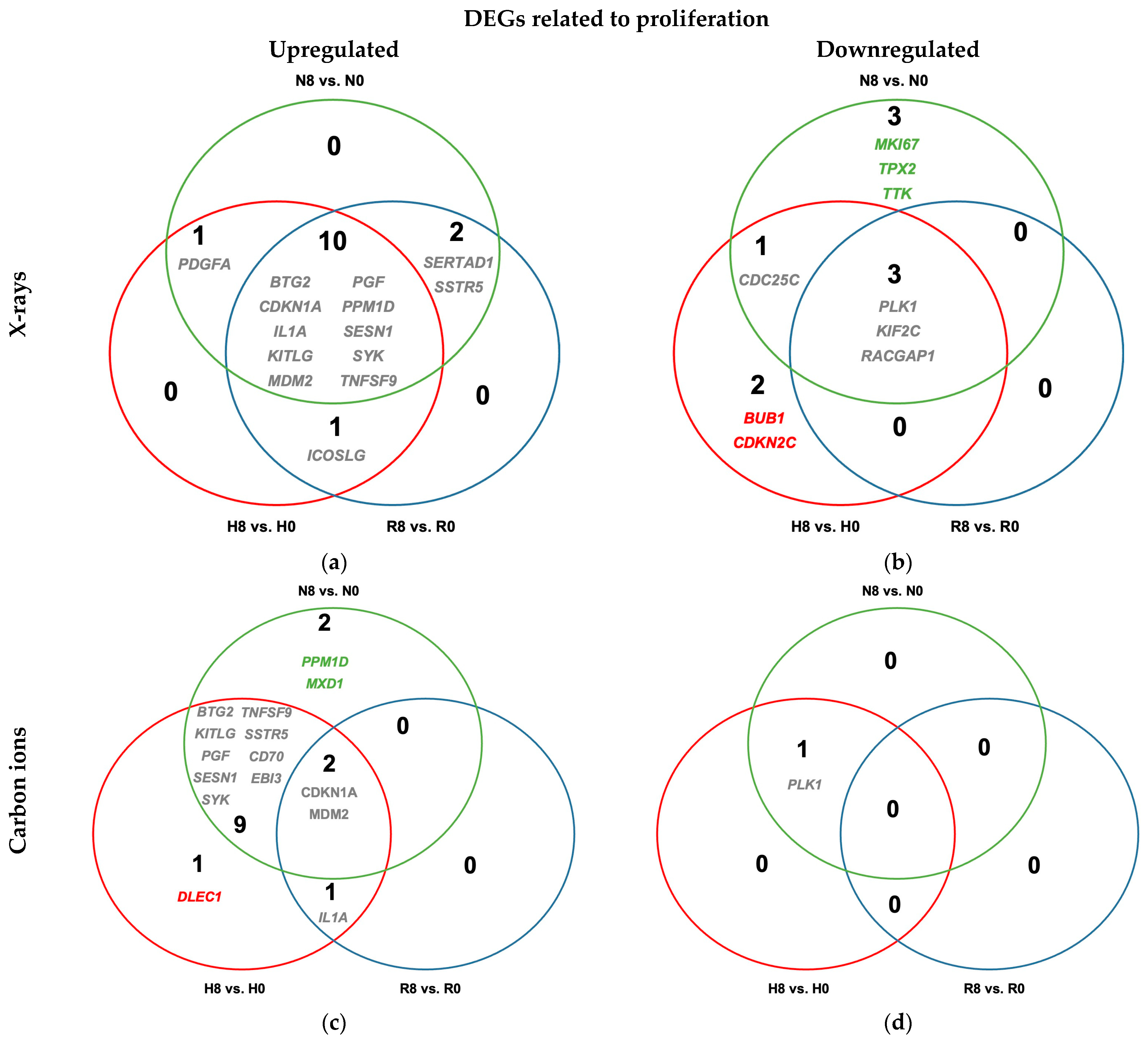
2.4.1. Reoxygenation Reversed Hypoxia-Regulated Expression of Proliferation Genes in Irradiated and Unirradiated A549 Cells
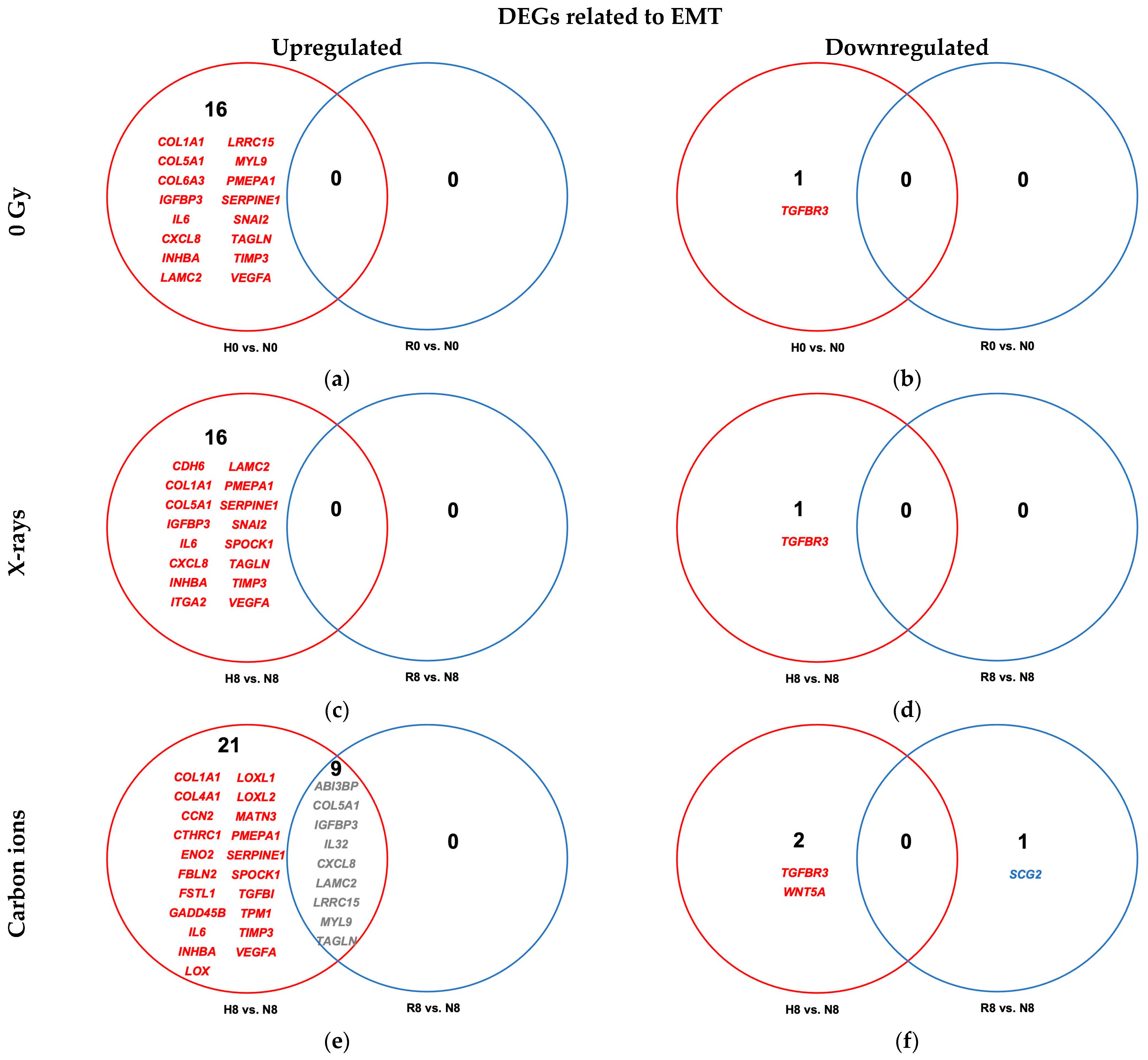
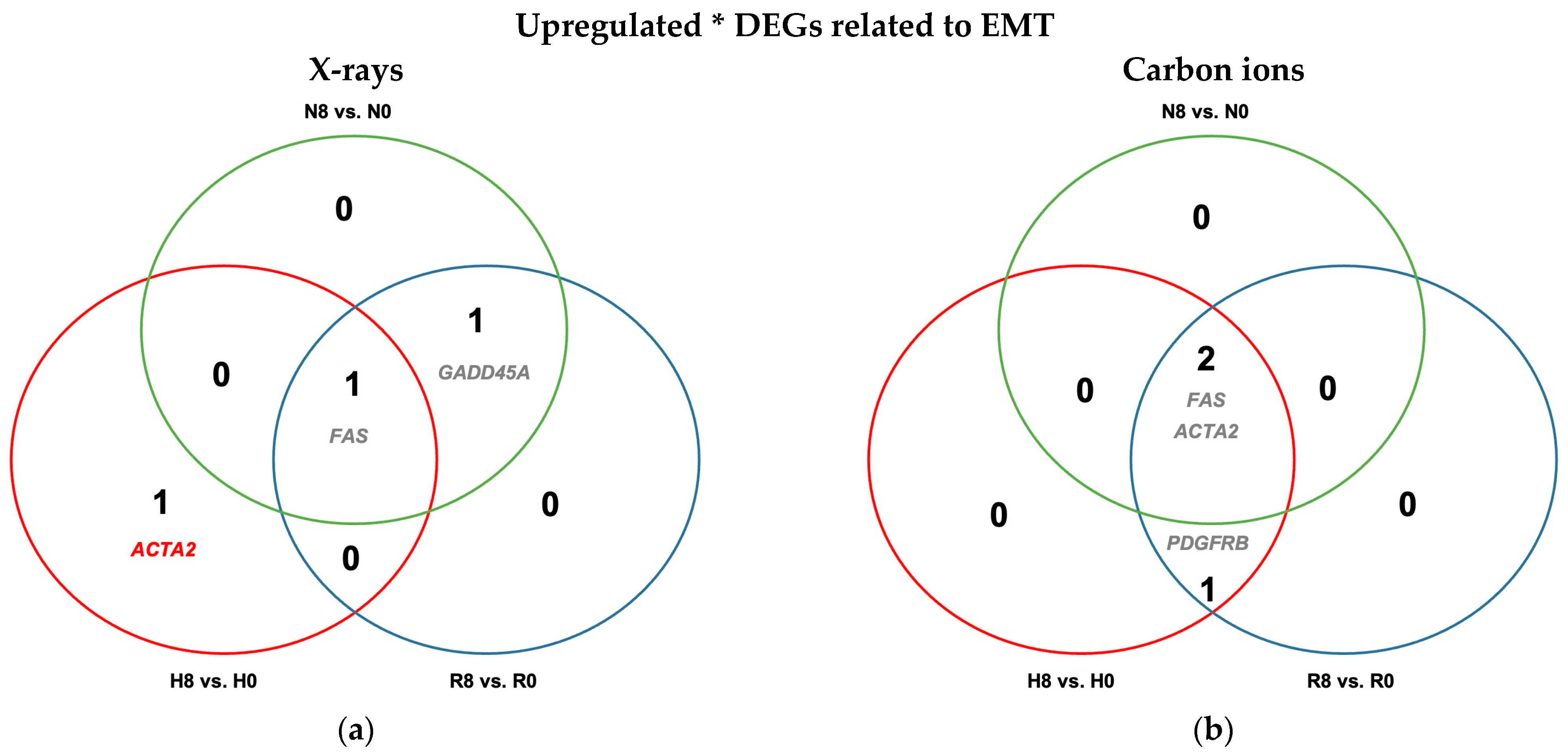
2.4.2. Reoxygenation Reversed Hypoxia-Regulated Expression of EMT Genes in Unirradiated and Irradiated A549 Cells
2.4.3. Antiproliferative Signaling Post-Irradiation Is Observed Regardless of Oxygenation Status in A549 Cells
2.4.4. Minimal Effect of Irradiation on EMT Regulation Regardless of Oxygenation Status in A549 Cells
3. Discussion
3.1. Reoxygenation After Irradiation Has No Impact on Survival in A549 Cells
3.2. Reoxygenation After Irradiation Has No Impact on Cell Cycle Dynamics in A549 Cells
3.3. Reoxygenation After Irradiation Has No Impact on IL-6 and IL-8 Secretion in A549 Cells
3.4. Reoxygenation After Irradiation Reversed Hypoxia-Induced Transcriptional Changes
3.4.1. Reoxygenation and Proliferative Signaling
3.4.2. Reoxygenation and EMT-Related Signaling
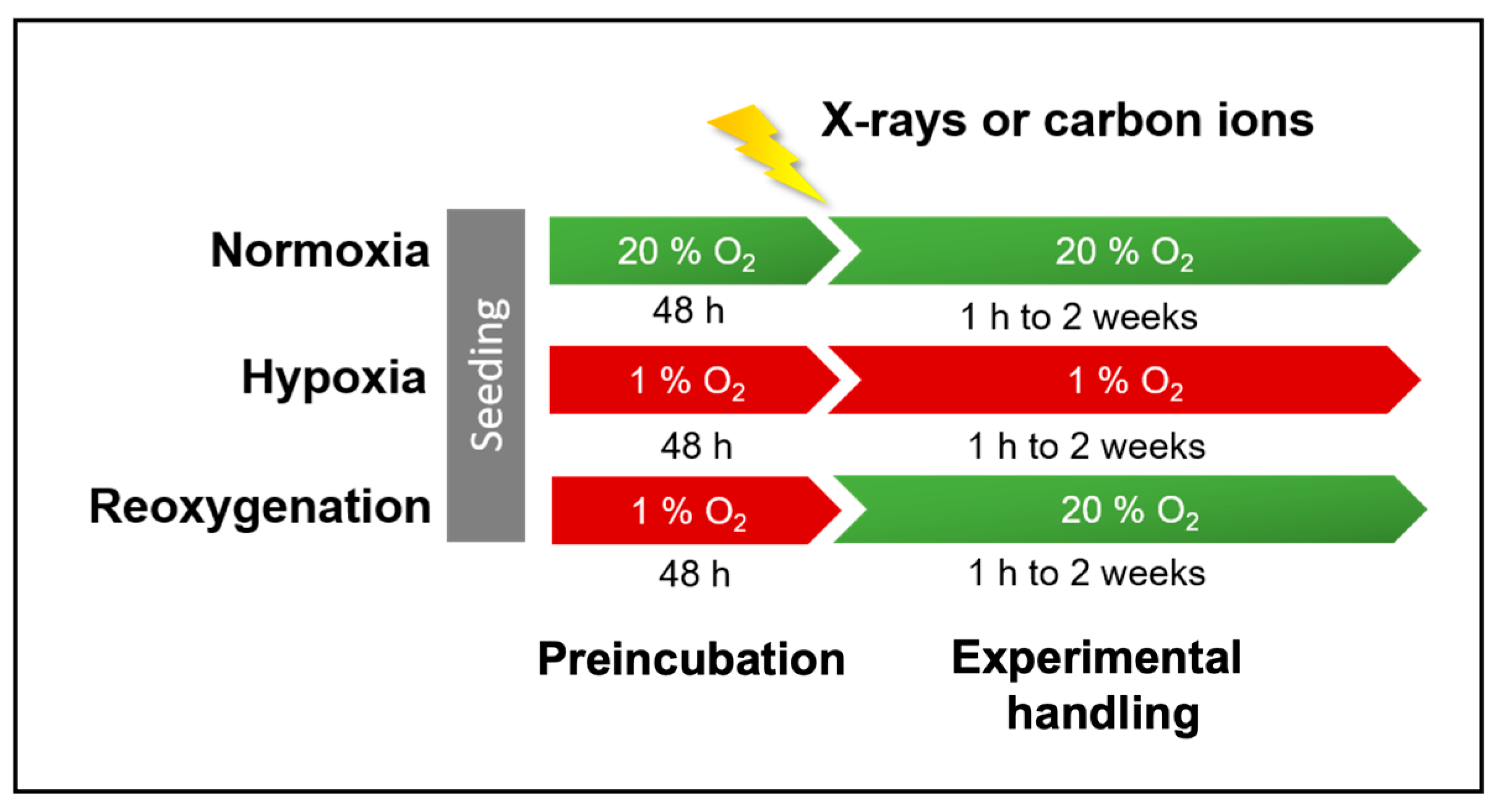
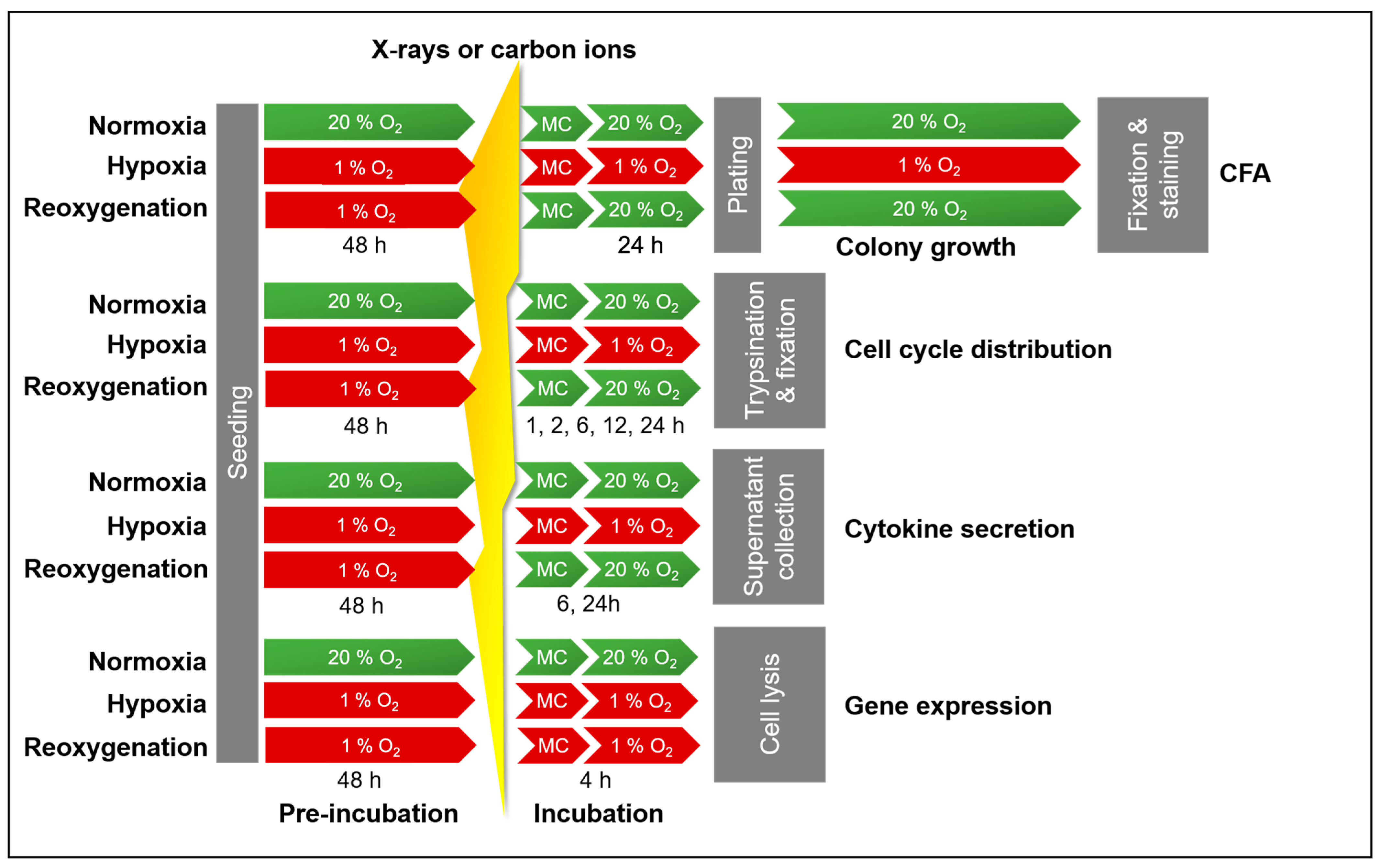
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Irradiation
4.3. Cell Survival Analysis Following Irradiation Under Normoxia and Hypoxia
4.4. Analysis of Cell Cycle Response Following X-Ray Exposure Under Normoxia and Hypoxia
4.5. Quantification of Cytokine Secretion Following X-Ray Exposure Under Normoxia and Hypoxia
4.6. Gene Expression Analysis Following X-Ray Exposure Under Normoxia and Hypoxia
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| α-MEM | Alpha-Minimally Essential Medium |
| ANOVA | Analysis of Variance |
| ARE | Antioxidative Response Element |
| CFA | Colony-Forming Ability |
| DAPI | 4′,6-Diamidino-2-Phenylindole |
| DSMZ | German Collection of Microorganisms and Cell Cultures GmbH |
| ECM | Extracellular Matrix |
| EDTA | Ethylene Diamine Tetra Acetic Acid |
| EGFR | Epidermal Growth Factor Receptor |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| EMT | Epithelial–Mesenchymal Transition |
| FBS | Fetal Bovine Serum |
| FDR | false discovery rate |
| FUCCI | Fluorescent Ubiquitination-based Cell Cycle Indicator |
| HIFs | hypoxia-inducible factors |
| hTERT | human telomerase reverse transcriptase |
| LET | Linear Energy Transfer |
| NF-κB | Nuclear Factor κB |
| NSCLC | Non-Small Cell Lung Cancer |
| OER | Oxygen Enhancement Ratio |
| PBS | Phosphate-buffered saline |
| PMMA | Polymethyl methacrylate |
| RBE | Relative Biological Effectiveness |
| RNA | Ribonucleic acid |
| ROS | Reactive Oxygen Species |
| RPE-1 | Human Retinal Pigment Epithelial-1 |
| RIN | RNA Integrity Numbers |
| TGF-β | transforming growth factor β |
| TMB | 3,3′,5,5′-Tetramethylbenzidine |
| TME | Tumor Microenvironment |
Appendix A
Appendix A.1. Cell Survival—Characteristics
| Radiation Quality | Plating Time | D10 (Gy) (±SE) | p-Values | ||||
|---|---|---|---|---|---|---|---|
| Normoxia (N) | Hypoxia (H) | Reoxygenation (R: H→N) | Normoxia vs. Hypoxia | Normoxia vs. Reoxygenation | Reoxygenation vs. Hypoxia | ||
| X-rays | LP | 4.16 ± 0.40 | 5.47 ± 0.27 | 5.52 ± 0.26 | 0.039 | 0.029 | ns |
| Carbon ions | LP | 1.89 ± 0.25 | 1.76 ± 0.38 | 1.79 ± 0.14 | ns | ns | ns |
| Radiation Quality | Type of Assay | RBE (±SE) | p-Values | ||||
|---|---|---|---|---|---|---|---|
| Normoxia (N) | Hypoxia (H) | Reoxygenation (R: H→N) | N vs. H | N vs. R | R vs. H | ||
| Carbon ions compared to X-rays | LP | 2.27 ± 0.18 | 3.49 ± 0.46 | 3.13 ± 0.15 | 0.002 | 0.039 | ns |
| Radiation Quality | Type of Assay | OER (±SE) | p-Values | |
|---|---|---|---|---|
| Hypoxia (H) | Reoxygenation (R: H→N) | H vs. R | ||
| X-rays | Late plating | 1.34 a ± 0.08 | 1.35 b ± 0.08 | ns |
| Carbon ions | Late plating | 0.96 ± 0.12 | 0.97 ± 0.07 | ns |
Appendix A.2. Cell Cycle Progression

Appendix A.3. Cytokine Secretion
Appendix A.4. Differential Gene Expression
| NCBI Gene Symbol | Log2 Fold Change | NCBI Gene Name | Log2 Fold Change | ||
|---|---|---|---|---|---|
| H0 1 vs. N0 | R0 vs. N0 | H0 1 vs. N0 | R0 vs. N0 | ||
| BMPR2 | 1.12 | 0.16 | IL1A | 2.63 | 0.93 |
| BNC1 | 1.15 | 0.31 | IL1B | 1.80 | 0.43 |
| CD274 | 0.91 | 1.03 | IL6 | 1.23 | 0.67 |
| CHRM1 | −3.02 | −0.01 | IL6R | −1.36 | −0.36 |
| CSF1 | −1.08 | −0.39 | KITLG | −1.10 | −0.19 |
| CTF1 | 1.23 | 0.42 | TNFSF9 | −1.09 | −0.10 |
| EHF | −1.40 | −0.01 | VEGFA | 1.00 | 0.19 |
| FGF18 | −1.05 | −0.86 | |||
| NCBI Gene Symbol | Log2 Fold Change | |||
|---|---|---|---|---|
| H8 1 vs. N8 | R8 vs. N8 | |||
| X-Rays | Carbon Ions | X-Rays | Carbon Ions | |
| ADRA1D | −1.03 | −1.19 | −0.32 | −0.78 |
| ALOX12 | −1.24 | −0.30 | −0.21 | −0.06 |
| BMPR2 | 1.25 | 1.15 | 0.04 | 0.29 |
| BNC1 | 1.87 | 1.04 | 0.49 | 0.96 |
| CD274 | 1.11 | 1.05 | 0.59 | 1.56 |
| CHRM1 | −5.36 | −3.60 | −0.99 | −2.92 |
| CTF1 | 1.81 | 1.09 | 1.49 | −0.02 |
| DLEC1 | −1.19 | 0.10 | −0.02 | −0.25 |
| ETS1 | 1.05 | 0.76 | 0.30 | 0.56 |
| FABP6 | 0.13 | 1.17 | −0.52 | 1.20 |
| FIGF | −1.38 | −0.79 | −0.37 | −0.23 |
| IGFBP7 | 0.69 | 1.29 | 0.09 | 1.09 |
| IL1A | 2.81 | 3.46 | 1.13 | 2.40 |
| IL1B | 2.58 | 0.05 | 0.96 | N/A |
| IL31RA | 1.08 | 0.58 | 0.22 | 0.32 |
| IL6 | 1.18 | 1.28 | 0.50 | 0.96 |
| IL6R | −1.18 | −1.05 | −0.24 | −0.51 |
| LMO1 | 0.40 | 1.56 | 0.00 | 1.17 |
| PTK2B | 1.01 | 0.31 | 0.25 | 0.13 |
| SPOCK1 | 1.05 | 1.06 | 0.30 | 0.81 |
| SSTR5 | −1.64 | −1.36 | 0.04 | −1.12 |
| TNFSF9 | −1.05 | −0.95 | −0.16 | −0.58 |
| VEGFA | 1.09 | 1.04 | 0.10 | −0.09 |
| NCBI Gene Symbol | Log2 Fold Change | NCBI Gene Name | Log2 Fold Change | ||
|---|---|---|---|---|---|
| H0 1 vs. N0 | R0 vs. N0 | H0 1 vs. N0 | R0 vs. N0 | ||
| COL1A1 | 1.80 | 0.35 | MYL9 | 1.18 | 0.67 |
| COL5A1 | 1.43 | 0.54 | PMEPA1 | 1.18 | 0.17 |
| COL6A3 | 1.13 | 0.35 | SERPINE1 | 1.64 | 0.43 |
| IGFBP3 | 1.73 | 0.75 | SNAI2 | 1.04 | 0.60 |
| IL6 | 1.23 | 0.67 | TAGLN | 1.21 | 0.56 |
| CXCL8 | 1.31 | 0.84 | TGFBR3 | −1.52 | −0.32 |
| INHBA | 1.41 | 0.16 | TIMP3 | 1.35 | 0.70 |
| LAMC2 | 1.64 | 0.44 | VEGFA | 1.00 | 0.19 |
| LRRC15 | 1.58 | 0.32 | |||
| NCBI Gene Symbol | Log2 Fold Change | |||
|---|---|---|---|---|
| H8 1 vs. N8 | R8 vs. N8 | |||
| X-Rays | Carbon Ions | X-Rays | Carbon Ions | |
| ABI3BP | 0.41 | 1.21 | 0.11 | 1.09 |
| CDH6 | 1.33 | 0.88 | 0.42 | 0.10 |
| COL1A1 | 1.75 | 2.36 | 0.32 | 0.89 |
| COL4A1 | 0.89 | 1.02 | 0.03 | 0.54 |
| COL5A1 | 1.51 | 1.95 | 0.24 | 1.35 |
| CCN2 | 1.30 | 1.18 | -0.16 | 0.19 |
| CTHRC1 | 0.11 | 1.37 | -0.29 | 0.80 |
| ENO2 | 0.56 | 1.08 | 0.01 | 0.51 |
| FBLN2 | 0.40 | 1.07 | -0.10 | 0.72 |
| FSTL1 | 0.91 | 1.01 | 0.09 | 0.74 |
| GADD45B | 0.93 | 1.19 | 0.21 | 0.66 |
| IGFBP3 | 1.36 | 2.82 | 0.32 | 1.95 |
| IL32 | 0.72 | 1.40 | 0.12 | 1.45 |
| IL6 | 1.18 | 1.28 | 0.50 | 0.96 |
| CXCL8 | 1.49 | 1.41 | 0.65 | 1.27 |
| INHBA | 1.55 | 1.55 | 0.24 | 0.74 |
| ITGA2 | 1.61 | 0.69 | 0.25 | 0.50 |
| LAMC2 | 2.16 | 1.42 | 0.37 | 1.03 |
| LOX | 0.37 | 1.07 | 0.08 | 0.37 |
| LOXL1 | 0.53 | 1.25 | 0.14 | 1.08 |
| LOXL2 | 0.70 | 1.18 | 0.18 | 0.87 |
| LRRC15 | 0.97 | 2.81 | -0.08 | 1.55 |
| MATN3 | 0.54 | 1.09 | 0.25 | 0.89 |
| MYL9 | 0.73 | 1.87 | 0.12 | 1.59 |
| PMEPA1 | 1.25 | 1.25 | 0.09 | 0.40 |
| SCG2 | −0.53 | −0.99 | −0.03 | −1.07 |
| SERPINE1 | 1.84 | 1.91 | 0.29 | 0.98 |
| SNAI2 | 1.03 | 0.95 | 0.44 | 0.72 |
| SPOCK1 | 1.05 | 1.06 | 0.30 | 0.81 |
| TAGLN | 1.15 | 2.02 | 0.04 | 1.71 |
| TGFBI | 0.88 | 1.36 | 0.02 | 0.81 |
| TGFBR3 | −1.19 | −1.36 | −0.17 | −0.62 |
| TPM1 | 0.44 | 1.22 | −0.02 | 0.98 |
| TIMP3 | 1.47 | 2.26 | 0.19 | 1.30 |
| VEGFA | 1.09 | 1.04 | 0.10 | −0.09 |
| WNT5A | −0.42 | −1.10 | 0.06 | −0.65 |
| NCBI Gene Symbol | Log2 Fold Change | |||||
|---|---|---|---|---|---|---|
| N8 1 vs. N0 | H8 vs. H0 | R8 vs. R0 | ||||
| X-Rays | Carbon Ions | X-Rays | Carbon Ions | X-Rays | Carbon Ions | |
| BTG2 | 2.70 | 1.97 | 2.71 | 1.83 | 2.74 | 1.88 |
| CDKN1A | 2.63 | 2.61 | 2.62 | 2.38 | 2.56 | 2.50 |
| IL1A | 1.37 | 0.43 | 1.54 | 1.26 | 1.57 | 1.90 |
| KITLG | 1.01 | 1.36 | 1.38 | 1.46 | 1.06 | 1.27 |
| MDM2 | 2.47 | 2.14 | 2.65 | 2.16 | 2.37 | 2.12 |
| PGF | 2.46 | 2.59 | 2.65 | 3.05 | 2.50 | 2.94 |
| PLK1 | −1.73 | −1.59 | −2.01 | −1.61 | −1.57 | −1.65 |
| PPM1D | 2.03 | 1.47 | 1.90 | 1.23 | 1.91 | 1.25 |
| SESN1 | 2.31 | 2.06 | 2.32 | 1.95 | 2.33 | 2.07 |
| SYK | 1.40 | 1.58 | 1.06 | 1.26 | 1.37 | 0.87 |
| TNFSF9 | 1.53 | 1.39 | 1.57 | 1.52 | 1.47 | 0.91 |
| SSTR5 | 1.38 | 1.62 | 0.64 | 1.16 | 1.64 | 0.72 |
| BUB1 | −0.95 | −0.81 | −1.04 | −0.99 | −0.79 | −0.97 |
| CD70 | 0.70 | 1.06 | 0.91 | 1.10 | 0.77 | 1.13 |
| CDC25C | −1.03 | −0.62 | −1.03 | −0.69 | −0.77 | −0.75 |
| CDKN2C | −0.76 | −0.98 | −1.08 | −0.66 | −0.83 | −0.88 |
| DLEC1 | 0.11 | 0.47 | −0.22 | 1.44 | 0.19 | 0.33 |
| EBI3 | 0.60 | 1.04 | 0.81 | 1.45 | 0.16 | 1.02 |
| ICOSLG | 0.79 | 0.97 | 1.17 | 0.88 | 1.08 | 0.83 |
| KIF2C | −1.27 | −0.98 | −1.23 | −0.86 | −1.16 | −1.04 |
| MKI67 | −1.00 | −0.68 | −0.89 | −0.71 | −0.98 | −0.72 |
| MXD1 | 0.47 | 1.00 | 0.64 | 0.96 | 0.40 | 0.78 |
| PDGFA | 1.00 | 0.94 | 1.12 | 0.45 | 0.99 | 0.77 |
| POU3F2 | 1.98 | 0.69 | 2.89 | 0.93 | 0.95 | 0.26 |
| RACGAP1 | −1.18 | −0.93 | −1.11 | −0.86 | −1.06 | −0.82 |
| SERTAD1 | 1.08 | 0.72 | 0.83 | 0.50 | 1.01 | 0.54 |
| TPX2 | −1.01 | −0.87 | −0.99 | −0.87 | −0.91 | −0.78 |
| TTK | −1.05 | −0.74 | −0.99 | −0.74 | −0.89 | −0.65 |
| NCBI Gene Symbol | Log2 Fold Change | |||||
|---|---|---|---|---|---|---|
| N8 1 vs. N0 | H8 vs. H0 | R8 vs. R0 | ||||
| X-Rays | Carbon Ions | X-Rays | Carbon Ions | X-Rays | Carbon Ions | |
| ACTA2 | 0.98 | 1.55 | 1.74 | 2.23 | 0.99 | 2.17 |
| FAS | 2.06 | 2.31 | 2.75 | 2.85 | 2.16 | 2.64 |
| GADD45A | 1.11 | 0.77 | 0.95 | 0.55 | 1.02 | 0.91 |
| PDGFRB | 0.90 | 1.04 | 1.21 | 1.96 | 1.30 | 1.81 |
Appendix B
| NCBI Gene Symbol | Name | Primary Function (NCBI Gene Summary) |
|---|---|---|
| ABI3BP | ABI Family Member 3 Binding Protein | Enables actin filament binding activity. Predicted to be involved in extracellular matrix organization, positive regulation of cell–substrate adhesion, and regulation of post-synapse organization. |
| ACTA2 | Actin Alpha 2 | Encodes one of six actin proteins. Actins are involved in cell motility, structure, integrity, and intercellular signaling. The encoded protein is a smooth muscle actin that is involved in vascular contractility and blood pressure homeostasis. |
| ADRA1D | Adrenoceptor Alpha 1D | Alpha-1-adrenergic receptors (alpha-1-ARs) are members of the G protein-coupled receptor superfamily. They activate mitogenic responses and regulate growth and proliferation of many cells. |
| ALDOC | Aldolase, Fructose-Bisphosphate C | Encodes a member of the class I fructose-biphosphate aldolase gene family. Expressed specifically in the brain. It is a glycolytic enzyme that catalyzes the reversible cleavage of fructose-1,6-biphosphate and fructose 1-phosphate to dihydroxyacetone phosphate and either glyceraldehyde-3-phosphate or glyceraldehyde, respectively. |
| ALOX12 | Arachidonate 12-Lipoxygenase, 12S Type | Encodes a member of the lipoxygenase protein family. It acts on different polyunsaturated fatty acid substrates to generate bioactive lipid mediators, including eicosanoids and lipoxins. It has been shown to regulate platelet function. |
| BMPR2 | Bone Morphogenetic Protein Receptor Type 2 | Encodes a member of the bone morphogenetic protein (BMP) receptor family of transmembrane serine/threonine kinases. The ligands of this receptor are members of the TGF-beta superfamily. BMPs are involved in endochondral bone formation and embryogenesis. |
| BNC1 | Basonuclin Zinc Finger Protein 1 | Encodes a zinc finger protein; thought to play a regulatory role in keratinocyte proliferation, and it may also be a regulator for rRNA transcription. Disruption of this gene has been implicated in premature ovarian failure as well as premature testicular aging. |
| BPGM | Bisphosphoglycerate Mutase | Encodes an enzyme that catalyzes 2,3-DPG synthesis via its synthetase activity and 2,3-DPG degradation via its phosphatase activity. Deficiency of this enzyme increases the affinity of cells for oxygen. Multiple alternatively spliced variants, encoding the same protein, have been identified. |
| BTG2 | BTG Anti-Proliferation Factor 2 | Encodes a member of the BTG/Tob family. This family has anti-proliferative properties. The encoded protein is involved in the regulation of the G1/S transition of the cell cycle. |
| BUB1 | BUB1 Mitotic Checkpoint Serine/Threonine Kinase | Encodes a serine/threonine kinase that functions by activating the spindle checkpoint. This protein also plays a role in inhibiting the activation of the anaphase-promoting complex/cyclosome. It may also function in the DNA damage response. Mutations have been associated with aneuploidy and several forms of cancer. |
| CCN2 | Cellular Communication Network Factor 2 | Encodes a mitogen that is secreted by vascular endothelial cells. Plays a role in chondrocyte proliferation and differentiation and cell adhesion in many cell types and is related to platelet-derived growth factor. |
| CD274 | CD274 Molecule | Encodes an immune inhibitory receptor ligand that is expressed by hematopoietic and non-hematopoietic cells, such as T cells and B cells and various types of tumor cells. Interaction of this ligand with its receptor inhibits T-cell activation and cytokine production. In tumor microenvironments, this interaction provides an immune escape for tumor cells through cytotoxic T-cell inactivation. |
| CD70 | CD70 Molecule | Encodes a cytokine that belongs to the tumor necrosis factor (TNF) ligand family. It is a ligand for TNFRSF27/CD27. It induces proliferation of co-stimulated T cells, enhances the generation of cytolytic T cells, and contributes to T cell activation. This cytokine is also reported to play a role in regulating B-cell activation, the cytotoxic function of natural killer cells, and immunoglobulin synthesis. |
| CDC25C | Cell Division Cycle 25C | Encodes a protein that directs dephosphorylation of cyclin B-bound CDC2 and triggers entry into mitosis. It also suppresses p53-induced growth arrest. |
| CDH6 | Cadherin 6 | Encodes a type II cadherin. Cadherins are membrane glycoproteins that mediate homophilic cell–cell adhesion and play critical roles in cell differentiation and morphogenesis. It may play a role in kidney development as well as endometrium and placenta formation. Decreased expression may be associated with tumor growth and metastasis. |
| CDKN1A | Cyclin-Dependent Kinase Inhibitor 1A | Encodes a potent cyclin-dependent kinase inhibitor, which binds to and inhibits CDK2 and CDK4 complexes, and thus functions as a regulator of cell cycle progression at G1. The expression of this gene is tightly controlled by the tumor suppressor protein p53, through which this protein mediates the p53-dependent cell cycle G1 phase arrest in response to a variety of stress stimuli. This protein can interact with PCNA, a DNA polymerase accessory factor, and plays a regulatory role in DNA replication and DNA damage repair. CDKN1A was reported to be cleaved by CASP3-like caspases, which leads to activation of CDK2 and may be instrumental in the execution of apoptosis. |
| CDKN2C | Cyclin-Dependent Kinase Inhibitor 2C | Encodes a member of the INK4 family of cyclin-dependent kinase inhibitors. It binds to and inhibits CDK4 or CDK6, thus regulating cell growth by controlling cell cycle G1 progression. Ectopic expression of this gene was shown to suppress the growth of human cells in a manner that appears to correlate with the presence of a wild-type RB1 function. It may have a role in regulating spermatogenesis, as well as in suppressing tumorigenesis. |
| CHRM1 | Cholinergic Receptor Muscarinic 1 | Involved in the mediation of bronchoconstriction and in the acid secretion of the gastrointestinal tract. The functional diversity of such receptors includes cellular responses such as adenylate cyclase inhibition, phosphoinositide degeneration, and potassium channel mediation. |
| COL1A1 | Collagen Type I Alpha 1 Chain | Encodes the pro-alpha1 chains of type I collagen found in most connective tissues. Reciprocal translocations between chromosomes 17 and 22, where this gene and the gene for PDGF-beta are located, are associated with a skin tumor called dermatofibrosarcoma protuberans, resulting from unregulated expression of the growth factor. |
| COL4A1 | Collagen Type IV Alpha 1 Chain | Encodes a type IV collagen alpha protein, an integral component of basement membranes. The protein interacts with other extracellular matrix components such as perlecans, proteoglycans, and laminins. In addition, proteolytic cleavage of the non-collagenous carboxy-terminal domain results in a biologically active fragment known as arresten, which has anti-angiogenic and tumor suppressor properties. |
| COL5A1 | Collagen Type V Alpha 1 Chain | Encodes an alpha chain for one of the low-abundance fibrillar collagens. Type V collagen is found in tissues containing type I collagen and appears to regulate the assembly of heterotypic fibers composed of both type I and type V collagen. This gene product is closely related to type XI collagen. |
| COL6A3 | Collagen Type VI Alpha 3 Chain | Encodes the alpha-3 chain of type VI collagen, a beaded filament collagen found in most connective tissues. Its domains have been shown to bind extracellular matrix proteins, an interaction that explains the importance of this collagen in organizing matrix components. |
| CSF1 | Colony-Stimulating Factor 1 | Encodes a cytokine that controls the production, differentiation, and function of macrophages. |
| CTF1 | Cardiotrophin 1 | Encodes a secreted cytokine that induces cardiac myocyte hypertrophy in vitro. |
| CTHRC1 | Collagen Triple Helix Repeat-Containing 1 | Encodes a protein that may play a role in the cellular response to arterial injury through involvement in vascular remodeling. |
| CXCL2 | C-X-C Motif Chemokine Ligand 2 | Encodes a secreted CXC-chemokine involved in immunoregulatory and inflammatory processes. It may suppress hematopoietic progenitor cell proliferation. |
| CXCL8 | C-X-C Motif Chemokine Ligand 8 | Encodes a CXC-chemokine family and is a major mediator of the inflammatory response. It is secreted by mononuclear macrophages, neutrophils, eosinophils, T lymphocytes, epithelial cells, and fibroblasts. It functions as a chemotactic factor for neutrophils. Plays a role in the pathogenesis of the lower respiratory tract infection bronchiolitis, a common respiratory tract disease. The overproduction of this pro-inflammatory protein is thought to cause the lung inflammation associated with cystic fibrosis. This protein is also secreted by tumor cells and promotes tumor migration, invasion, angiogenesis, and metastasis. It is also a potent angiogenic factor. |
| DLEC1 | DLEC1 Cilia and Flagella-Associated Protein | It is located in a region that is commonly deleted in a variety of malignancies. Down-regulation of this gene has been observed in several human cancers, including lung cancer. |
| EBI3 | Epstein–Barr Virus-Induced 3 | Encodes a secreted glycoprotein belonging to the hematopoietin receptor family and heterodimerizes with a 28 kDa protein to form interleukin 27 (IL-27). IL-27 regulates T cell and inflammatory responses. |
| EHF | ETS Homologous Factor | Encodes a protein that belongs to an ETS transcription factor subfamily characterized by epithelial-specific expression (ESEs). The encoded protein acts as a transcriptional repressor and may be involved in epithelial differentiation and carcinogenesis. |
| ENO2 | Enolase 2 | Encodes one of the three enolase isoenzymes involved in glycolysis found in mammals. This isoenzyme, a homodimer, is found in mature neurons and cells of neuronal origin. |
| ETS1 | ETS Proto-Oncogene 1, Transcription Factor | This gene encodes a member of the ETS family of transcription factors and functions either as transcriptional activators or repressors of numerous genes and is involved in stem cell development, cell senescence and death, and tumorigenesis. |
| FABP6 | Fatty Acid Binding Protein 6 | Encodes the ileal fatty acid binding protein that binds long-chain fatty acids and other hydrophobic ligands. FABP’s roles include fatty acid uptake, transport, and metabolism. |
| FAS | Fas Cell Surface Death Receptor | The protein encoded by this gene is a member of the TNF-receptor superfamily. This receptor contains a death domain. It has been shown to play a central role in the physiological regulation of programmed cell death and has been implicated in the pathogenesis of various malignancies and diseases of the immune system. This receptor has also been shown to activate NF-κB, MAPK3/ERK1, and MAPK8/JNK and is found to be involved in transducing the proliferating signals in normal diploid fibroblast and T cells. |
| FBLN2 | Fibulin 2 | Encodes an extracellular matrix protein, which belongs to the fibulin family. This protein binds various extracellular ligands and calcium. It may play a role during organ development, in particular, during the differentiation of heart, skeletal, and neuronal structures. |
| FGF18 | Fibroblast Growth Factor 18 | Encodes a member of the FGF family, which possesses broad mitogenic and cell survival activities and is involved in a variety of biological processes, including embryonic development, cell growth, morphogenesis, tissue repair, tumor growth, and invasion. This protein is a pleiotropic growth factor that stimulates proliferation in a number of tissues, most notably the liver and small intestine. |
| FIGF (VEGFD) | Vascular Endothelial Growth Factor D | Encodes a member of the PDGF/VEGF family and is active in angiogenesis, lymphangiogenesis, and endothelial cell growth. It binds and activates VEGFR-2 and VEGFR-3. It is structurally and functionally similar to VEGFC. |
| FSTL1 | Follistatin Like 1 | Encodes a protein with similarity to follistatin, an activin-binding protein. It is an autoantigen associated with rheumatoid arthritis. |
| GADD45A and GADD45B | Growth Arrest and DNA Damage Inducible Alpha and Beta | They are members of a group of genes that are upregulated following stressful growth arrest conditions and treatment with DNA-damaging agents. Mediate activation of the p38/JNK pathway via MTK1/MEKK4 kinase. Their upregulation is mediated by both p53-dependent and -independent mechanisms. |
| HK2 | Hexokinase2 | Hexokinases phosphorylate glucose to produce glucose-6-phosphate, the first step in most glucose metabolism pathways. HK2 is the predominant form found in skeletal muscle. |
| ICOSLG | Inducible T Cell Costimulator Ligand | Enables identical protein binding activity. Predicted to be involved in the T cell receptor signaling pathway and positive regulation of interleukin-4 production. Located in intracellular membrane-bounded organelles and plasma membrane. |
| IGFBP3 | Insulin Like Growth Factor Binding Protein 3 | A member of the insulin-like growth factor binding protein (IGFBP) family. It circulates in the plasma, prolonging the half-life of IGFs and altering their interaction with cell surface receptors. |
| IGFBP7 | Insulin Like Growth Factor Binding Protein 7 | Encodes a member of the IGFBP family and regulates IGF availability in body fluids and tissues and modulates IGF binding to its receptors. This protein binds IGF-I and IGF-II with relatively low affinity and belongs to a subfamily of low-affinity IGFBPs. It also stimulates prostacyclin production and cell adhesion. |
| IL1A | Interleukin 1 Alpha | Encodes a member of the interleukin 1 cytokine family. It is a pleiotropic cytokine involved in various immune responses, inflammatory processes, and hematopoiesis. This cytokine is produced by monocytes and macrophages as a proprotein, which is proteolytically processed and released in response to cell injury, and thus induces apoptosis. |
| IL1B | Interleukin 1 Beta | Encodes a member of the interleukin 1 cytokine family. This cytokine is produced by activated macrophages as a proprotein, which is proteolytically processed to its active form by caspase 1 (CASP1/ICE). This cytokine is an important mediator of the inflammatory response and is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis. Patients with severe Coronavirus Disease 2019 (COVID-19) present elevated levels of pro-inflammatory cytokines such as IL-1B in bronchial alveolar lavage fluid samples. |
| IL31RA | Interleukin 31 Receptor A | Encodes a protein of the type I cytokine receptor family expressed on monocytes and is involved in IL-31 signaling via activation of STAT-3 and STAT-5. It functions either as a monomer or as part of a receptor complex with oncostatin M receptor (OSMR). |
| IL32 | Interleukin 32 | Expression of this protein is increased after the activation of T-cells by mitogens or the activation of NK cells by IL-2. This protein induces the production of TNFα from macrophage cells. |
| IL6 | Interleukin 6 | Encodes a cytokine that functions in inflammation and the maturation of B cells. It is an endogenous pyrogen. It is primarily produced at sites of acute and chronic inflammation, where it is secreted into the serum and induces a transcriptional inflammatory response through interleukin 6 receptor, alpha. Elevated levels of the encoded protein have been found in virus infections, including COVID-19. |
| IL6R | Interleukin 6 Receptor | Encodes a subunit of the interleukin 6 (IL6) receptor complex. Dysregulated production of IL6 and this receptor are implicated in the pathogenesis of many diseases, such as multiple myeloma, autoimmune diseases, and prostate cancer. |
| INHBA | Inhibin Subunit Beta A | Encodes a member of the TGF-beta superfamily of proteins. Inhibits follicle-stimulating hormone secretion. It also plays a role in eye, tooth, and testis development. Elevated expression of this gene may be associated with cancer cachexia in human patients. |
| ITGA2 | Integrin Subunit Alpha 2 | Encodes the alpha subunit of a transmembrane receptor for collagens and related proteins. The encoded protein forms a heterodimer with a beta subunit and mediates the adhesion of platelets and other cell types to the extracellular matrix. |
| KIF2C | Kinesin Family Member 2C | Encodes a kinesin-like protein that functions as a microtubule-dependent molecular motor. The encoded protein can depolymerize microtubules at the plus end, thereby promoting mitotic chromosome segregation. |
| KITLG | KIT Ligand | Encodes the ligand of the tyrosine-kinase receptor encoded by the KIT locus. This ligand is a pleiotropic factor that acts in utero in germ cell and neural cell development and hematopoiesis, all believed to reflect a role in cell migration. |
| LAMC2 | Laminin Subunit Gamma 2 | This gene encodes the gamma chain isoform laminin, gamma 2. It is expressed in several fetal tissues but differently from gamma 1 and is specifically localized to epithelial cells in skin, lung, and kidney. The gamma 2 chain together with the alpha 3 and beta 3 chains constitute laminin 5 (earlier known as kalinin), which is an integral part of the anchoring filaments that connect epithelial cells to the underlying basement membrane. Laminins are the major non-collagenous constituent of basement membranes. They are implicated in a wide variety of biological processes, including cell adhesion, differentiation, migration, signaling, neurite outgrowth, and metastasis. |
| LDHA | Lactate Dehydrogenase A | Encodes the A subunit of LDH, which catalyzes the reversible conversion of pyruvate to lactate with the concomitant oxidation of NADH to NAD in anaerobic glycolysis. |
| LMO1 | LIM Domain Only 1 | This locus encodes a transcriptional regulator that contains two cysteine-rich LIM domains but lacks a DNA-binding domain. LIM domains may play a role in protein interactions; thus, the encoded protein may regulate transcription by competitively binding to specific DNA-binding transcription factors. |
| LOX | Lysyl Oxidase | This gene encodes a member of the lysyl oxidase family of proteins. The copper-dependent amine oxidase activity of this enzyme functions in the crosslinking of collagens and elastin. |
| LOXL1 | Lysyl Oxidase Like 1 | This gene encodes a member of the lysyl oxidase family of proteins. The prototypic member of the family is essential to the biogenesis of connective tissue, encoding an extracellular copper-dependent amine oxidase that catalyzes the first step in the formation of crosslinks in collagen and elastin. The encoded preproprotein is proteolytically processed to generate the mature enzyme. |
| LOXL2 | Lysyl Oxidase Like 2 | This gene encodes a member of the lysyl oxidase gene family. The prototypic member of the family is essential to the biogenesis of connective tissue, encoding an extracellular copper-dependent amine oxidase that catalyzes the first step in the formation of crosslinks in collagens and elastin. |
| LRRC15 | Leucine-Rich Repeat Containing 15 | Enables several functions, including fibronectin binding activity, laminin binding activity, and protein sequestering activity. Involved in several processes, including negative regulation of protein localization to the plasma membrane; negative regulation of viral entry into the host cell; and receptor-mediated virion attachment to host cell. Located in the collagen-containing extracellular matrix and the plasma membrane. It is active in the apical plasma membrane. |
| MATN3 | Matrilin 3 | This gene encodes a member of the Von Willebrand factor A domain-containing protein family. This family of proteins is thought to be involved in the formation of filamentous networks in the extracellular matrices of various tissues. This protein contains two von Willebrand factor A domains; it is present in the cartilage extracellular matrix and has a role in the development and homeostasis of cartilage and bone. Mutations in this gene result in multiple epiphyseal dysplasia. |
| MDM2 | MDM2 Proto-Oncogene | This gene encodes a nuclear-localized E3 ubiquitin ligase. The encoded protein can promote tumor formation by targeting tumor suppressor proteins, such as p53, for proteasomal degradation. This gene is itself transcriptionally regulated by p53. Overexpression or amplification of this locus is detected in a variety of different cancers. There is a pseudogene for this gene on chromosome 2. Alternative splicing results in a multitude of transcript variants, many of which may be expressed only in tumor cells. |
| MKI67 | Marker of Proliferation Ki-67 | Enables RNA binding activity. Involved in regulation of chromosome segregation and regulation of mitotic nuclear division. Located in condensed chromosomes, nuclear bodies, and nucleoli. Implicated in several diseases, including Crohn’s disease, colorectal cancer, endocrine gland cancer (multiple), graft-versus-host disease, and human immunodeficiency virus infectious disease. Biomarker of several diseases, including Barrett’s esophagus; autoimmune disease of the musculoskeletal system (multiple); endocrine gland cancer (multiple); gastrointestinal system cancer (multiple); and lung cancer (multiple). |
| MXD1 | MAX Dimerization Protein 1 | This gene encodes a member of the MYC/MAX/MAD network of basic helix-loop-helix leucine zipper transcription factors. The MYC/MAX/MAD transcription factors mediate cellular proliferation, differentiation, and apoptosis. The encoded protein antagonizes MYC-mediated transcriptional activation of target genes by competing for the binding partner MAX and recruiting repressor complexes containing histone deacetylases. Mutations in this gene may play a role in acute leukemia, and the encoded protein is a potential tumor suppressor. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. |
| MYL9 | Myosin Light Chain 9 | Myosin, a structural component of muscle, consists of two heavy chains and four light chains. The protein encoded by this gene is a myosin light chain that may regulate muscle contraction by modulating the ATPase activity of myosin heads. The encoded protein binds calcium and is activated by myosin light chain kinase. Two transcript variants encoding different isoforms have been found for this gene. |
| PDGFA | Platelet-Derived Growth Factor Subunit A | This gene encodes a member of the protein family comprising both platelet-derived growth factors (PDGF) and vascular endothelial growth factors (VEGF). The encoded preproprotein is proteolytically processed to generate platelet-derived growth factor subunit A, which can homodimerize or, alternatively, heterodimerize with the related platelet-derived growth factor subunit B. These proteins bind and activate PDGF receptor tyrosine kinases, which play a role in a wide range of developmental processes. Alternative splicing results in multiple transcript variants. |
| PDGFRB | Platelet-Derived Growth Factor Receptor Beta | The protein encoded by this gene is a cell surface tyrosine kinase receptor for members of the platelet-derived growth factor family. These growth factors are mitogens for cells of mesenchymal origin. The identity of the growth factor bound to a receptor monomer determines whether the functional receptor is a homodimer (PDGFB or PDGFD) or a heterodimer (PDGFA and PDGFB). This gene is essential for normal development of the cardiovascular system and aids in rearrangement of the actin cytoskeleton. This gene is flanked on chromosome 5 by the genes for granulocyte-macrophage colony-stimulating factor and macrophage-colony stimulating factor receptor; all three genes may be implicated in the 5-q syndrome. A translocation between chromosomes 5 and 12 that fuses this gene to that of the ETV6 gene results in chronic myeloproliferative disorder with eosinophilia. |
| PGF | Placental Growth Factor | Enables growth factor activity. Involved in positive regulation of cell population proliferation. Predicted to be located in the extracellular region. Predicted to be active in the extracellular space. Implicated in several diseases, including brain ischemia, diabetic neuropathy, glioblastoma, myocardial infarction, and pancreatic endocrine carcinoma. Biomarker of several diseases, including artery disease (multiple), autoimmune disease of musculoskeletal system (multiple), epilepsy (multiple), limited scleroderma, and pancreatic endocrine carcinoma. |
| PGK1 | Phosphoglycerate Kinase 1 | The protein encoded by this gene is a glycolytic enzyme that catalyzes the conversion of 1,3-diphosphoglycerate to 3-phosphoglycerate. The encoded protein may also act as a cofactor for polymerase alpha. Additionally, this protein is secreted by tumor cells, where it participates in angiogenesis by functioning to reduce disulfide bonds in the serine protease, plasmin, which consequently leads to the release of the tumor blood vessel inhibitor angiostatin. The encoded protein has been identified as a moonlighting protein based on its ability to perform mechanistically distinct functions. Deficiency of the enzyme is associated with a wide range of clinical phenotypes, including hemolytic anemia and neurological impairment. Pseudogenes of this gene have been defined on chromosomes 19, 21, and the X chromosome. |
| PLK1 | Polo Like Kinase 1 | The Ser/Thr protein kinase encoded by this gene belongs to the CDC5/Polo subfamily. It is highly expressed during mitosis, and elevated levels are found in many different types of cancer. Depletion of this protein in cancer cells dramatically inhibited cell proliferation and induced apoptosis; hence, it is a target for cancer therapy. |
| PMEPA1 | Prostate Transmembrane Protein, Androgen Induced 1 | This gene encodes a transmembrane protein that contains a Smad interacting motif (SIM). Expression of this gene is induced by androgens and transforming growth factor beta, and the encoded protein suppresses the androgen receptor and transforming growth factor beta signaling pathways through interactions with Smad proteins. Overexpression of this gene may play a role in multiple types of cancer. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. |
| POU3F2 | POU Class 3 Homeobox 2 | This gene encodes a member of the POU-III class of neural transcription factors. The encoded protein is involved in neuronal differentiation and enhances the activation of corticotropin-releasing hormone-regulated genes. Overexpression of this protein is associated with an increase in the proliferation of melanoma cells. |
| PPM1D | Protein Phosphatase, Mg2+/Mn2+ Dependent 1D | The protein encoded by this gene is a member of the PP2C family of Ser/Thr protein phosphatases. PP2C family members are known to be negative regulators of cell stress response pathways. The expression of this gene is induced in a p53-dependent manner in response to various environmental stresses. While being induced by tumor suppressor protein TP53/p53, this phosphatase negatively regulates the activity of p38 MAP kinase, MAPK/p38, through which it reduces the phosphorylation of p53, and in turn suppresses p53-mediated transcription and apoptosis. This phosphatase thus mediates a feedback regulation of p38-p53 signaling that contributes to growth inhibition and the suppression of stress-induced apoptosis. This gene is located in a chromosomal region known to be amplified in breast cancer. The amplification of this gene has been detected in both breast cancer cell lines and primary breast tumors, which suggests a role of this gene in cancer development. |
| PTK2B | Protein Tyrosine Kinase 2 Beta | This gene encodes a cytoplasmic protein tyrosine kinase, which is involved in calcium-induced regulation of ion channels and activation of the map kinase signaling pathway. The encoded protein may represent an important signaling intermediate between neuropeptide-activated receptors or neurotransmitters that increase calcium flux and the downstream signals that regulate neuronal activity. The encoded protein undergoes rapid tyrosine phosphorylation and activation in response to increases in the intracellular calcium concentration, nicotinic acetylcholine receptor activation, membrane depolarization, or protein kinase C activation. This protein has been shown to bind CRK-associated substrate, nephrocystin, GTPase regulator associated with FAK, and the SH2 domain of GRB2. The encoded protein is a member of the FAK subfamily of protein tyrosine kinases but lacks significant sequence similarity to kinases from other subfamilies. Four transcript variants encoding two different isoforms have been found for this gene. |
| RACGAP1 | Rac GTPase Activating Protein 1 | This gene encodes a GTPase-activating protein (GAP) that is a component of the central spindlin complex. This protein binds activated forms of Rho GTPases and stimulates GTP hydrolysis, which results in negative regulation of Rho-mediated signals. This protein plays a regulatory role in cytokinesis, cell growth, and differentiation. Alternatively spliced transcript variants have been found for this gene. There is a pseudogene for this gene on chromosome 12. |
| SCG2 | Secretogranin II | The protein encoded by this gene is a member of the chromogranin/secretogranin family of neuroendocrine secretory proteins. Studies in rodents suggest that the full-length protein, secretogranin II, is involved in the packaging or sorting of peptide hormones and neuropeptides into secretory vesicles. The full-length protein is cleaved to produce the active peptide secretoneurin, which exerts chemotaxic effects on specific cell types, and EM66, whose function is unknown. |
| SERPINE1 | Serpin Family E Member 1 | Encodes a member of the serpin superfamily. This member is the principal inhibitor of tissue plasminogen activator (tPA) and urokinase (uPA) and hence is an inhibitor of fibrinolysis. |
| SERTAD1 | SERTA Domain Containing 1 | Acts upstream of or within negative regulation of cell growth. Located in cytoplasm and nucleus. |
| SESN1 | Sestrin 1 | This gene encodes a member of the sestrin family. Sestrins are induced by the p53 tumor suppressor protein and play a role in the cellular response to DNA damage and oxidative stress. The encoded protein mediates p53 inhibition of cell growth by activating AMP-activated protein kinase, which results in the inhibition of the mammalian target of rapamycin protein. The encoded protein also plays a critical role in antioxidant defense by regenerating overoxidized peroxiredoxins, and the expression of this gene is a potential marker for exposure to radiation. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. |
| SNAI2 | Snail Family Transcriptional Repressor 2 | This gene encodes a member of the Snail family of C2H2-type zinc finger transcription factors. The encoded protein acts as a transcriptional repressor that binds to E-box motifs and is also likely to repress E-cadherin transcription in breast carcinoma. This protein is involved in epithelial–mesenchymal transitions and has antiapoptotic activity. Mutations in this gene may be associated with sporadic cases of neural tube defects. |
| SPOCK1 | SPARC (Osteonectin), Cwcv, and Kazal-Like Domains Proteoglycan 1 | This gene encodes the protein core of a seminal plasma proteoglycan containing chondroitin- and heparan-sulfate chains. The protein’s function is unknown, although similarity to thyropin-type cysteine protease inhibitors suggests its function may be related to protease inhibition. |
| SSTR5 | Somatostatin Receptor 5 | The protein encoded by this gene is one of the SSTRs, which is a multi-pass membrane protein. The activity of this receptor is mediated by G proteins, which inhibit adenylyl cyclase, and different regions of this receptor molecule are required for the activation of different signaling pathways. |
| SYK | Spleen Associated Tyrosine Kinase | Encodes a member of the family of non-receptor-type Tyr protein kinases. This protein is widely expressed in hematopoietic cells and is involved in coupling activated immunoreceptors to downstream signaling events that mediate diverse cellular responses, including proliferation, differentiation, and phagocytosis. It is thought to be a modulator of epithelial cell growth and a potential tumor suppressor in human breast carcinomas. |
| TAGLN | Transgelin | Encodes a shape-change- and transformation-sensitive actin-binding protein that belongs to the calponin family. It is an early marker of smooth muscle differentiation. Involved in calcium-independent smooth muscle contraction. It acts as a tumor suppressor, and the loss of its expression is an early event in cell transformation and the development of some tumors. |
| TGFBI | Transforming Growth Factor Beta Induced | Encodes an RGD-containing protein that binds to type I, II, and IV collagens. The RGD motif is found in many ECM proteins modulating cell adhesion and serves as a ligand recognition sequence for several integrins. Plays a role in cell-collagen interactions. Induced by transforming growth factor-beta and acts to inhibit cell adhesion. |
| TGFBR3 | Transforming Growth Factor Beta Receptor 3 | The encoded receptor is a membrane proteoglycan that often functions as a co-receptor with other TGF-beta receptor superfamily members. Ectodomain shedding produces soluble TGFBR3, which may inhibit TGFB signaling. Decreased expression of this receptor has been observed in various cancers. |
| TIMP3 | TIMP Metallopeptidase Inhibitor 3 | The family encodes inhibitors of the MMPs. Induced in response to mitogenic stimulation. |
| TNFSF9 | TNF Superfamily Member 9 | Encodes a cytokine that belongs to the TNF ligand family that acts as a ligand for TNFRSF9/4-1BB, which is a costimulatory receptor molecule in T lymphocytes. This cytokine and its receptor are involved in the antigen presentation process and in the generation of cytotoxic T cells. It has been shown to reactivate anergic T lymphocytes in addition to promoting T lymphocyte proliferation. It is expressed in carcinoma cell lines and is thought to be involved in T cell–tumor cell interaction. |
| TPM1 | Tropomyosin 1 | A member of the tropomyosin family of actin-binding proteins involved in the contractile system of striated and smooth muscles. In non-muscle cells, it is implicated in stabilizing cytoskeleton actin filaments. |
| TPX2 | TPX2 Microtubule Nucleation Factor | Involved in activation of protein kinase activity, microtubule cytoskeleton organization, and negative regulation of microtubule depolymerization. Located in the intercellular bridge, nucleoplasm, and spindle. |
| TTK | TTK Protein Kinase | Encodes a dual specificity protein kinase associated with cell proliferation; this protein is essential for chromosome alignment at the centromere during mitosis and is required for centrosome duplication. It has been found to be a critical mitotic checkpoint protein for accurate segregation of chromosomes during mitosis. Tumorigenesis may occur when this protein fails to degrade and produces excess centrosomes, resulting in aberrant mitotic spindles. |
| VEGFA | Vascular Endothelial Growth Factor A | Encodes a member of the PDGF/VEGF growth factor family. This growth factor induces proliferation and migration of vascular endothelial cells and is essential for both physiological and pathological angiogenesis. This gene is upregulated in many known tumors, and its expression is correlated with tumor stage and progression. The levels of VEGF are increased during infection with SARS-CoV-2, thus promoting inflammation by facilitating recruitment of inflammatory cells and by increasing the level of angiopoietin II (Ang II). In turn, Ang II facilitates the elevation of VEGF, thus forming a vicious cycle in the release of inflammatory cytokines. |
| WNT5A | WNT Family Member 5 A | Encodes a secreted signaling protein that has been implicated in embryogenesis as well as in oncogenesis. It decreases proliferation, migration, invasiveness, and clonogenicity of carcinoma cells and may act as a tumor suppressor. |
References
- Thariat, J.; Hannoun-Levi, J.-M.; Sun Myint, A.; Vuong, T.; Gérard, J.-P. Past, Present, and Future of Radiotherapy for the Benefit of Patients. Nat. Rev. Clin. Oncol. 2013, 10, 52–60. [Google Scholar] [CrossRef]
- Grimes, D.R.; Partridge, M. A mechanistic investigation of the oxygen fixation hypothesis and oxygen enhancement ratio. Biomed. Phys. Eng. Express 2015, 1, 045209. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Liu, X.; Zhang, C.; Zhang, Q. Tumor Hypoxia: From Basic Knowledge to Therapeutic Implications. Semin. Cancer Biol. 2023, 88, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Ravanat, J.-L.; Douki, T. UV and Ionizing Radiations Induced DNA Damage, Differences and Similarities. Radiat. Phys. Chem. 2016, 128, 92–102. [Google Scholar] [CrossRef]
- Hirayama, R.; Uzawa, A.; Obara, M.; Takase, N.; Koda, K.; Ozaki, M.; Noguchi, M.; Matsumoto, Y.; Li, H.; Yamashita, K.; et al. Determination of the Relative Biological Effectiveness and Oxygen Enhancement Ratio for Micronuclei Formation Using High-LET Radiation in Solid Tumor Cells: An In Vitro and In Vivo Study. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 793, 41–47. [Google Scholar] [CrossRef]
- Jones, B. The Influence of Hypoxia on LET and RBE Relationships with Implications for Ultra-High Dose Rates and FLASH Modelling. Phys. Med. Biol. 2022, 67, 125011. [Google Scholar] [CrossRef]
- Lee, P.; Chandel, N.S.; Simon, M.C. Cellular Adaptation to Hypoxia through Hypoxia Inducible Factors and Beyond. Nat. Rev. Mol. Cell Biol. 2020, 21, 268–283. [Google Scholar] [CrossRef]
- Barsoum, I.B.; Smallwood, C.A.; Siemens, D.R.; Graham, C.H. A Mechanism of Hypoxia-Mediated Escape from Adaptive Immunity in Cancer Cells. Cancer Res. 2014, 74, 665–674. [Google Scholar] [CrossRef]
- Yeo, C.D.; Kang, N.; Choi, S.Y.; Kim, B.N.; Park, C.K.; Kim, J.W.; Kim, Y.K.; Kim, S.J. The Role of Hypoxia on the Acquisition of Epithelial-Mesenchymal Transition and Cancer Stemness: A Possible Link to Epigenetic Regulation. Korean J. Intern. Med. 2017, 32, 589–599. [Google Scholar] [CrossRef]
- Mitchell, G. The Rationale for Fractionation in Radiotherapy. Clin. J. Oncol. Nurs. 2013, 17, 412–417. [Google Scholar] [CrossRef]
- Hong, B.-J.; Kim, J.; Jeong, H.; Bok, S.; Kim, Y.-E.; Ahn, G.-O. Tumor Hypoxia and Reoxygenation: The Yin and Yang for Radiotherapy. Radiat. Oncol. J. 2016, 34, 239–249. [Google Scholar] [CrossRef]
- Kryston, T.B.; Georgiev, A.B.; Pissis, P.; Georgakilas, A.G. Role of Oxidative Stress and DNA Damage in Human Carcinogenesis. Mutat. Res. 2011, 711, 193–201. [Google Scholar] [CrossRef]
- Goto, T.; Kaida, A.; Miura, M. Visualizing Cell-Cycle Kinetics after Hypoxia/Reoxygenation in HeLa Cells Expressing Fluorescent Ubiquitination-Based Cell Cycle Indicator (Fucci). Exp. Cell Res. 2015, 339, 389–396. [Google Scholar] [CrossRef]
- Michiels, C.; Tellier, C.; Feron, O. Cycling Hypoxia: A Key Feature of the Tumor Microenvironment. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2016, 1866, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.-M.; Zhou, J. The Role of PI3K/AKT Signaling Pathway in Myocardial Ischemia-Reperfusion Injury. Int. Immunopharmacol. 2023, 123, 110714. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, J.; Gong, N. Role of the PI3K/Akt Signaling Pathway in Liver Ischemia Reperfusion Injury: A Narrative Review. Ann. Palliat. Med. 2022, 11, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.S.; Kwon, C.H.; Kim, J.H.; Woo, J.S.; Jung, J.S.; Kim, Y.K. Signal Transduction of MEK/ERK and PI3K/Akt Activation by Hypoxia/Reoxygenation in Renal Epithelial Cells. Eur. J. Cell Biol. 2006, 85, 1189–1199. [Google Scholar] [CrossRef]
- Nisar, H.; Labonté, F.M.; Roggan, M.D.; Schmitz, C.; Chevalier, F.; Konda, B.; Diegeler, S.; Baumstark-Khan, C.; Hellweg, C.E. Hypoxia Modulates Radiosensitivity and Response to Different Radiation Qualities in A549 Non-Small Cell Lung Cancer (NSCLC) Cells. Int. J. Mol. Sci. 2024, 25, 1010. [Google Scholar] [CrossRef]
- Nisar, H.; González, P.; Labonté, F.; Schmitz, C.; Roggan, M.; Kronenberg, J.; Konda, B.; Chevalier, F.; Hellweg, C. NF-κB in the Radiation Response of A549 Non-Small Cell Lung Cancer Cells to X-Rays and Carbon Ions Under Hypoxia. Int. J. Mol. Sci. 2024, 25, 4495. [Google Scholar] [CrossRef]
- Nisar, H.; Brauny, M.; Labonté, F.M.; Schmitz, C.; Konda, B.; Hellweg, C.E. DNA Damage and Inflammatory Response of P53 Null H358 Non-Small Cell Lung Cancer Cells to X-Ray Exposure Under Chronic Hypoxia. Int. J. Mol. Sci. 2024, 25, 12590. [Google Scholar] [CrossRef]
- Zhao, C.; Luo, S.; Shao, Q.; Li, P.; Huang, L.; Meng, L.; Cheng, H.; Zhang, A.; Gong, X. Mechanisms Underlying Radioresistance and Reversal Strategies in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2025, 26, 6559. [Google Scholar] [CrossRef] [PubMed]
- Ancel, J.; Perotin, J.-M.; Dewolf, M.; Launois, C.; Mulette, P.; Nawrocki-Raby, B.; Dalstein, V.; Gilles, C.; Deslée, G.; Polette, M.; et al. Hypoxia in Lung Cancer Management: A Translational Approach. Cancers 2021, 13, 3421. [Google Scholar] [CrossRef]
- Kabakov, A.E.; Yakimova, A.O. Hypoxia-Induced Cancer Cell Responses Driving Radioresistance of Hypoxic Tumors: Approaches to Targeting and Radiosensitizing. Cancers 2021, 13, 1102. [Google Scholar] [CrossRef]
- Hanley, R.; Pagliari, F.; Garcia-Calderón, D.; Fernandes Guerreiro, J.; Genard, G.; Jansen, J.; Nisticò, C.; Marafioti, M.G.; Tirinato, L.; Seco, J. Radio-Resistance of Hypoxic Tumors: Exploring the Effects of Oxygen and x-Ray Radiation on Non-Small Lung Cancer Cell Lines. Radiat. Oncol. 2023, 18, 81. [Google Scholar] [CrossRef]
- Morović, S. EBSCOhost|Tumor Hypoxia and Radioresistance. Available online: https://openurl.ebsco.com/contentitem/doi:10.55378%2Frv.48.2.6?sid=ebsco:plink:crawler&id=ebsco:doi:10.55378%2Frv.48.2.6 (accessed on 23 April 2025).
- Lee, S.; Ryu, H.; Son, A.; Seo, B.; Kim, J.; Jung, S.-Y.; Song, J.-Y.; Hwang, S.-G.; Ahn, J. TGF-β and Hypoxia/Reoxygenation Promote Radioresistance of A549 Lung Cancer Cells through Activation of Nrf2 and EGFR. Oxidative Med. Cell. Longev. 2016, 2016, 6823471. [Google Scholar] [CrossRef] [PubMed]
- Sokol, O.; Durante, M. Carbon Ions for Hypoxic Tumors: Are We Making the Most of Them? Cancers 2023, 15, 4494. [Google Scholar] [CrossRef]
- Subtil, F.S.B.; Wilhelm, J.; Bill, V.; Westholt, N.; Rudolph, S.; Fischer, J.; Scheel, S.; Seay, U.; Fournier, C.; Taucher-Scholz, G.; et al. Carbon Ion Radiotherapy of Human Lung Cancer Attenuates HIF-1 Signaling and Acts with Considerably Enhanced Therapeutic Efficiency. FASEB J. 2014, 28, 1412–1421. [Google Scholar] [CrossRef]
- Klein, C.; Dokic, I.; Mairani, A.; Mein, S.; Brons, S.; Häring, P.; Haberer, T.; Jäkel, O.; Zimmermann, A.; Zenke, F.; et al. Overcoming Hypoxia-Induced Tumor Radioresistance in Non-Small Cell Lung Cancer by Targeting DNA-Dependent Protein Kinase in Combination with Carbon Ion Irradiation. Radiat. Oncol. 2017, 12, 208. [Google Scholar] [CrossRef]
- Dadgar, S.; Troncoso, J.R.; Siegel, E.R.; Curry, N.M.; Griffin, R.J.; Dings, R.P.M.; Rajaram, N. Spectroscopic Investigation of Radiation-Induced Reoxygenation in Radiation-Resistant Tumors. Neoplasia 2021, 23, 49–57. [Google Scholar] [CrossRef]
- Saxena, K.; Jolly, M.K. Acute vs. Chronic vs. Cyclic Hypoxia: Their Differential Dynamics, Molecular Mechanisms, and Effects on Tumor Progression. Biomolecules 2019, 9, 339. [Google Scholar] [CrossRef] [PubMed]
- Druker, J.; Wilson, J.W.; Child, F.; Shakir, D.; Fasanya, T.; Rocha, S. Role of Hypoxia in the Control of the Cell Cycle. Int. J. Mol. Sci. 2021, 22, 4874. [Google Scholar] [CrossRef]
- Ortmann, B.; Druker, J.; Rocha, S. Cell Cycle Progression in Response to Oxygen Levels. Cell Mol. Life Sci. 2014, 71, 3569–3582. [Google Scholar] [CrossRef]
- Menegakis, A.; Klompmaker, R.; Vennin, C.; Arbusà, A.; Damen, M.; van den Broek, B.; Zips, D.; van Rheenen, J.; Krenning, L.; Medema, R.H. Resistance of Hypoxic Cells to Ionizing Radiation Is Mediated in Part via Hypoxia-Induced Quiescence. Cells 2021, 10, 610. [Google Scholar] [CrossRef]
- Kato, T.A.; Fujii, Y.; Junko, M.; Su, C.; Haskin, J.S.; Hirakawa, H.; Fujimori, A.; Wilson, P.F. Cell Cycle-Dependent Radiosensitivity of CHO DNA Repair Mutants Exposed to Accelerated Charged Particles. Biochem. Biophys. Res. Commun. 2025, 762, 151747. [Google Scholar] [CrossRef] [PubMed]
- Karger, C.P.; Peschke, P. RBE and Related Modeling in Carbon-Ion Therapy. Phys. Med. Biol. 2017, 63, 01TR02. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.J.; Giaccia, A.J. Radiosensitivity and Cell Age in the Mitotic Cycle. In Radiobiology for the Radiologist; Lippincott, Williams and Wilkins: Philadelphia, PA, USA, 2000. [Google Scholar]
- Burki, N.K.; Tetenta, S.U. Inflammatory Response to Acute Hypoxia in Humans. Pulm. Pharmacol. Ther. 2014, 27, 208–211. [Google Scholar] [CrossRef]
- Rašková, M.; Lacina, L.; Kejík, Z.; Venhauerová, A.; Skaličková, M.; Kolář, M.; Jakubek, M.; Rosel, D.; Smetana, K.; Brábek, J. The Role of IL-6 in Cancer Cell Invasiveness and Metastasis—Overview and Therapeutic Opportunities. Cells 2022, 11, 3698. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lan, T.; Zhang, C.; Zeng, C.; Hou, J.; Yang, Z.; Zhang, M.; Liu, J.; Liu, B. Reciprocal Activation between IL-6/STAT3 and NOX4/Akt Signalings Promotes Proliferation and Survival of Non-Small Cell Lung Cancer Cells. Oncotarget 2015, 6, 1031–1048. [Google Scholar] [CrossRef]
- Song, S.; Zhang, Y.; Duan, X.; Liu, C.; Du, Y.; Wang, X.; Luo, Y.; Cui, Y. HIF-1α/IL-8 Axis in Hypoxic Macrophages Promotes Esophageal Cancer Progression by Enhancing PD-L1 Expression. Cancer Gene Ther. 2023, 30, 358–367. [Google Scholar] [CrossRef]
- Mi, Y.; Mu, L.; Huang, K.; Hu, Y.; Yan, C.; Zhao, H.; Ma, C.; Li, X.; Tao, D.; Qin, J. Hypoxic Colorectal Cancer Cells Promote Metastasis of Normoxic Cancer Cells Depending on IL-8/P65 Signaling Pathway. Cell Death Dis. 2020, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Favaro, F.; Luciano-Mateo, F.; Moreno-Caceres, J.; Hernández-Madrigal, M.; Both, D.; Montironi, C.; Püschel, F.; Nadal, E.; Eldering, E.; Muñoz-Pinedo, C. TRAIL Receptors Promote Constitutive and Inducible IL-8 Secretion in Non-Small Cell Lung Carcinoma. Cell Death Dis. 2022, 13, 1046. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, N.C.; Shayakhmetov, D.M. Interleukin 1α and the Inflammatory Process. Nat. Immunol. 2016, 17, 906–913. [Google Scholar] [CrossRef]
- Korbecki, J.; Simińska, D.; Gąssowska-Dobrowolska, M.; Listos, J.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. Chronic and Cycling Hypoxia: Drivers of Cancer Chronic Inflammation through HIF-1 and NF-κB Activation: A Review of the Molecular Mechanisms. Int. J. Mol. Sci. 2021, 22, 10701. [Google Scholar] [CrossRef]
- Rattigan, Y.; Hsu, J.-M.; Mishra, P.J.; Glod, J.; Banerjee, D. Interleukin 6 Mediated Recruitment of Mesenchymal Stem Cells to the Hypoxic Tumor Milieu. Exp. Cell Res. 2010, 316, 3417–3424. [Google Scholar] [CrossRef]
- Tomassetti, C.; Insinga, G.; Gimigliano, F.; Morrione, A.; Giordano, A.; Giurisato, E. Insights into CSF-1R Expression in the Tumor Microenvironment. Biomedicines 2024, 12, 2381. [Google Scholar] [CrossRef]
- Verocq, C.; Decaestecker, C.; Rocq, L.; De Clercq, S.; Verrellen, A.; Mekinda, Z.; Ocak, S.; Compère, C.; Stanciu-Pop, C.; Salmon, I.; et al. The Daily Practice Reality of PD-L1 (CD274) Evaluation in Non-small Cell Lung Cancer: A Retrospective Study. Oncol. Lett. 2020, 19, 3400–3410. [Google Scholar] [CrossRef]
- Hirayama, A.; Tanaka, K.; Tsutsumi, H.; Nakanishi, T.; Yamashita, S.; Mizusaki, S.; Ishii, Y.; Ota, K.; Yoneshima, Y.; Iwama, E.; et al. Regulation of PD-L1 Expression in Non–Small Cell Lung Cancer by Interleukin-1β. Front. Immunol. 2023, 14, 1192861. [Google Scholar] [CrossRef]
- Abu Hejleh, T.; Furqan, M.; Ballas, Z.; Clamon, G. The Clinical Significance of Soluble PD-1 and PD-L1 in Lung Cancer. Crit. Rev. Oncol./Hematol. 2019, 143, 148–152. [Google Scholar] [CrossRef]
- Li, J.; Liu, T.; Tang, N.; Lin, S.; Zhang, F.; Yuan, W.; Zhang, T.; Deng, S.; Wu, D.; Xu, Y. Cyclin-Dependent Kinase Inhibitor 1A Inhibits Pyroptosis to Enhance Human Lung Adenocarcinoma Cell Radioresistance by Promoting DNA Repair. Heliyon 2024, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zhang, S.; Xue, A.; Feng, G.; Fan, S. Elevated BTG2 Improves the Radiosensitivity of Non-Small Cell Lung Cancer (NSCLC) through Apoptosis. Thorac. Cancer 2022, 13, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Levina, V.; Marrangoni, A.; Wang, T.; Parikh, S.; Su, Y.; Herberman, R.; Lokshin, A.; Gorelik, E. Elimination of Human Lung Cancer Stem Cells through Targeting of the Stem Cell Factor–c-Kit Autocrine Signaling Loop. Cancer Res. 2010, 70, 338–346. [Google Scholar] [CrossRef]
- Wang, T.; Liu, X.; Tian, Q.; Liang, T.; Chang, P. Reduced SPOCK1 Expression Inhibits Non-Small Cell Lung Cancer Cell Proliferation and Migration through Wnt/β-Catenin Signaling. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 637–644. [Google Scholar] [CrossRef]
- Gao, Y.; Yu, M.; Ma, M.; Zhuang, Y.; Qiu, X.; Zhao, Q.; Dai, J.; Cai, H.; Yan, X. SPOCK1 Contributes to the Third-Generation EGFR Tyrosine Kinase Inhibitors Resistance in Lung Cancer. J. Cell. Biochem. 2019, 120, 12566–12573. [Google Scholar] [CrossRef]
- Liu, Y.; Han, T.; Wu, J.; Zhou, J.; Guo, J.; Miao, R.; Xu, Z.; Xing, Y.; Bai, Y.; Hu, D. SPOCK1, as a Potential Prognostic and Therapeutic Biomarker for Lung Adenocarcinoma, Is Associated with Epithelial-Mesenchymal Transition and Immune Evasion. J. Transl. Med. 2023, 21, 909. [Google Scholar] [CrossRef]
- Sun, L.; Li, S.; Guo, Q.; Zhou, W.; Zhang, H. SPOCK1 Involvement in Epithelial-to-Mesenchymal Transition: A New Target in Cancer Therapy? Cancer Manag. Res. 2020, 12, 3561–3569. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tian, H.; Wang, X.; Ding, J. Expression and their clinical significance of SSTR2A, SSTR5 and EGFR in non-small cell lung cancer. Zhongguo Fei Ai Za Zhi 2007, 10, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xia, H.; Wang, Y.; Han, W.; Qin, J.; Gao, W.; Qu, X.; Wang, X. Targeted Paclitaxel-Octreotide Conjugates Inhibited the Growth of Paclitaxel-Resistant Human Non-Small Cell Lung Cancer A549 Cells In Vitro. Thorac. Cancer 2021, 12, 3053–3061. [Google Scholar] [CrossRef] [PubMed]
- Karaca, B.; Degirmenci, M.; Ozveren, A.; Atmaca, H.; Bozkurt, E.; Karabulut, B.; Sanli, U.A.; Uslu, R. Docetaxel in Combination with Octreotide Shows Synergistic Apoptotic Effect by Increasing SSTR2 and SSTR5 Expression Levels in Prostate and Breast Cancer Cell Lines. Cancer Chemother. Pharmacol. 2015, 75, 1273–1280. [Google Scholar] [CrossRef]
- Wang, J.; Ning, D.; Xie, D.; Chen, X.; Cao, X.; Wan, C. Functional Involvement of ADRA1D in Cutaneous Melanoma Progression and Angiogenesis. Cell. Mol. Biol. 2023, 69, 44–50. [Google Scholar] [CrossRef]
- Sentek, H.; Braun, A.; Budeus, B.; Klein, D. Non-Small Cell Lung Cancer Cells and Concomitant Cancer Therapy Induce a Resistance-Promoting Phenotype of Tumor-Associated Mesenchymal Stem Cells. Front. Oncol. 2024, 14, 1406268. [Google Scholar] [CrossRef]
- Liu, J.; Eischeid, A.N.; Chen, X.-M. Col1A1 Production and Apoptotic Resistance in TGF-Β1-Induced Epithelial-to-Mesenchymal Transition-Like Phenotype of 603B Cells. PLoS ONE 2012, 7, e51371. [Google Scholar] [CrossRef]
- Liu, M.; Cai, R.; Wang, T.; Yang, X.; Wang, M.; Kuang, Z.; Xie, Y.; Zhang, J.; Zheng, Y. LAMC2 Promotes the Proliferation of Cancer Cells and Induce Infiltration of Macrophages in Non-Small Cell Lung Cancer. Ann. Transl. Med. 2021, 9, 1392. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cai, J.; Du, S.; Wei, W.; Shen, X. LAMC2 Modulates the Acidity of Microenvironments to Promote Invasion and Migration of Pancreatic Cancer Cells via Regulating AKT-Dependent NHE1 Activity. Exp. Cell Res. 2020, 391, 111984. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Wang, A.; Zhang, J.; Xia, M.; Jiang, Z.; Jia, B.; Lu, C.; Chen, C.; Wang, S.; Zhang, Y.; et al. Hypoxia Promotes Histone H3K9 Lactylation to Enhance LAMC2 Transcription in Esophageal Squamous Cell Carcinoma. iScience 2024, 27, 110188. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-H.; Zhang, L.; Huang, S.-T.; Xu, J.; Zhou, Y.; Yu, X.-J.; Luo, R.-Z.; Wen, Z.-S.; Jia, W.-H.; Zheng, M. Expression and Prognostic Significance of MYL9 in Esophageal Squamous Cell Carcinoma. PLoS ONE 2017, 12, e0175280. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, L.; Feng, W.; Lin, Z.; Ning, Y.; Luo, X. High Expression of MYL9 Indicates Poor Clinical Prognosis of Epithelial Ovarian Cancer. Recent. Pat. Anti-Cancer Drug Discov. 2021, 16, 533–539. [Google Scholar] [CrossRef]
- Deng, S.; Cheng, D.; Wang, J.; Gu, J.; Xue, Y.; Jiang, Z.; Qin, L.; Mao, F.; Cao, Y.; Cai, K. MYL9 Expressed in Cancer-Associated Fibroblasts Regulate the Immune Microenvironment of Colorectal Cancer and Promotes Tumor Progression in an Autocrine Manner. J. Exp. Clin. Cancer Res. 2023, 42, 294. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Chen, Y.; Xia, L.; Yu, X.; Peng, Y.; Zhang, X.; Yang, Z. PMEPA1 Facilitates Non-Small Cell Lung Cancer Progression via Activating the JNK Signaling Pathway. Cancer Biomark. 2021, 31, 203–210. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, S.; Deng, J.; Shen, J.; Du, F.; Wu, X.; Chen, Y.; Li, M.; Chen, M.; Li, X.; et al. VEGF/VEGFR-Targeted Therapy and Immunotherapy in Non-Small Cell Lung Cancer: Targeting the Tumor Microenvironment. Int. J. Biol. Sci. 2022, 18, 3845–3858. [Google Scholar] [CrossRef]
- Annals of Surgical Oncology. TGFBI Expression in Cancer Stromal Cells Is Associated with Poor Prognosis and Hematogenous Recurrence in Esophageal Squamous Cell Carcinoma. Available online: https://link.springer.com/article/10.1245/s10434-014-4259-4 (accessed on 5 March 2025).
- Guo, H.; Tang, H.; Zhao, Y.; Zhao, Q.; Hou, X.; Ren, L. Molecular Typing of Gastric Cancer Based on Invasion-Related Genes and Prognosis-Related Features. Front. Oncol. 2022, 12, 848163. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-Generation Characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Dean, P.N.; Jett, J.H. Mathematical Analysis of DNA Distributions Derived from Flow Microfluorometry. J. Cell Biol. 1974, 60, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]

| Effect Being Studied | Abbreviation | Compared Groups |
|---|---|---|
| Effect of oxygenation status in unirradiated cells | H0 vs. N0 | Continuous hypoxia without irradiation vs. normoxia without irradiation |
| R0 vs. N0 | Reoxygenation after 48 h of hypoxia without irradiation vs. normoxia without irradiation | |
| Effect of oxygenation status in irradiated cells | H8 vs. N8 | Hypoxia before and after irradiation with 8 Gy vs. normoxia before and after irradiation with 8 Gy |
| R8 vs. N8 | Reoxygenation after 48 h of hypoxia and irradiation with 8 Gy vs. normoxia before and after irradiation with 8 Gy | |
| Effect of irradiation under different oxygen conditions | N8 vs. N0 | Normoxia before and after irradiation with 8 Gy vs. normoxia without irradiation |
| H8 vs. H0 | Hypoxia before and after irradiation with 8 Gy vs. hypoxia without irradiation | |
| R8 vs. R0 | Reoxygenation after 48 h of hypoxia and irradiation with 8 Gy vs. reoxygenation without irradiation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nisar, H.; Konda, B.; Hoffmann, M.D.; Labonté, F.M.; Arif, M.; Diegeler, S.; Schmitz, C.; Baumstark-Khan, C.; Chevalier, F.; Hellweg, C.E. Effect of Reoxygenation on Radioresistance of Chronically Hypoxic A549 Non-Small Cell Lung Cancer (NSCLC) Cells Following X-Ray and Carbon Ion Exposure. Int. J. Mol. Sci. 2025, 26, 9153. https://doi.org/10.3390/ijms26189153
Nisar H, Konda B, Hoffmann MD, Labonté FM, Arif M, Diegeler S, Schmitz C, Baumstark-Khan C, Chevalier F, Hellweg CE. Effect of Reoxygenation on Radioresistance of Chronically Hypoxic A549 Non-Small Cell Lung Cancer (NSCLC) Cells Following X-Ray and Carbon Ion Exposure. International Journal of Molecular Sciences. 2025; 26(18):9153. https://doi.org/10.3390/ijms26189153
Chicago/Turabian StyleNisar, Hasan, Bikash Konda, Marie Denise Hoffmann, Frederik M. Labonté, Maryam Arif, Sebastian Diegeler, Claudia Schmitz, Christa Baumstark-Khan, François Chevalier, and Christine E. Hellweg. 2025. "Effect of Reoxygenation on Radioresistance of Chronically Hypoxic A549 Non-Small Cell Lung Cancer (NSCLC) Cells Following X-Ray and Carbon Ion Exposure" International Journal of Molecular Sciences 26, no. 18: 9153. https://doi.org/10.3390/ijms26189153
APA StyleNisar, H., Konda, B., Hoffmann, M. D., Labonté, F. M., Arif, M., Diegeler, S., Schmitz, C., Baumstark-Khan, C., Chevalier, F., & Hellweg, C. E. (2025). Effect of Reoxygenation on Radioresistance of Chronically Hypoxic A549 Non-Small Cell Lung Cancer (NSCLC) Cells Following X-Ray and Carbon Ion Exposure. International Journal of Molecular Sciences, 26(18), 9153. https://doi.org/10.3390/ijms26189153








