Genome-Wide Identification of the KAN Gene Family and Expression Profiles During the Fruit Developmental Stages in Prunus mume
Abstract
1. Introduction
2. Results
2.1. Identification of KAN Family Members in P. mume Genome
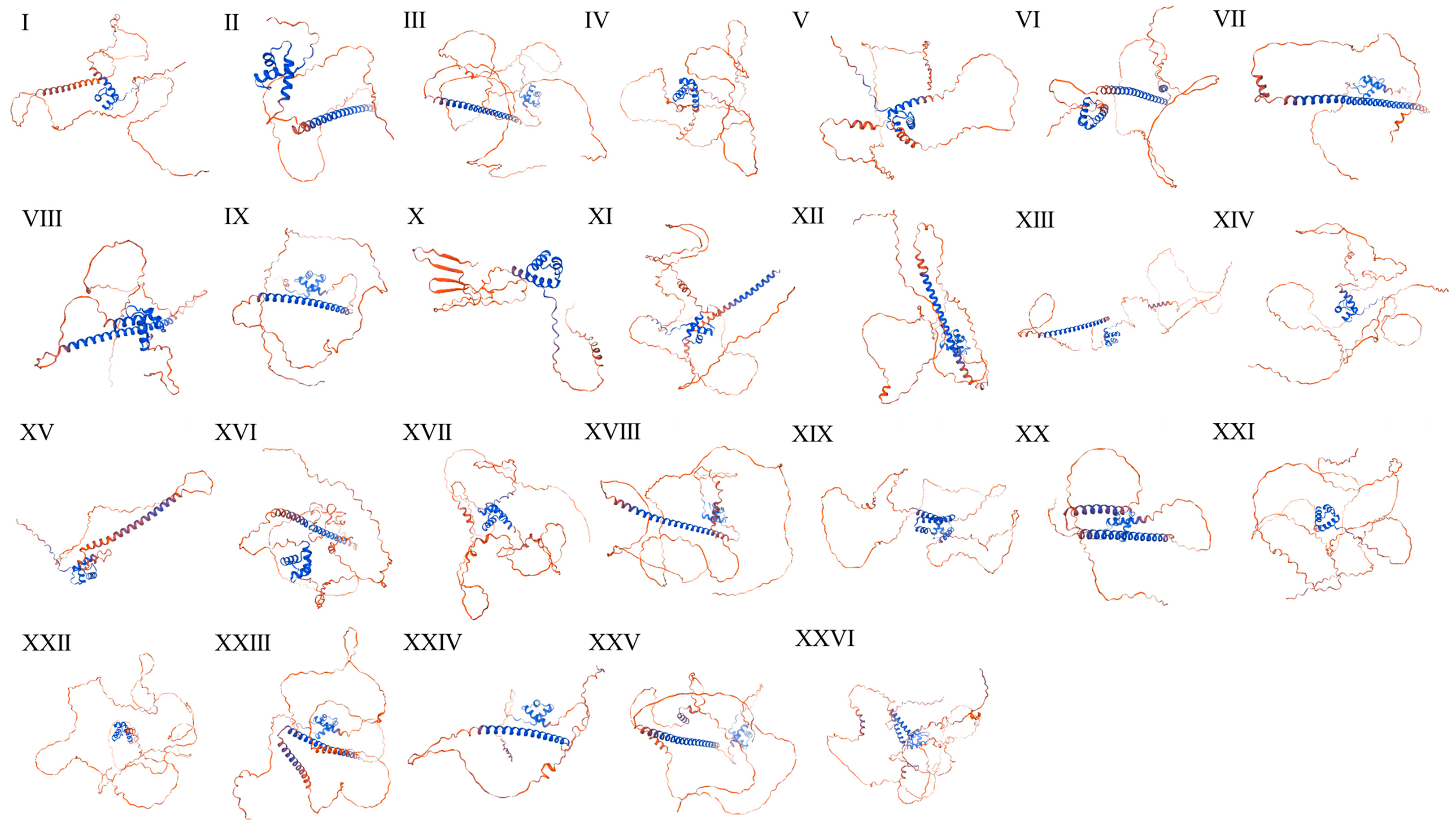
2.2. Secondary Structure Analysis of PmKAN Protein
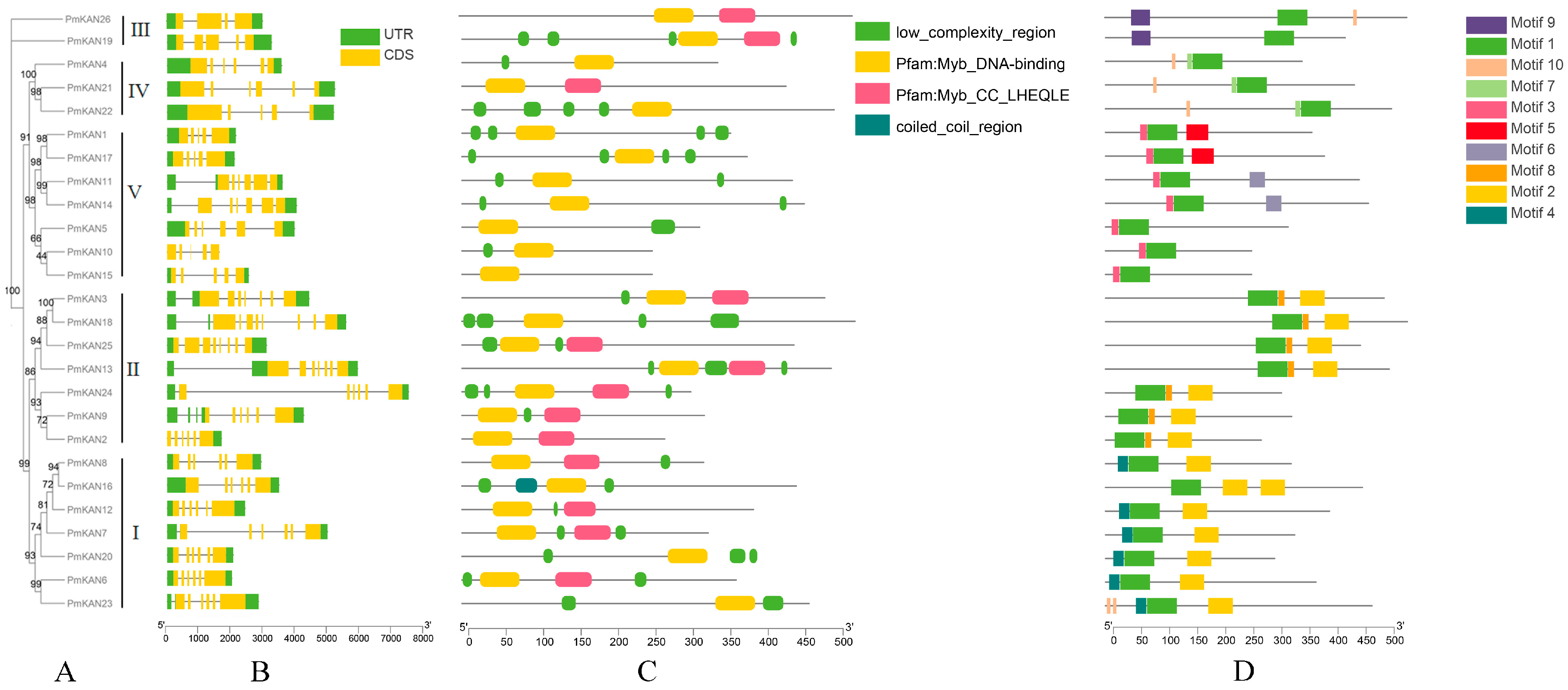
2.3. Gene Structures, Protein Conserved Domain and Motif Compositions of PmKAN Proteins
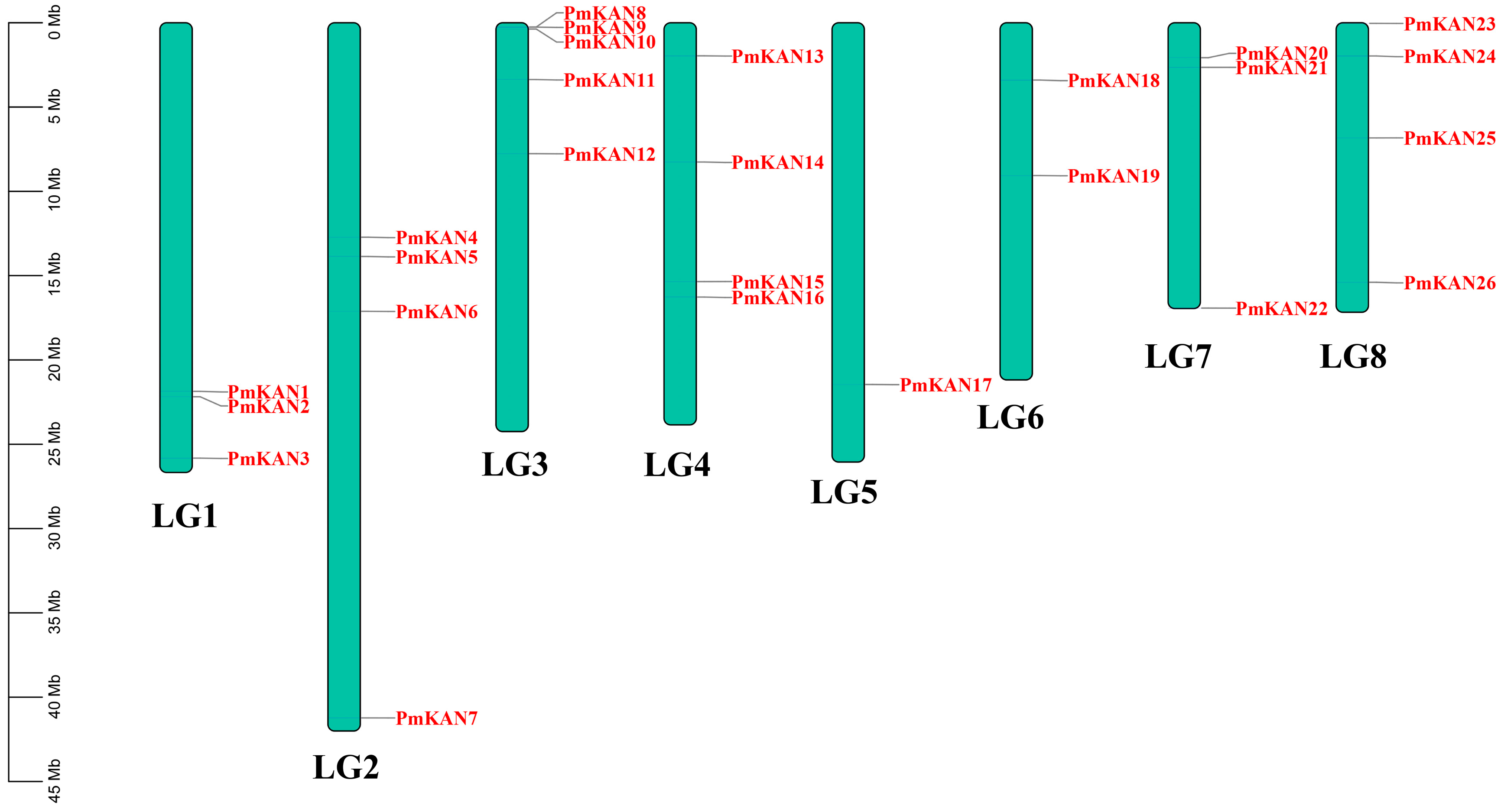
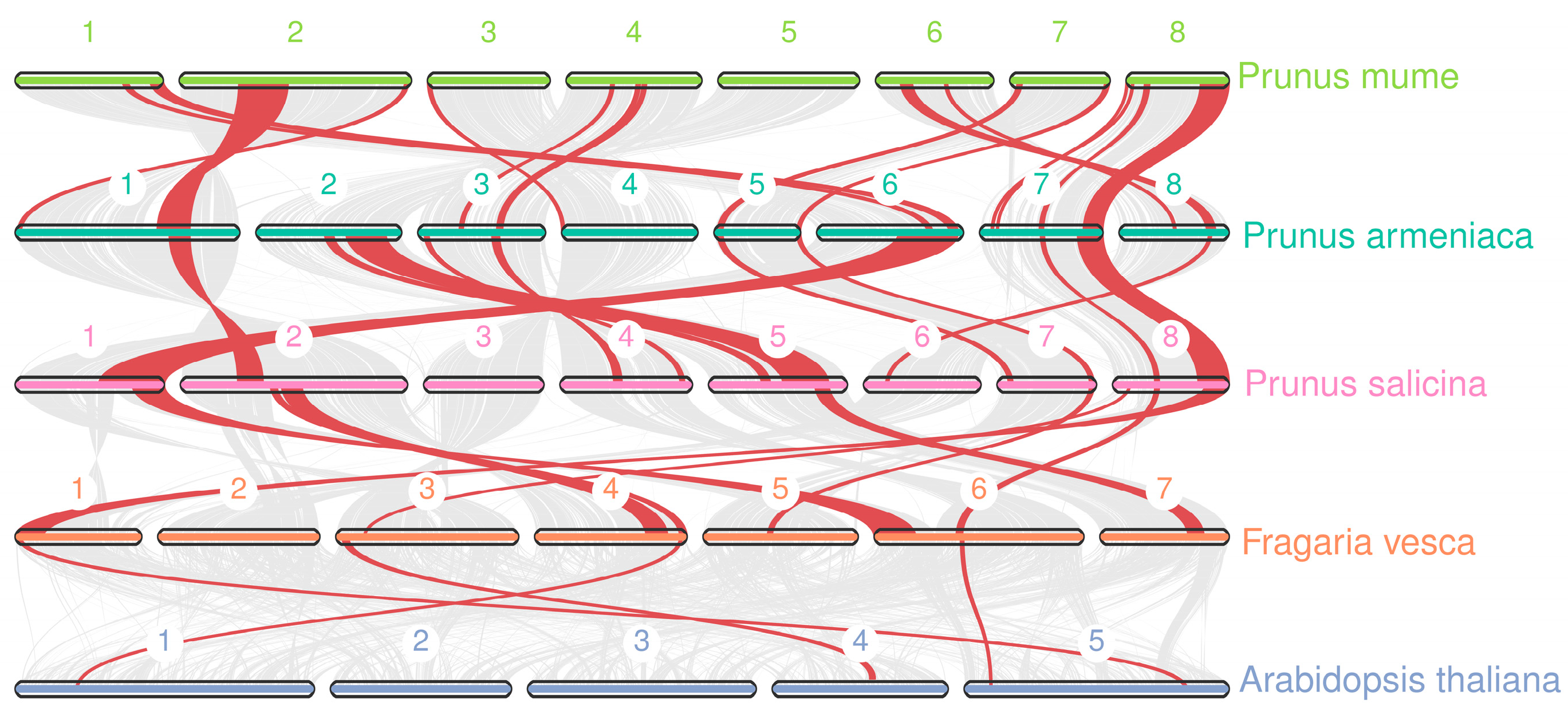
2.4. Chromosomal Distributions Analysis of PmKAN Genes
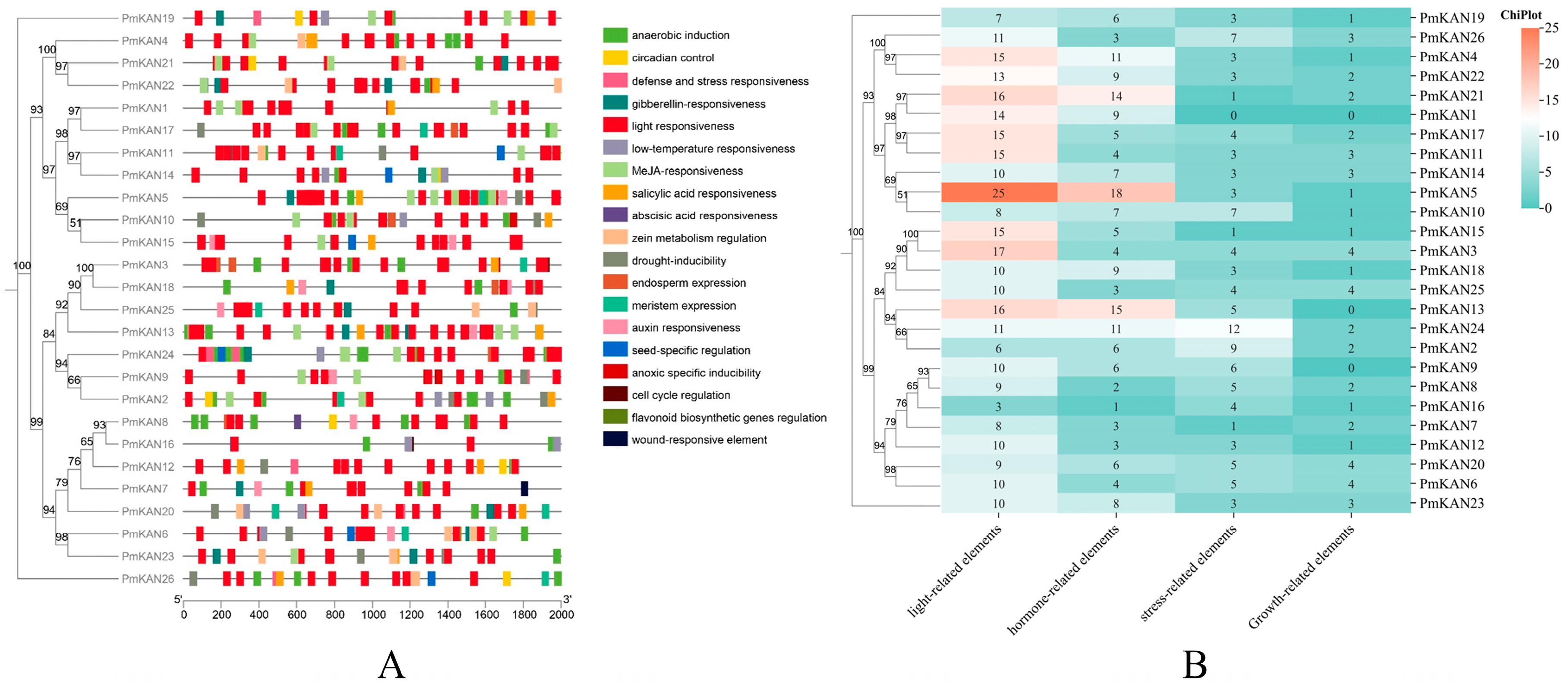
2.5. Cis-Acting Element Analysis in PmKAN Promoter Region
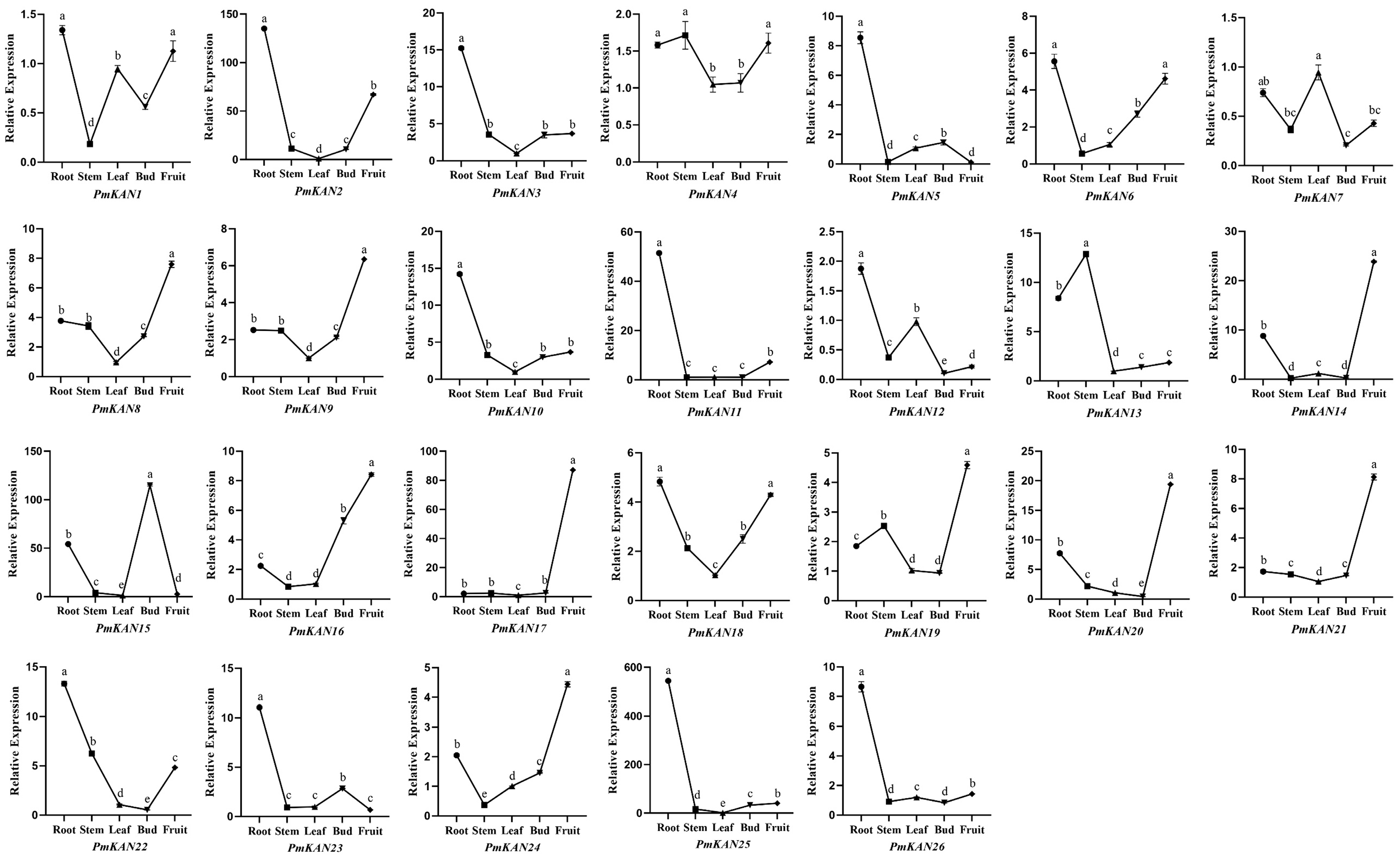
2.6. Expression Analysis of PmKAN Genes in Different Tissues
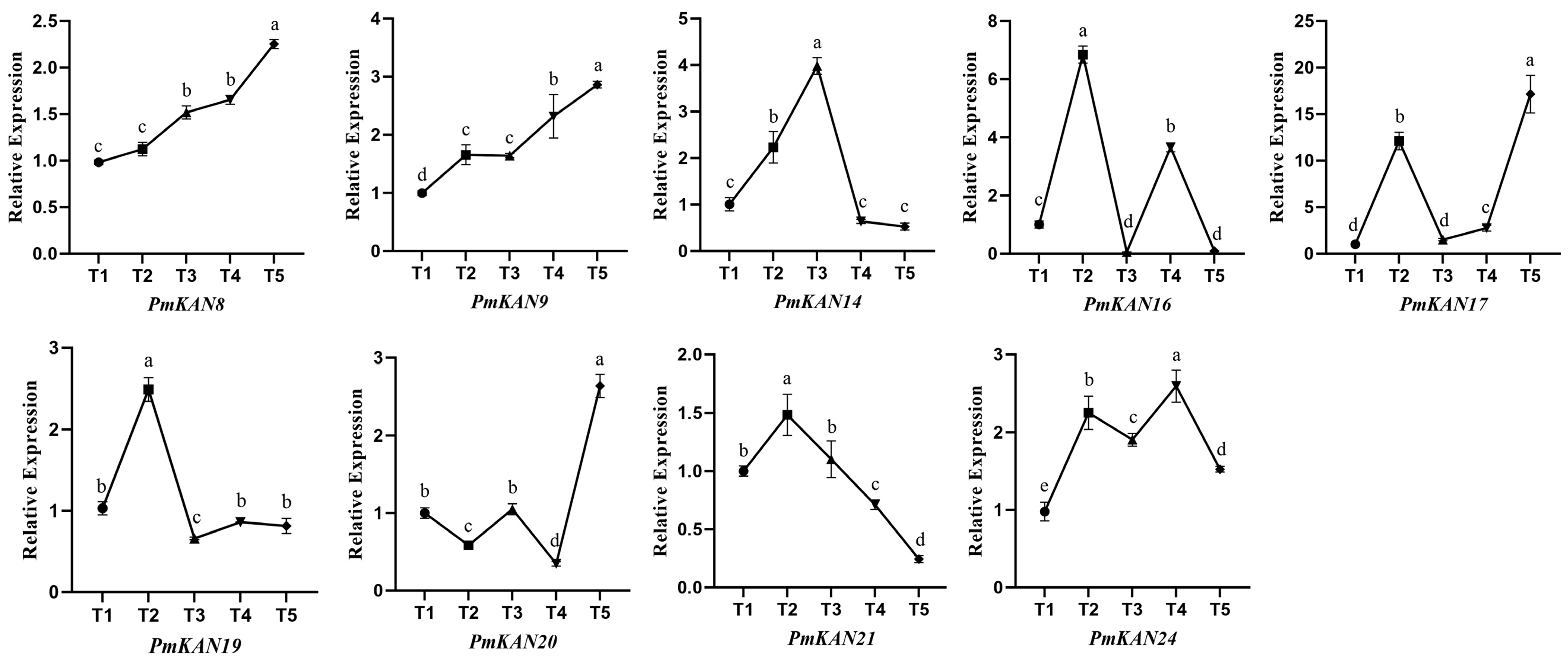
2.7. Expression Analysis at Different Developmental Stages of Fruit
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of PmKAN Family Members
4.3. Analysis of Structure and Conserved Motifs of PmKAN Gene in P. mume
4.4. Chromosomal Location and Syntenic Analysis
4.5. Analysis of Cis-Acting Elements in PmKAN Gene Upstream Promoter Region
4.6. Quantitative qRT-PCR Analysis of PmKAN Genes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| CTK | Cytokinin |
| FPKM | Fragments Per Kilobase of transcript per Million mapped fragments |
| GA | Gibberellic acid |
| HMM | Hidden Markov Model |
| JA | Jasmonic acid |
| KAN | KANADI |
| MeJA | Methyl jasmonate |
| MEME | Multiple Em for Motif Elicitation |
| ML | Maximum Likelihood |
| NCBI | National Center for Biotechnology Information |
| ORF | Open Reading Frame |
| P. mume | Prunus mume Sieb. et Zucc. |
| PFAM | Protein Family Database |
| SA | Salicylic acid |
| WGD | Whole-Genome Duplication |
References
- Eshed, Y.; Baum, S.F. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001, 11, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Kerstetter, R.A.; Bollman, K. KANADI regulates organ polarity in Arabidopsis. Nature 2001, 411, 706–709. [Google Scholar] [CrossRef]
- Henderson, D.C.; Zhang, X. RAGGED SEEDLING2 is required for expression of KANADI2 and REVOLUTA homologues in the maize shoot apex. Genesis 2006, 44, 372–382. [Google Scholar] [CrossRef]
- Zhang, G.H.; Xu, Q. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 2009, 21, 719–735. [Google Scholar] [CrossRef]
- Juanhong, W.; Qi, D. Cloning and expression analysis of KANADI gene in Nicotiana benthamiana. J. Agric. Biotechnol. 2013, 21, 1328–1336. [Google Scholar]
- Niu, M.; Zhang, H. Genome-Wide Analysis of the KANADI Gene Family and Its Expression Patterns under Different Nitrogen Concentrations Treatments in Populus trichocarpa. Phyton-Int. J. Exp. Bot. 2023, 92, 2001–2015. [Google Scholar] [CrossRef]
- Ahmad, R.; Liu, Y. GOLDEN2-LIKE Transcription Factors Regulate WRKY40 Expression in Response to Abscisic Acid. Plant Physiol. 2019, 179, 1844–1860. [Google Scholar] [CrossRef] [PubMed]
- Zumajo-Cardona, C.; Vasco, A. The Evolution of the KANADI Gene Family and Leaf Development in Lycophytes and Ferns. Plants 2019, 8, 313. [Google Scholar] [CrossRef]
- Zumajo-Cardona, C.; Ambrose, A.B. Phylogenetic analyses of key developmental genes provide insight into the complex evolution of seeds. Mol. Phylogenetics Evol. 2020, 147, 106778. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.R.; Arreola, A. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 2012, 139, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- McAbee, J.M.; Hill, T.A. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 2006, 46, 522–531. [Google Scholar] [CrossRef]
- Eshed, Y.; Izhaki, A. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 2004, 131, 2997–3006. [Google Scholar] [CrossRef]
- Candela, H.; Johnston, R. The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell 2008, 20, 2073–2087. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Cui, Y. AH2 encodes a MYB domain protein that determines hull fate and affects grain yield and quality in rice. Plant J. 2019, 100, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Si, F.; Yang, C. Control of OsARF3a by OsKANADI1 contributes to lemma development in rice. Plant J. 2022, 110, 1717–1730. [Google Scholar] [CrossRef]
- He, Q.; Wu, H. OsKANADI1 and OsYABBY5 regulate rice plant height by targeting GIBERELLIN 2-OXIDASE6. Plant Cell 2024, 37, koae276. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.Y.; Fang, J.G. Culture of Prunus mume. J. Beijing For. Univ. 2001, 3, 47–49. [Google Scholar]
- Lin, X.; Zhou, P. Genome-Wide Identification of the ALMT Gene Family in Nine Rosaceae Species and Functional Analysis Associated with Organic Acid Accumulation in Prunus mume. Horticulturae 2024, 10, 1305. [Google Scholar] [CrossRef]
- Wei, L.; Wang, W. Genome-wide identification of the CsPAL gene family and functional analysis for strengthening green mold resistance in citrus fruit. Postharvest Biol. Technol. 2023, 196, 112178. [Google Scholar] [CrossRef]
- Wang, M.M. Functional Study of KANADI in Compound Leaf Development in Medicago truncatula. Ph.D. Thesis, Shandong University, Jinan, China, 2021. [Google Scholar]
- Zhao, Y.; Yu, D. Nuclear phylogenomics provide evidence to clarify key morphological evolution and whole-genome duplication across rosids. J. Integr. Plant Biol. 2025, 1–27. [Google Scholar] [CrossRef]
- Leon-Kloosterziel, K.M.; Keijzer, C.J. A Seed Shape Mutant of Arabidopsis That Is Affected in Integument Development. Plant Cell 1994, 6, 385–392. [Google Scholar] [CrossRef]
- Ilegems, M.; Douet, V. Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development 2010, 137, 975–984. [Google Scholar] [CrossRef]
- Ishibashi, N.; Kanamaru, K. ASYMMETRIC-LEAVES2 and an ortholog of eukaryotic NudC domain proteins repress expression of AUXIN-RESPONSE-FACTOR and class 1 KNOX homeobox genes for development of flat symmetric leaves in Arabidopsis. Biol. Open 2012, 1, 197–207. [Google Scholar] [CrossRef]
- Yao, D.; Ni, X. Identification and Tissue Expression Analysis of NAC Gene Family in Prunus mume. J. Nucl. Agric. Sci. 2019, 33, 14. [Google Scholar]
- Wang, B.; Xiao, Y. Gene Duplication and Functional Diversification of MADS-Box Genes in Malus × domestica following WGD: Implications for Fruit Type and Floral Organ Evolution. Int. J. Mol. Sci. 2024, 25, 8962. [Google Scholar] [CrossRef] [PubMed]
- Pires, H.R.; Monfared, M.M. ULTRAPETALA trxG genes interact with KANADI transcription factor genes to regulate Arabidopsis gynoecium patterning. Plant Cell 2014, 26, 4345–4361. [Google Scholar] [CrossRef]
- Xie, Y.; Straub, D. Meta-Analysis of Arabidopsis KANADI1 Direct Target Genes Identifies a Basic Growth-Promoting Module Acting Upstream of Hormonal Signaling Pathways. Plant Physiol. 2015, 169, 1240–1253. [Google Scholar] [CrossRef]
- Bhatia, N.; Åhl, H. Quantitative analysis of auxin sensing in leaf primordia argues against proposed role in regulating leaf dorsoventrality. Elife 2019, 8, e39298. [Google Scholar] [CrossRef]
- Ram, H.; Sahadevan, S. An integrated analysis of cell-type specific gene expression reveals genes regulated by REVOLUTA and KANADI1 in the Arabidopsis shoot apical meristem. PLoS Genet. 2020, 16, e1008661. [Google Scholar] [CrossRef]
- Izhaki, A.; Bowman, J.L. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 2007, 19, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Pekker, I.; Alvarez, J.P. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef]
- Hawker, N.P.; Bowman, J.L. Roles for Class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 2004, 135, 2261–2270. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Roy, S. Phytohormonal crosstalk modulates the expression of miR166/165s, target Class III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana. Sci. Rep. 2017, 7, 3408. [Google Scholar] [CrossRef]
- Zhou, Y.; Xiong, J. The telomere-to-telomere genome of Fragaria vesca reveals the genomic evolution of Fragaria and the origin of cultivated octoploid strawberry. Hortic. Res. 2023, 10, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, W. The genome of Prunus mume. Nat. Commun. 2012, 3, 1318. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Høie, M.H.; Kiehl, E.N. NetSurfP-3.0: Accurate and fast prediction of protein structural features by protein language models and deep learning. Nucleic Acids Res. 2022, 50, W510–W515. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H. Genome-wide analysis of VQ motif-containing proteins in Moso bamboo (Phyllostachys edulis). Planta 2017, 246, 165–181. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, L. Phytohormone and integrated mRNA and miRNA transcriptome analyses and differentiation of male between hermaphroditic floral buds of andromonoecious Diospyros kaki Thunb. BMC Genom. 2021, 22, 203. [Google Scholar] [CrossRef] [PubMed]




| Sequence ID | Gene ID | Protein ID | Number of Amino Acid | Molecular Weight (Da) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|---|---|
| PmKAN1 | LOC103341420 | XP_008243158.1 | 339 | 38,330.28 | 7.22 | 54.77 | 66.17 | −0.996 |
| PmKAN2 | LOC103342254 | XP_008244092.2 | 256 | 28,978.03 | 9.16 | 54.78 | 78.55 | −0.628 |
| PmKAN3 | LOC103318708 | XP_008218346.1 | 458 | 51,491.82 | 5.09 | 60.69 | 62.14 | −0.941 |
| PmKAN4 | LOC103320783 | XP_008220727.1 | 323 | 35,643.63 | 8.38 | 40.90 | 63.72 | −0.729 |
| PmKAN5 | LOC103321009 | XP_008220974.1 | 300 | 33,444.13 | 8.86 | 62.06 | 62.43 | −0.782 |
| PmKAN6 | LOC103321568 | XP_008221605.1 | 346 | 38,907.83 | 7.68 | 46.67 | 74.62 | −0.810 |
| PmKAN7 | LOC103324237 | XP_008224497.1 | 311 | 33,616.20 | 6.11 | 52.42 | 81.03 | −0.352 |
| PmKAN8 | LOC103324390 | XP_008224659.1 | 305 | 33,185.51 | 6.50 | 44.14 | 71.97 | −0.551 |
| PmKAN9 | LOC103324391 | XP_008224662.1 | 306 | 33,843.00 | 6.22 | 58.46 | 73.66 | −0.744 |
| PmKAN10 | LOC103324465 | XP_008224747.1 | 240 | 26,712.18 | 8.25 | 51.90 | 75.96 | −0.642 |
| PmKAN11 | LOC103324956 | XP_008225297.1 | 417 | 48,261.83 | 8.84 | 60.77 | 58.39 | −1.06 |
| PmKAN12 | LOC103325588 | XP_008226001.1 | 368 | 40,994.31 | 8.55 | 55.89 | 65.49 | −0.787 |
| PmKAN13 | LOC103327465 | XP_008228027.1 | 466 | 51,018.80 | 5.58 | 75.71 | 69.10 | −0.688 |
| PmKAN14 | LOC103328312 | XP_008228924.1 | 432 | 49,087.78 | 8.70 | 52.45 | 68.56 | −0.811 |
| PmKAN15 | LOC103328941 | XP_016649340.1 | 240 | 27,684.77 | 6.60 | 48.97 | 60.92 | −0.890 |
| PmKAN16 | LOC103329105 | XP_008229756.1 | 422 | 47,845.56 | 5.09 | 52.08 | 77.16 | −0.646 |
| PmKAN17 | LOC103332813 | XP_008233790.1 | 360 | 40,640.07 | 6.77 | 49.14 | 61.17 | −0.946 |
| PmKAN18 | LOC103334420 | XP_016650539.1 | 496 | 54,847.79 | 5.44 | 61.42 | 70.58 | −0.626 |
| PmKAN19 | LOC103335407 | XP_008236633.1 | 394 | 44,002.27 | 7.61 | 58.71 | 62.64 | −0.831 |
| PmKAN20 | LOC103336798 | XP_008238120.1 | 278 | 31,421.01 | 8.30 | 37.42 | 62.12 | −0.981 |
| PmKAN21 | LOC103336839 | XP_008238177.1 | 409 | 45,952.91 | 7.39 | 70.15 | 65.82 | −0.796 |
| PmKAN22 | LOC103338908 | XP_008240401.1 | 470 | 52,192.55 | 6.78 | 54.37 | 57.94 | −0.829 |
| PmKAN23 | LOC103338947 | XP_008240441.1 | 438 | 49,015.82 | 7.36 | 49.74 | 69.06 | −0.817 |
| PmKAN24 | LOC103339100 | XP_008240600.1 | 289 | 31,469.29 | 7.69 | 53.84 | 65.78 | −0.792 |
| PmKAN25 | LOC103339429 | XP_008240946.2 | 419 | 47,156.13 | 7.66 | 70.79 | 64.25 | −0.819 |
| PmKAN26 | LOC103341001 | XP_008242700.1 | 496 | 54,334.02 | 7.15 | 62.68 | 56.94 | −0.882 |
| Protein Name | Coil Regions | 310 Helix (%) | Alpha Helix (%) | Random Coil (%) | Beta Strand (%) | Beta Turn (%) |
|---|---|---|---|---|---|---|
| PmKAN1 | 86.14 | 0.88 | 10.62 | 0.88 | 0.88 | 0.59 |
| PmKAN2 | 70.70 | 0.78 | 26.56 | 1.95 | 0.00 | 0.00 |
| PmKAN3 | 82.53 | 0.66 | 15.94 | 0.87 | 0.00 | 0.00 |
| PmKAN4 | 87.00 | 0.93 | 10.22 | 0.62 | 1.24 | 0.00 |
| PmKAN5 | 86.00 | 1.00 | 11.00 | 1.00 | 0.33 | 0.67 |
| PmKAN6 | 72.83 | 0.58 | 25.43 | 1.16 | 0.00 | 0.00 |
| PmKAN7 | 68.49 | 0.64 | 28.62 | 1.93 | 0.32 | 0.00 |
| PmKAN8 | 69.84 | 0.66 | 26.89 | 1.97 | 0.66 | 0.00 |
| PmKAN9 | 71.24 | 0.98 | 26.14 | 1.63 | 0.00 | 0.00 |
| PmKAN10 | 80.42 | 1.25 | 13.33 | 1.67 | 2.50 | 0.83 |
| PmKAN11 | 88.97 | 0.72 | 8.39 | 0.72 | 0.72 | 0.48 |
| PmKAN12 | 78.26 | 0.54 | 20.11 | 1.09 | 0.00 | 0.00 |
| PmKAN13 | 80.90 | 0.00 | 17.38 | 1.72 | 0.00 | 0.00 |
| PmKAN14 | 90.05 | 0.69 | 7.41 | 0.69 | 0.69 | 0.46 |
| PmKAN15 | 82.08 | 0.83 | 13.33 | 2.08 | 0.83 | 0.83 |
| PmKAN16 | 72.75 | 0.71 | 25.12 | 0.95 | 0.47 | 0.00 |
| PmKAN17 | 86.39 | 0.83 | 10.00 | 0.83 | 1.39 | 0.56 |
| PmKAN18 | 81.25 | 0.40 | 17.34 | 1.01 | 0.00 | 0.00 |
| PmKAN19 | 73.10 | 1.02 | 23.35 | 1.27 | 1.27 | 0.00 |
| PmKAN20 | 68.71 | 0.72 | 27.70 | 2.16 | 0.72 | 0.00 |
| PmKAN21 | 89.98 | 0.73 | 7.82 | 0.00 | 0.98 | 0.49 |
| PmKAN22 | 90.85 | 0.64 | 6.81 | 0.43 | 0.85 | 0.43 |
| PmKAN23 | 77.63 | 0.46 | 20.55 | 1.14 | 0.23 | 0.00 |
| PmKAN24 | 69.90 | 0.69 | 27.34 | 2.08 | 0.00 | 0.00 |
| PmKAN25 | 79.24 | 0.48 | 18.85 | 1.43 | 0.00 | 0.00 |
| PmKAN26 | 79.84 | 0.60 | 17.14 | 0.81 | 1.61 | 0.00 |
| Gene Name | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| PmKAN 1 | TTCTCGCGGATCATCAAGGC | GAGGTTTGTTACCTGAAGCTGG |
| PmKAN 2 | ACTTACGACGTTGCACCGAA | TCGTTTTCCACAGAAGGCAA |
| PmKAN 3 | GCAGAAGGCACAAGAAACCG | GGCTGGGTACCAGACTCAAC |
| PmKAN 4 | CCGCAATCAAATGGCACCAA | ACAAGGACAATAATGCGCACAC |
| PmKAN 5 | CCTTCTGAGTATGACCAGCGA | AGGCAATAGAGGTGACGCAG |
| PmKAN 6 | TGGAAGTGCAGAGGAAACTTCA | AGAACTCTGGGTTTGTTGGCA |
| PmKAN 7 | GCATGAGCAGCTGGAGGTTA | GGTGGTTTTTGGTCATGCGG |
| PmKAN 8 | GTCAGCTTGGCGGTTCATCT | CATCGCCCATTTCCTTCCCT |
| PmKAN 9 | GCGACTGCACGAGCAATTAG | AGAGCCAGGTGCTTCTGAAC |
| PmKAN 10 | GCTGCTATGTCTCTCTGGGC | CTGCACTCACTAGCGACACA |
| PmKAN 11 | ACTTGCCTCAATCCTTCTTCCA | CTCTCCCCTTTCACCGCATC |
| PmKAN 12 | GAGGTTGCTTTCCGAGCCTA | TTTTCGGTTTGCCTTGACGA |
| PmKAN 13 | TCATCAGCTGGTGTATCGCC | GGTGGCTCAGATTCACTGCT |
| PmKAN 14 | GTCTTGTCAAGTCGTGTAAGC | CGCAAAAGCAAGGACTAAAATGC |
| PmKAN 15 | CAAAAGAGGAAGCAGCAGGC | TCAGTTGCAGACCACGACAG |
| PmKAN 16 | CCTCCTAGGCCTGGACAGTT | CACGAATAACTCCCCCAGCA |
| PmKAN 17 | CTTAGGTGTTGCTCCTGCAC | TTAATTTGCCTCTGCCCAGC |
| PmKAN 18 | GGGGGTATCTGCAGCAATGT | GACACCTGTTGGGAGTCAGG |
| PmKAN 19 | AATAGGGCTGCTGGAAGAGC | TTATCTTCCTTCCTCGCCGC |
| PmKAN 20 | CGGACTTGGGAGTTGCTTCT | GGTGATCAATTTGTCGTTTTGGC |
| PmKAN 21 | AGGAGTGAGAGAGTAGAGCTTG | GCGCTGGACTACCACTCTTC |
| PmKAN 22 | GGAGGGGCTGACTTTCATGG | ACAAGAGGAACATGAGGGCG |
| PmKAN 23 | GGAGTAGCCTCTGCATTGGA | AACCAGCTAGGAGGAGCAGA |
| PmKAN 24 | GAAAAGCCCAACGCCTTCTG | GACGAAGCGGTCATGGAGAT |
| PmKAN 25 | AGCACTTTCATATTGCCGAGG | TGAGCTGCTTCCCTTGTTCTT |
| PmKAN 26 | GGTTGACGGTTTGACCAACG | ATGTTTCATGCTGCCAGTGC |
| PmACTIN | TGAAGCATACACCTATGATGATGAAG | CTTTGACAGCACCAGTAGATTCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Huang, X.; Lin, X.; Wang, Z.; Gao, F.; Gao, Z. Genome-Wide Identification of the KAN Gene Family and Expression Profiles During the Fruit Developmental Stages in Prunus mume. Int. J. Mol. Sci. 2025, 26, 9121. https://doi.org/10.3390/ijms26189121
Li M, Huang X, Lin X, Wang Z, Gao F, Gao Z. Genome-Wide Identification of the KAN Gene Family and Expression Profiles During the Fruit Developmental Stages in Prunus mume. International Journal of Molecular Sciences. 2025; 26(18):9121. https://doi.org/10.3390/ijms26189121
Chicago/Turabian StyleLi, Minglu, Xiao Huang, Ximeng Lin, Ziqi Wang, Feng Gao, and Zhihong Gao. 2025. "Genome-Wide Identification of the KAN Gene Family and Expression Profiles During the Fruit Developmental Stages in Prunus mume" International Journal of Molecular Sciences 26, no. 18: 9121. https://doi.org/10.3390/ijms26189121
APA StyleLi, M., Huang, X., Lin, X., Wang, Z., Gao, F., & Gao, Z. (2025). Genome-Wide Identification of the KAN Gene Family and Expression Profiles During the Fruit Developmental Stages in Prunus mume. International Journal of Molecular Sciences, 26(18), 9121. https://doi.org/10.3390/ijms26189121






