Genome-Wide Characterization and Expression Pattern Analysis Insights into Plant NBS-LRR Gene Family of Salvia miltiorrhiza
Abstract
1. Introduction
2. Results
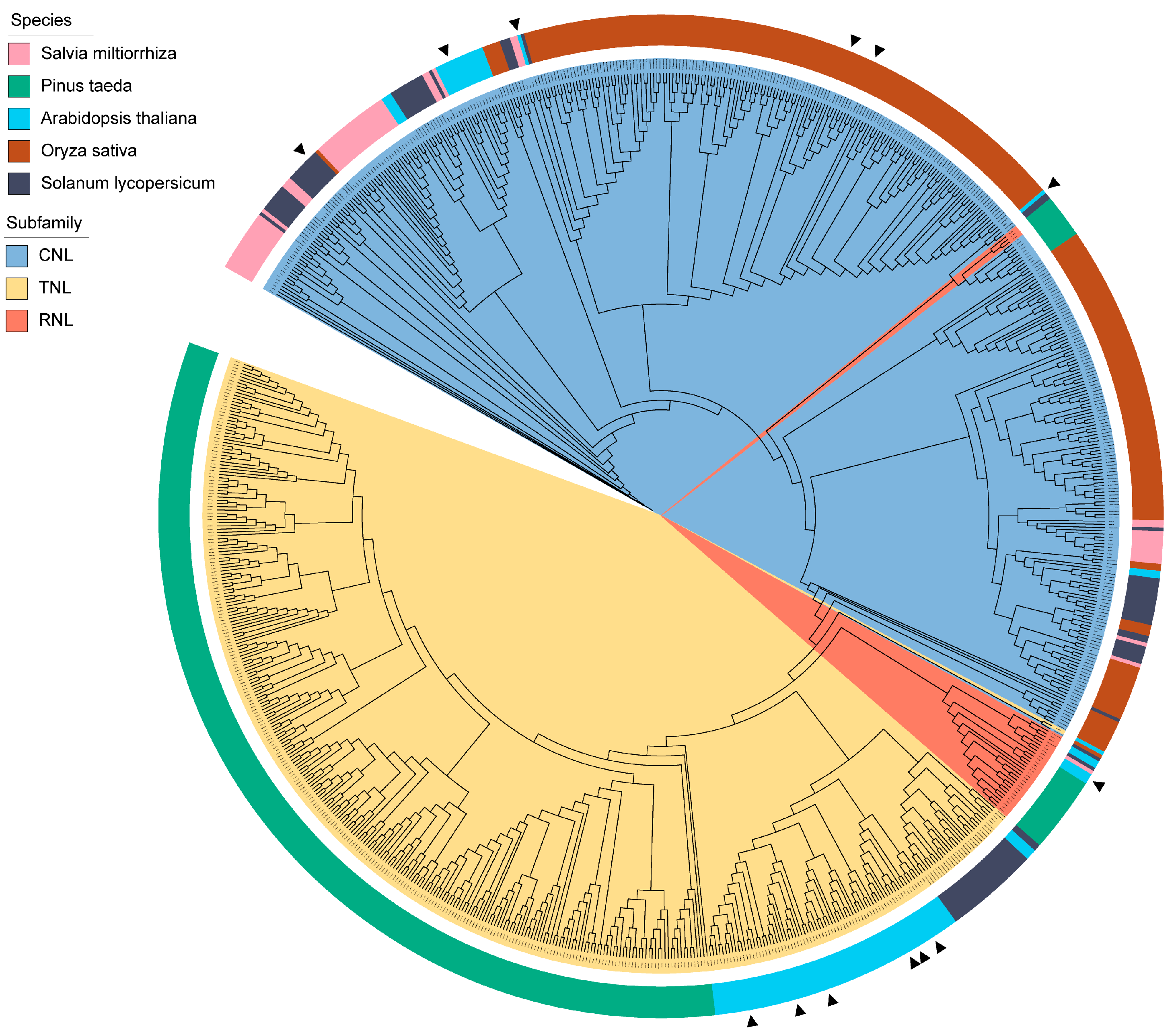
2.1. Structural and Phylogenetic Characterization of SmNBS-LRR Resistance Genes
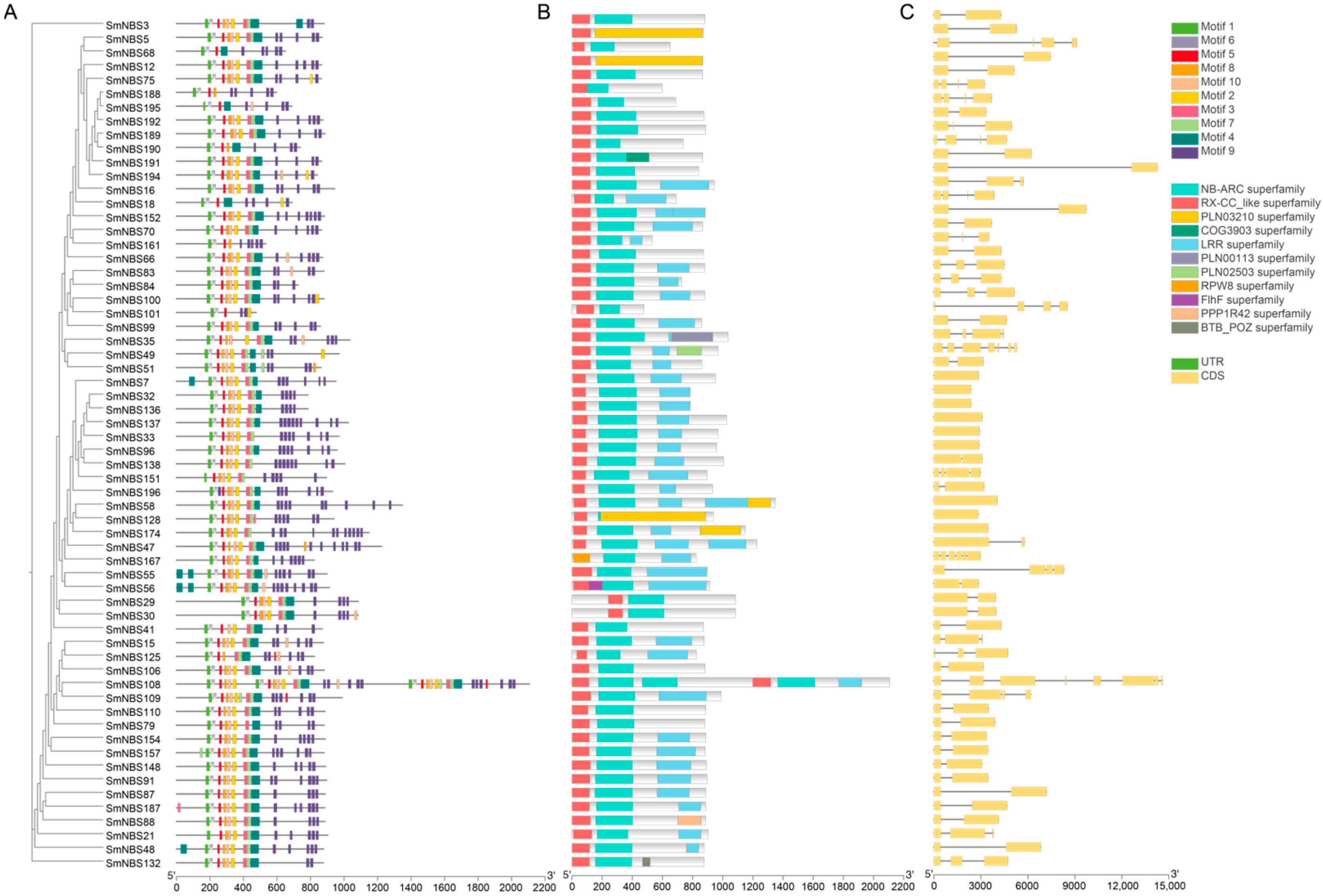
2.2. Gene Structure and Cis-Regulatory Element Analysis of SmNBS-LRRs
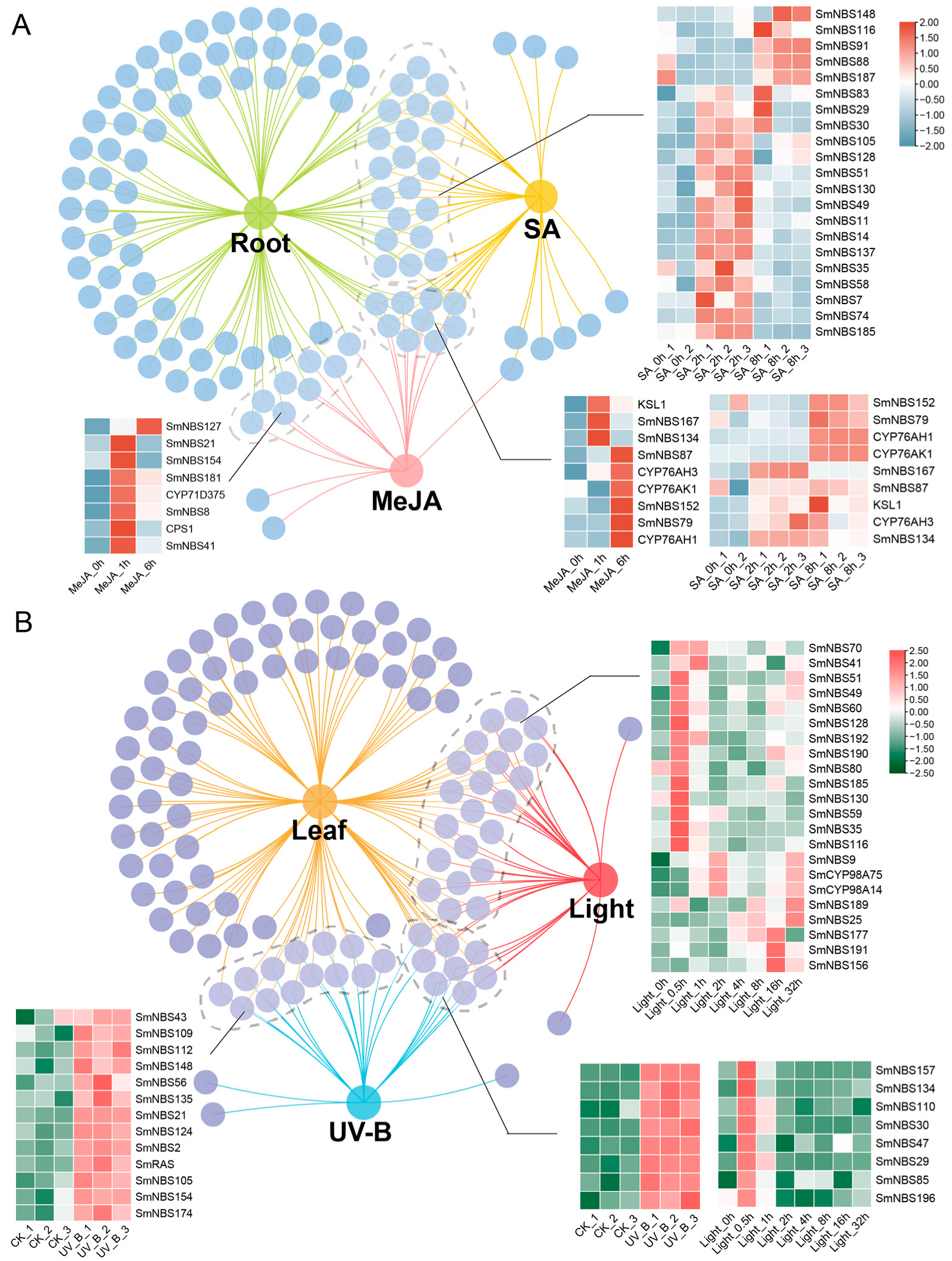
2.3. Expression Patterns and Structural Analysis of SmNBS-LRR Genes Across Different Phylogenetic Clades
3. Discussion
4. Materials and Methods
4.1. Identification and Classification of SmNBS-LRR
4.2. Phylogenetic Analysis and Classification of NBS-LRR
4.3. Gene Structure, Conserved Motif and cis-Regulatory Elements Analysis
4.4. Expression Profiling of NBS-LRR Across Different Tissues and Different Stress Based on RNA-Seq Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, Q.; Chen, X.; Tian, X.; Zhang, J.; Xue, S.; Jiang, Y.; Liu, T.; Wang, X.; Sun, Q.; Hong, Y.; et al. Tanshinone I inhibits doxorubicin-induced cardiotoxicity by regulating Nrf2 signaling pathway. Phytomedicine 2022, 106, 154439. [Google Scholar] [CrossRef]
- Li, R.; Wang, J.; Han, Y.; Dai, Z. Compound Danshen Yeast 1.0. Sci. Tradit. Chin. Med. 2024, 2, 303–311. [Google Scholar] [CrossRef]
- Chen, X.; Yu, J.; Zhong, B.; Lu, J.; Lu, J.J.; Li, S.; Lu, Y. Pharmacological activities of dihydrotanshinone I, a natural product from Salvia miltiorrhiza Bunge. Pharmacol. Res. 2019, 145, 104254. [Google Scholar] [CrossRef] [PubMed]
- Sa, R.; He, S.; Han, D.; Liu, M.; Yu, Y.; Shang, R.; Song, M. Isolation and identification of a new biocontrol bacteria against Salvia miltiorrhiza root rot and optimization of culture conditions for antifungal substance production using response surface methodology. BMC Microbiol. 2022, 22, 231. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Deng, Y.; Ning, Y.; He, Z.; Wang, G.L. Exploiting Broad-Spectrum Disease Resistance in Crops: From Molecular Dissection to Breeding. Annu. Rev. Plant Biol. 2020, 71, 575–603. [Google Scholar] [CrossRef]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef]
- Han, G.Z. Origin and evolution of the plant immune system. New Phytol. 2019, 222, 70–83. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Remick, B.C.; Gaidt, M.M.; Vance, R.E. Effector-Triggered Immunity. Annu. Rev. Immunol. 2023, 41, 453–481. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.M.; Tang, Y.; Chen, J.Q.; Shao, Z.Q. The evolution of plant NLR immune receptors and downstream signal components. Curr. Opin. Plant Biol. 2023, 73, 102363. [Google Scholar] [CrossRef]
- Gao, A.; Hu, M.; Gong, Y.; Dong, R.; Jiang, Y.; Zhu, S.; Ji, J.; Zhang, D.; Li, S.; He, H. Pm21 CC domain activity modulated by intramolecular interactions is implicated in cell death and disease resistance. Mol. Plant Pathol. 2020, 21, 975–984. [Google Scholar] [CrossRef]
- Song, W.; Wang, B.; Li, X.; Wei, J.; Chen, L.; Zhang, D.; Zhang, W.; Li, R. Identification of Immune Related LRR-Containing Genes in Maize (Zea mays L.) by Genome-Wide Sequence Analysis. Int. J. Genom. 2015, 2015, 231358. [Google Scholar]
- Yang, X.; Wang, J. Genome-Wide Analysis of NBS-LRR Genes in Sorghum Genome Revealed Several Events Contributing to NBS-LRR Gene Evolution in Grass Species. Evol. Bioinform. Online 2016, 12, 9–21. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Xue, J.Y.; Wu, P.; Zhang, Y.M.; Wu, Y.; Hang, Y.Y.; Wang, B.; Chen, J.Q. Large-Scale Analyses of Angiosperm Nucleotide-Binding Site-Leucine-Rich Repeat Genes Reveal Three Anciently Diverged Classes with Distinct Evolutionary Patterns. Plant Physiol. 2016, 170, 2095–2109. [Google Scholar] [CrossRef]
- Glowacki, S.; Macioszek, V.K.; Kononowicz, A.K. R proteins as fundamentals of plant innate immunity. Cell Mol. Biol. Lett. 2011, 16, 1–24. [Google Scholar] [CrossRef]
- Tameling, W.I.; Elzinga, S.D.; Darmin, P.S.; Vossen, J.H.; Takken, F.L.; Haring, M.A.; Cornelissen, B.J. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 2002, 14, 2929–2939. [Google Scholar] [CrossRef]
- Yue, J.X.; Meyers, B.C.; Chen, J.Q.; Tian, D.; Yang, S. Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol. 2012, 193, 1049–1063. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, Y.; Ruan, M.; Ye, Q.; Wang, R.; Yao, Z.; Zhou, G.; Liu, C.; Wan, H. Phylogenetic, Structural, and Evolutionary Insights into Pepper NBS-LRR Resistance Genes. Int. J. Mol. Sci. 2025, 26, 1828. [Google Scholar] [CrossRef] [PubMed]
- McDowell, J.M.; Woffenden, B.J. Plant disease resistance genes: Recent insights and potential applications. Trends Biotechnol. 2003, 21, 178–183. [Google Scholar] [CrossRef]
- Bent, A.F.; Kunkel, B.N.; Dahlbeck, D.; Brown, K.L.; Schmidt, R.; Giraudat, J.; Leung, J.; Staskawicz, B.J. RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 1994, 265, 1856–1860. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Wang, X.; Wang, F.; Zhao, Z.; Li, G.; Zhu, X.; Su, J.; Chen, L. The Fungal Effector Avr-Pita Suppresses Innate Immunity by Increasing COX Activity in Rice Mitochondria. Rice 2021, 14, 12. [Google Scholar] [CrossRef]
- Matsushima, N.; Miyashita, H. Leucine-Rich Repeat (LRR) Domains Containing Intervening Motifs in Plants. Biomolecules 2012, 2, 288–311. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Locci, F.; Wanke, F.; Zhang, L.; Saile, S.C.; Joe, A.; Karelina, D.; Hua, C.; Frohlich, K.; Wan, W.L.; et al. The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 2021, 598, 495–499. [Google Scholar] [CrossRef]
- Ma, Y.; Cui, G.; Chen, T.; Ma, X.; Wang, R.; Jin, B.; Yang, J.; Kang, L.; Tang, J.; Lai, C.; et al. Expansion within the CYP71D subfamily drives the heterocyclization of tanshinones synthesis in Salvia miltiorrhiza. Nat. Commun. 2021, 12, 685. [Google Scholar] [CrossRef]
- Pan, X.; Chang, Y.; Li, C.; Qiu, X.; Cui, X.; Meng, F.; Zhang, S.; Li, X.; Lu, S. Chromosome-level genome assembly of Salvia miltiorrhiza with orange roots uncovers the role of Sm2OGD3 in catalyzing 15,16-dehydrogenation of tanshinones. Hortic Res. 2023, 10, uhad069. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Lin, C.; Xing, P.; Fen, Y.; Jin, H.; Zhou, C.; Gu, Y.Q.; Wang, J.; Li, X. A high-quality reference genome sequence of Salvia miltiorrhiza provides insights into tanshinone synthesis in its red rhizomes. Plant Genome 2020, 13, e20041. [Google Scholar] [CrossRef]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zeng, Z.; Zhang, Y.M.; Li, Q.; Jiang, X.M.; Jiang, Z.; Tang, J.H.; Chen, D.; Wang, Q.; Chen, J.Q.; et al. An angiosperm NLR Atlas reveals that NLR gene reduction is associated with ecological specialization and signal transduction component deletion. Mol. Plant 2021, 14, 2015–2031. [Google Scholar] [CrossRef]
- McDowell, J.M.; Dhandaydham, M.; Long, T.A.; Aarts, M.G.; Goff, S.; Holub, E.B.; Dangl, J.L. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 1998, 10, 1861–1874. [Google Scholar] [CrossRef] [PubMed]
- Stahl, E.A.; Dwyer, G.; Mauricio, R.; Kreitman, M.; Bergelson, J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 1999, 400, 667–671. [Google Scholar] [CrossRef]
- Chen, T.; Liu, D.; Niu, X.; Wang, J.; Qian, L.; Han, L.; Liu, N.; Zhao, J.; Hong, Y.; Liu, Y. Antiviral Resistance Protein Tm-2(2) Functions on the Plasma Membrane. Plant Physiol. 2017, 173, 2399–2410. [Google Scholar] [CrossRef]
- Zhang, Q.; Guo, N.; Zhang, Y.; Yu, Y.; Liu, S. Genome-Wide Characterization and Expression Analysis of Pathogenesis-Related 1 (PR-1) Gene Family in Tea Plant (Camellia sinensis (L.) O. Kuntze) in Response to Blister-Blight Disease Stress. Int. J. Mol. Sci. 2022, 23, 1292. [Google Scholar] [CrossRef]
- Fan, H.; Zhou, J.; Wang, Q.; Zhang, M.; Huang, Z.; Han, J.; Ying, Y.; Yu, Z.; Kai, G. Comprehensive Genome-Wide Analysis and Expression Profiling of Pathogenesis-Related Protein 1 (PR-1) Genes in Salvia miltiorrhiza. Food Sci. Nutr. 2025, 13, e70117. [Google Scholar] [CrossRef]
- Akhgari, A.; Laakso, I.; Maaheimo, H.; Choi, Y.H.; Seppanen-Laakso, T.; Oksman-Caldentey, K.M.; Rischer, H. Methyljasmonate Elicitation Increases Terpenoid Indole Alkaloid Accumulation in Rhazya stricta Hairy Root Cultures. Plants 2019, 8, 534. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Xue, J.Y.; Wang, Q.; Wang, B.; Chen, J.Q. Revisiting the Origin of Plant NBS-LRR Genes. Trends Plant Sci. 2019, 24, 9–12. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Zhang, Y.M.; Hang, Y.Y.; Xue, J.Y.; Zhou, G.C.; Wu, P.; Wu, X.Y.; Wu, X.Z.; Wang, Q.; Wang, B.; et al. Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: Understanding gained from and beyond the legume family. Plant Physiol. 2014, 166, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yuan, Y.; Zhang, Y.; Yang, S.; Zhang, X. Extreme expansion of NBS-encoding genes in Rosaceae. BMC Genet. 2015, 16, 48. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.J.; Nepal, M.P.; Purintun, J.M.; Nelson, D.; Mermigka, G.; Sarris, P.F. Wheat Disease Resistance Genes and Their Diversification Through Integrated Domain Fusions. Front. Genet. 2020, 11, 898. [Google Scholar] [CrossRef]
- Xue, J.Y.; Zhao, T.; Liu, Y.; Liu, Y.; Zhang, Y.X.; Zhang, G.Q.; Chen, H.; Zhou, G.C.; Zhang, S.Z.; Shao, Z.Q. Genome-Wide Analysis of the Nucleotide Binding Site Leucine-Rich Repeat Genes of Four Orchids Revealed Extremely Low Numbers of Disease Resistance Genes. Front. Genet. 2019, 10, 1286. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhang, Y.; Hu, Q.; Chen, J.; Li, K.; Lu, C.; Liu, H.; Wang, W.; Kuang, H. Dynamic nucleotide-binding site and leucine-rich repeat-encoding genes in the grass family. Plant Physiol. 2012, 159, 197–210. [Google Scholar] [CrossRef]
- Song, W.; Liu, L.; Yu, D.; Bernardy, H.; Jirschitzka, J.; Huang, S.; Jia, A.; Jemielniak, W.; Acker, J.; Laessle, H.; et al. Substrate-induced condensation activates plant TIR domain proteins. Nature 2024, 627, 847–853. [Google Scholar] [CrossRef]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, eaav5870. [Google Scholar] [CrossRef]
- Jacob, P.; Kim, N.H.; Wu, F.; El-Kasmi, F.; Chi, Y.; Walton, W.G.; Furzer, O.J.; Lietzan, A.D.; Sunil, S.; Kempthorn, K.; et al. Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science 2021, 373, 420–425. [Google Scholar] [CrossRef]
- Baggs, E.L.; Monroe, J.G.; Thanki, A.S.; O’Grady, R.; Schudoma, C.; Haerty, W.; Krasileva, K.V. Convergent Loss of an EDS1/PAD4 Signaling Pathway in Several Plant Lineages Reveals Coevolved Components of Plant Immunity and Drought Response. Plant Cell 2020, 32, 2158–2177. [Google Scholar] [CrossRef]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef]
- Bonardi, V.; Tang, S.; Stallmann, A.; Roberts, M.; Cherkis, K.; Dangl, J.L. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA 2011, 108, 16463–16468, Erratum in Proc. Natl. Acad. Sci. USA 2017, 114, E108. [Google Scholar] [CrossRef] [PubMed]
- Collier, S.M.; Hamel, L.P.; Moffett, P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 2011, 24, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Karasov, T.L.; Kniskern, J.M.; Gao, L.; DeYoung, B.J.; Ding, J.; Dubiella, U.; Lastra, R.O.; Nallu, S.; Roux, F.; Innes, R.W.; et al. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 2014, 512, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Traw, M.B.; Chen, J.Q.; Kreitman, M.; Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 2003, 423, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Contreras, M.P.; Harant, A.; Wu, C.H.; Derevnina, L.; Sakai, T.; Duggan, C.; Moratto, E.; Bozkurt, T.O.; Maqbool, A.; et al. An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. eLife 2019, 8, e49956. [Google Scholar] [CrossRef]
- Abdul Malik, N.A.; Kumar, I.S.; Nadarajah, K. Elicitor and Receptor Molecules: Orchestrators of Plant Defense and Immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, D.; Zhou, L.; Yu, X.; Yan, X.; Zhang, Q.; Li, B.; Liu, Y.; Zhou, W.; Cao, X.; et al. JA-Responsive Transcription Factor SmMYB97 Promotes Phenolic Acid and Tanshinone Accumulation in Salvia miltiorrhiza. J. Agric. Food Chem. 2020, 68, 14850–14862. [Google Scholar] [CrossRef] [PubMed]
- Musetti, R.; Buxa, S.V.; De Marco, F.; Loschi, A.; Polizzotto, R.; Kogel, K.H.; van Bel, A.J. Phytoplasma-triggered Ca2+ influx is involved in sieve-tube blockage. Mol. Plant Microbe Interact. 2013, 26, 379–386. [Google Scholar] [CrossRef]
- Wan, L.; Essuman, K.; Anderson, R.G.; Sasaki, Y.; Monteiro, F.; Chung, E.H.; Osborne Nishimura, E.; DiAntonio, A.; Milbrandt, J.; Dangl, J.L.; et al. TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 2019, 365, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.T.; Anderson, R.G.; Cherkis, K.A.; Law, T.F.; Liu, Q.L.; Machius, M.; Nimchuk, Z.L.; Yang, L.; Chung, E.H.; El Kasmi, F.; et al. TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E2053–E2062. [Google Scholar] [CrossRef]
- Ding, M.; Zhou, Y.; Becker, D.; Yang, S.; Krischke, M.; Scherzer, S.; Yu-Strzelczyk, J.; Mueller, M.J.; Hedrich, R.; Nagel, G.; et al. Probing plant signal processing optogenetically by two channelrhodopsins. Nature 2024, 633, 872–877. [Google Scholar] [CrossRef]
- Rahman, M.A.; Hasan, M.M.; Corpas, F.J. Leveraging light-gated channelrhodopsins for strengthening plant physiological responses. Trends Plant Sci. 2025, 30, 681–683. [Google Scholar] [CrossRef]
- Zhou, Z.; Feng, J.; Huo, J.; Qiu, S.; Zhang, P.; Wang, Y.; Li, Q.; Li, Y.; Han, C.; Feng, X.; et al. Versatile CYP98A enzymes catalyse meta-hydroxylation reveals diversity of salvianolic acids biosynthesis. Plant Biotechnol. J. 2024, 22, 1536–1548. [Google Scholar] [CrossRef]
- Chen, T.; Yang, M.; Cui, G.; Tang, J.; Shen, Y.; Liu, J.; Yuan, Y.; Guo, J.; Huang, L. IMP: Bridging the gap for medicinal plant genomics. Nucleic Acids Res. 2024, 52, D1347–D1354. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534, Erratum in Mol. Biol. Evol. 2020, 37, 2461. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Mu, H.; Chen, J.; Huang, W.; Huang, G.; Deng, M.; Hong, S.; Ai, P.; Gao, C.; Zhou, H. OmicShare tools: A zero-code interactive online platform for biological data analysis and visualization. Imeta 2024, 3, e228. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, L.; Wang, J.; Cui, G.; Tang, J.; Ma, Y.; Jin, B.; Su, P.; Zhang, Y.; Wang, Y.; Chen, T.; et al. Genome-Wide Characterization and Expression Pattern Analysis Insights into Plant NBS-LRR Gene Family of Salvia miltiorrhiza. Int. J. Mol. Sci. 2025, 26, 9063. https://doi.org/10.3390/ijms26189063
Luo L, Wang J, Cui G, Tang J, Ma Y, Jin B, Su P, Zhang Y, Wang Y, Chen T, et al. Genome-Wide Characterization and Expression Pattern Analysis Insights into Plant NBS-LRR Gene Family of Salvia miltiorrhiza. International Journal of Molecular Sciences. 2025; 26(18):9063. https://doi.org/10.3390/ijms26189063
Chicago/Turabian StyleLuo, Linglong, Jian Wang, Guanghong Cui, Jinfu Tang, Ying Ma, Baolong Jin, Ping Su, Yifeng Zhang, Yanan Wang, Tong Chen, and et al. 2025. "Genome-Wide Characterization and Expression Pattern Analysis Insights into Plant NBS-LRR Gene Family of Salvia miltiorrhiza" International Journal of Molecular Sciences 26, no. 18: 9063. https://doi.org/10.3390/ijms26189063
APA StyleLuo, L., Wang, J., Cui, G., Tang, J., Ma, Y., Jin, B., Su, P., Zhang, Y., Wang, Y., Chen, T., Guo, J., & Huang, L. (2025). Genome-Wide Characterization and Expression Pattern Analysis Insights into Plant NBS-LRR Gene Family of Salvia miltiorrhiza. International Journal of Molecular Sciences, 26(18), 9063. https://doi.org/10.3390/ijms26189063





