Update on Structure and Function of SH2 Domains: Mechanisms and Emerging Targeting Strategies
Abstract
1. Introduction
| Protein Name | Function of Lipid Association | Lipid Moiety | Ref |
|---|---|---|---|
| SYK | PIP3-dependent membrane binding is required for the activation of SYK scaffolding function, leading to noncatalytic activation of STAT3/5 | PIP3 | [7] |
| ZAP70 | Lipids are essential for facilitating and sustenance of ZAP70 Interactions with TCR-ζ n | PIP3 | [7] |
| LCK | Modulates the interaction of LCK with its binding partners in the TCR signaling complex | PIP2, PIP3 | [9] |
| ABL | Membrane recruitment and Modulation of Abl activity | PIP2 interaction | [4,10] |
| VAV2 | Modulates the interaction of VAV2 with membrane receptors, e.g., EphA2 | PIP2, PIP3 | [11] |
| C1-Ten/Tensin2 | Regulation of Abl activity and the phosphorylation of IRS-1 in insulin signaling pathways | PIP3 | [5] |
2. SH2 Domain Structure and Binding to pY-Peptide Ligands
2.1. SH2 Domain Structure
2.2. Specificity Determinants of SH2 Domain Binding to pY-Peptides
2.3. Contextual Peptide Specificity
2.4. Contribution of Loop Regions on SH2 Domain/pY-Peptide Selectivity
3. Plasticity in SH2 Domain Binding and Allostery
4. Emerging Trends in Modeling SH2 Domain/pY-Peptide Binding
4.1. Search Methods for Computational Analysis of Peptide Binding Conformations
4.2. Machine Learning and Neural Networks
5. Categories of SH2 Domain Inhibitors
5.1. Peptidomimetics
5.2. Covalent Inhibitors
5.3. Proteolysis Targeting Chimeras (PROTAC)
5.4. Competitive Orthosteric and Allosteric Small-Molecule Inhibitors
6. Small-Molecule Screening and Lead Compound Identification of SH2 Inhibitors
6.1. Structure-Based and High-Throughput Screens
6.2. Computer-Aided Methods
7. Focus on Targeting SH2-Containing Transcription Factors
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gopalasingam, P.; Quill, L.; Jeeves, M.; Overduin, M. SH2 Domain Structures and Interactions. In SH Domains: Structure, Mechanisms and Applications; Kurochkina, N., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 159–185. [Google Scholar]
- Kaneko, T.; Joshi, R.; Feller, S.M.; Li, S.S.C. Phosphotyrosine recognition domains: The typical, the atypical and the versatile. Cell Commun. Signal. 2012, 10, 32. [Google Scholar] [CrossRef]
- Leonard, T.A.; Hurley, J.H. Regulation of protein kinases by lipids. Curr. Opin. Struct. Biol. 2011, 21, 785–791. [Google Scholar] [CrossRef]
- Sipeki, S.; Koprivanacz, K.; Takács, T.; Kurilla, A.; László, L.; Vas, V.; Buday, L. Novel Roles of SH2 and SH3 Domains in Lipid Binding. Cells 2021, 10, 1191. [Google Scholar] [CrossRef]
- Kim, E.; Kim, D.-H.; Singaram, I.; Jeong, H.; Koh, A.; Lee, J.; Cho, W.; Ryu, S.H. Cellular phosphatase activity of C1-Ten/Tensin2 is controlled by Phosphatidylinositol-3,4,5-triphosphate binding through the C1-Ten/Tensin2 SH2 domain. Cell. Signal. 2018, 51, 130–138. [Google Scholar] [CrossRef]
- Park, M.-J.; Sheng, R.; Silkov, A.; Jung, D.-J.; Wang, Z.-G.; Xin, Y.; Kim, H.; Thiagarajan-Rosenkranz, P.; Song, S.; Yoon, Y.; et al. SH2 Domains Serve as Lipid-Binding Modules for pTyr-Signaling Proteins. Mol. Cell 2016, 62, 7–20. [Google Scholar] [CrossRef]
- Singaram, I.; Sharma, A.; Pant, S.; Lihan, M.; Park, M.-J.; Pergande, M.; Buwaneka, P.; Hu, Y.; Mahmud, N.; Kim, Y.-M.; et al. Targeting lipid-protein interaction to treat Syk-mediated acute myeloid leukemia. Nat. Chem. Biol. 2023, 19, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wang, F.; Zhang, J.; Zhang, Z.; Qin, L.; Peng, J.; Li, F.; Liu, J.; Lu, G.; Gong, Q.; et al. Identification and structural basis for a novel interaction between Vav2 and Arap3. J. Struct. Biol. 2012, 180, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Sheng, R.; Jung, D.J.; Silkov, A.; Kim, H.; Singaram, I.; Wang, Z.G.; Xin, Y.; Kim, E.; Park, M.J.; Thiagarajan-Rosenkranz, P.; et al. Lipids Regulate Lck Protein Activity through Their Interactions with the Lck Src Homology 2 Domain. J. Biol. Chem. 2016, 291, 17639–17650. [Google Scholar] [CrossRef]
- Tokonzaba, E.; Capelluto, D.G.; Kutateladze, T.G.; Overduin, M. Phosphoinositide, phosphopeptide and pyridone interactions of the Abl SH2 domain. Chem. Biol. Drug Des. 2006, 67, 230–237. [Google Scholar] [CrossRef]
- Ge, L.; Wu, B.; Zhang, Y.; Wang, J.; Zhao, H.; Wang, J. Biochemical and NMR characterization of the interactions of Vav2-SH2 domain with lipids and the EphA2 juxtamembrane region on membrane. Biochem. J. 2020, 477, 3791–3801. [Google Scholar] [CrossRef]
- Deng, Y.; Efremov, A.K.; Yan, J. Modulating binding affinity, specificity, and configurations by multivalent interactions. Biophys. J. 2022, 121, 1868–1880. [Google Scholar] [CrossRef]
- Jimenez Salinas, A.; Lee, Y.K. In unity, there is strength: Phase separation controls receptor tyrosine kinase signal transduction. Mol. Cell 2022, 82, 1081–1083. [Google Scholar] [CrossRef]
- Wen, Y.; Ma, J. Phase separation drives the formation of biomolecular condensates in the immune system. Front. Immunol. 2022, 13, 986589. [Google Scholar] [CrossRef]
- Ditlev, J.A.; Vega, A.R.; Köster, D.V.; Su, X.; Tani, T.; Lakoduk, A.M.; Vale, R.D.; Mayor, S.; Jaqaman, K.; Rosen, M.K. A composition-dependent molecular clutch between T cell signaling condensates and actin. eLife 2019, 8, e42695. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.E.; Bhatt, A.; Erdmann, P.S.; Hou, Z.; Maier, J.; Pirkuliyeva, S.; Engelke, M.; Becker, S.; Plitzko, J.; Wienands, J.; et al. Tripartite phase separation of two signal effectors with vesicles priming B cell responsiveness. Nat. Commun. 2020, 11, 848. [Google Scholar] [CrossRef]
- Bilal, M.Y.; Houtman, J.C. GRB2 Nucleates T Cell Receptor-Mediated LAT Clusters That Control PLC-γ1 Activation and Cytokine Production. Front. Immunol. 2015, 6, 141. [Google Scholar]
- Jaqaman, K.; Ditlev, J.A. Biomolecular condensates in membrane receptor signaling. Curr. Opin. Cell Biol. 2021, 69, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, I.; Stone, J.C.; Pawson, T. A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol. Cell Biol. 1986, 6, 4396–4408. [Google Scholar]
- Bajusz, D.; Pándy-Szekeres, G.; Takács, Á.; de Araujo, E.D.; Keserű, G.M. SH2db, an information system for the SH2 domain. Nucleic Acids Research 2023, 51, W542–W552. [Google Scholar] [CrossRef]
- Al-Lazikani, B.; Sheinerman, F.B.; Honig, B. Combining multiple structure and sequence alignments to improve sequence detection and alignment: Application to the SH2 domains of Janus kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 14796–14801. [Google Scholar] [CrossRef]
- Jaber Chehayeb, R.; Boggon, T.J. SH2 Domain Binding: Diverse FLVRs of Partnership. Front. Endocrinol. 2020, 11, 575220. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Hua, J.; Kimura, R.; Headd, J.J.; Fu, X.-y.; Chin, Y.E. Identification of the Linker-SH2 Domain of STAT as the Origin of the SH2 Domain Using Two-dimensional Structural Alignment*. Mol. Cell. Proteom. 2004, 3, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Nioche, P.; Liu, W.-Q.; Broutin, I.; Charbonnier, F.; Latreille, M.-T.; Vidal, M.; Roques, B.; Garbay, C.; Ducruix, A. Crystal structures of the SH2 domain of grb2: Highlight on the binding of a new high-affinity inhibitor11Edited by R. Huber. J. Mol. Biol. 2002, 315, 1167–1177. [Google Scholar] [CrossRef]
- Eck, M.J.; Shoelson, S.E.; Harrison, S.C. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature 1993, 362, 87–91. [Google Scholar] [CrossRef]
- Kaneko, T.; Huang, H.; Zhao, B.; Li, L.; Liu, H.; Voss, C.K.; Wu, C.; Schiller, M.R.; Li, S.S.-C. Loops Govern SH2 Domain Specificity by Controlling Access to Binding Pockets. Sci. Signal. 2010, 3, ra34. [Google Scholar] [CrossRef]
- Huang, H.; Li, L.; Wu, C.; Schibli, D.; Colwill, K.; Ma, S.; Li, C.; Roy, P.; Ho, K.; Songyang, Z.; et al. Defining the Specificity Space of the Human Src Homology 2 Domain*. Mol. Cell. Proteom. 2008, 7, 768–784. [Google Scholar] [CrossRef]
- Liu, B.A.; Nash, P.D. Evolution of SH2 domains and phosphotyrosine signalling networks. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2556–2573. [Google Scholar] [CrossRef]
- Petsalaki, E.; Russell, R.B. Peptide-mediated interactions in biological systems: New discoveries and applications. Curr. Opin. Biotechnol. 2008, 19, 344–350. [Google Scholar] [CrossRef]
- Diop, A.; Santorelli, D.; Malagrinò, F.; Nardella, C.; Pennacchietti, V.; Pagano, L.; Marcocci, L.; Pietrangeli, P.; Gianni, S.; Toto, A. SH2 Domains: Folding, Binding and Therapeutical Approaches. Int. J. Mol. Sci. 2022, 23, 15944. [Google Scholar] [CrossRef] [PubMed]
- Ivarsson, Y.; Jemth, P. Affinity and specificity of motif-based protein–protein interactions. Curr. Opin. Struct. Biol. 2019, 54, 26–33. [Google Scholar] [CrossRef]
- Liu, B.A.; Jablonowski, K.; Shah, E.E.; Engelmann, B.W.; Jones, R.B.; Nash, P.D. SH2 domains recognize contextual peptide sequence information to determine selectivity. Mol. Cell Proteom. 2010, 9, 2391–2404. [Google Scholar] [CrossRef] [PubMed]
- Amanchy, R.; Kandasamy, K.; Mathivanan, S.; Periaswamy, B.; Reddy, R.; Yoon, W.H.; Joore, J.; Beer, M.A.; Cope, L.; Pandey, A. Identification of Novel Phosphorylation Motifs Through an Integrative Computational and Experimental Analysis of the Human Phosphoproteome. J. Proteom. Bioinform. 2011, 4, 22–35. [Google Scholar] [CrossRef]
- Visconti, L.; Toto, A.; Jarvis, J.A.; Troilo, F.; Malagrinò, F.; De Simone, A.; Gianni, S. Demonstration of Binding Induced Structural Plasticity in a SH2 Domain. Front. Mol. Biosci. 2020, 7, 89. [Google Scholar] [CrossRef]
- Kay, L.E.; Muhandiram, D.R.; Wolf, G.; Shoelson, S.E.; Forman-Kay, J.D. Correlation between binding and dynamics at SH2 domain interfaces. Nat. Struct. Biol. 1998, 5, 156–163. [Google Scholar] [CrossRef]
- Huang, W.Y.C.; Ditlev, J.A.; Chiang, H.-K.; Rosen, M.K.; Groves, J.T. Allosteric Modulation of Grb2 Recruitment to the Intrinsically Disordered Scaffold Protein, LAT, by Remote Site Phosphorylation. J. Am. Chem. Soc. 2017, 139, 18009–18015. [Google Scholar] [CrossRef]
- Sanches, K.; Caruso, I.P.; Almeida, F.C.L.; Melo, F.A. The dynamics of free and phosphopeptide-bound Grb2-SH2 reveals two dynamically independent subdomains and an encounter complex with fuzzy interactions. Sci. Rep. 2020, 10, 13040. [Google Scholar] [CrossRef]
- Kuriyan, J.; Eisenberg, D. The origin of protein interactions and allostery in colocalization. Nature 2007, 450, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhao, W.C.; Fu, X.Q.; Zheng, Q.C. Exploring the Allosteric Mechanism of Src Homology-2 Domain-Containing Protein Tyrosine Phosphatase 2 (SHP2) by Molecular Dynamics Simulations. Front. Chem. 2020, 8, 597495. [Google Scholar] [CrossRef]
- Duarte, D.P.; Lamontanara, A.J.; La Sala, G.; Jeong, S.; Sohn, Y.K.; Panjkovich, A.; Georgeon, S.; Kükenshöner, T.; Marcaida, M.J.; Pojer, F.; et al. Btk SH2-kinase interface is critical for allosteric kinase activation and its targeting inhibits B-cell neoplasms. Nat. Commun. 2020, 11, 2319. [Google Scholar] [CrossRef] [PubMed]
- Register, A.C.; Leonard, S.E.; Maly, D.J. SH2-catalytic domain linker heterogeneity influences allosteric coupling across the SFK family. Biochemistry 2014, 53, 6910–6923. [Google Scholar] [CrossRef]
- Mertens, C.; Haripal, B.; Klinge, S.; Darnell, J.E. Mutations in the linker domain affect phospho-STAT3 function and suggest targets for interrupting STAT3 activity. Proc. Natl. Acad. Sci. USA 2015, 112, 14811–14816. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Karki, N.; Zoltowski, B.D.; Matthews, D.A. Allosteric regulation in STAT3 interdomains is mediated by a rigid core: SH2 domain regulation by CCD in D170A variant. PLoS Comput. Biol. 2022, 18, e1010794. [Google Scholar] [CrossRef] [PubMed]
- Gangopadhyay, K.; Manna, B.; Roy, S.; Kumari, S.; Debnath, O.; Chowdhury, S.; Ghosh, A.; Das, R. An allosteric hot spot in the tandem-SH2 domain of ZAP-70 regulates T-cell signaling. Biochem. J. 2020, 477, 1287–1308. [Google Scholar] [CrossRef]
- Fraser, J.S.; Murcko, M.A. Structure is beauty, but not always truth. Cell 2024, 187, 517–520. [Google Scholar] [CrossRef]
- Holton, J.M.; Classen, S.; Frankel, K.A.; Tainer, J.A. The R-factor gap in macromolecular crystallography: An untapped potential for insights on accurate structures. FEBS J. 2014, 281, 4046–4060. [Google Scholar] [CrossRef]
- Torrens-Fontanals, M.; Stepniewski, T.M.; Aranda-García, D.; Morales-Pastor, A.; Medel-Lacruz, B.; Selent, J. How Do Molecular Dynamics Data Complement Static Structural Data of GPCRs. Int. J. Mol. Sci. 2020, 21, 5933. [Google Scholar] [CrossRef]
- Dölker, N.; Górna, M.W.; Sutto, L.; Torralba, A.S.; Superti-Furga, G.; Gervasio, F.L. The SH2 domain regulates c-Abl kinase activation by a cyclin-like mechanism and remodulation of the hinge motion. PLoS Comput. Biol. 2014, 10, e1003863. [Google Scholar] [CrossRef]
- Suenaga, A.; Hatakeyama, M.; Ichikawa, M.; Yu, X.; Futatsugi, N.; Narumi, T.; Fukui, K.; Terada, T.; Taiji, M.; Shirouzu, M.; et al. Molecular dynamics, free energy, and SPR analyses of the interactions between the SH2 domain of Grb2 and ErbB phosphotyrosyl peptides. Biochemistry 2003, 42, 5195–5200. [Google Scholar] [CrossRef]
- Sutto, L.; Mereu, I.; Gervasio, F.L. A Hybrid All-Atom Structure-Based Model for Protein Folding and Large Scale Conformational Transitions. J. Chem. Theory Comput. 2011, 7, 4208–4217. [Google Scholar] [CrossRef] [PubMed]
- Anselmi, M.; Hub, J.S. The loops of the N-SH2 binding cleft do not serve as allosteric switch in SHP2 activation. Proc. Natl. Acad. Sci. USA 2021, 118, e2025107118. [Google Scholar] [CrossRef]
- Caldararu, O.; Ekberg, V.; Logan, D.T.; Oksanen, E.; Ryde, U. Exploring ligand dynamics in protein crystal structures with ensemble refinement. Acta Crystallogr. D Struct. Biol. 2021, 77, 1099–1115. [Google Scholar] [CrossRef]
- Matsuura, Y. High-resolution structural analysis shows how different crystallographic environments can induce alternative modes of binding of a phosphotyrosine peptide to the SH2 domain of Fer tyrosine kinase. Protein Sci. 2019, 28, 2011–2019. [Google Scholar]
- Nachman, J.; Gish, G.; Virag, C.; Pawson, T.; Pomès, R.; Pai, E. Conformational determinants of phosphotyrosine peptides complexed with the Src SH2 domain. PLoS ONE 2010, 5, e11215. [Google Scholar] [CrossRef] [PubMed]
- Kasembeli, M.M.; Xu, X.; Tweardy, D.J. SH2 domain binding to phosphopeptide ligands: Potential for drug targeting. Front. Biosci. 2009, 14, 1010–1022. [Google Scholar] [CrossRef]
- Kundu, K.; Costa, F.; Huber, M.; Reth, M.; Backofen, R. Semi-supervised prediction of SH2-peptide interactions from imbalanced high-throughput data. PLoS ONE 2013, 8, e62732. [Google Scholar] [CrossRef] [PubMed]
- AlQuraishi, M.; Koytiger, G.; Jenney, A.; MacBeath, G.; Sorger, P.K. A multiscale statistical mechanical framework integrates biophysical and genomic data to assemble cancer networks. Nat. Genet. 2014, 46, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Ronan, T.; Garnett, R.; Naegle, K.M. New analysis pipeline for high-throughput domain-peptide affinity experiments improves SH2 interaction data. J. Biol. Chem. 2020, 295, 11346–11363. [Google Scholar] [CrossRef]
- Kundu, K.; Backofen, R. An Efficient Semi-supervised Learning Approach to Predict SH2 Domain Mediated Interactions. Methods Mol. Biol. 2017, 1555, 83–97. [Google Scholar]
- Schoepfer, J.; Gay, B.; Caravatti, G.; Garcia-Echeverria, C.; Fretz, H.; Rahuel, J.; Furet, P. Structure-based design of peptidomimetic ligands of the Grb2-SH2 domain. Bioorg Med. Chem. Lett. 1998, 8, 2865–2870. [Google Scholar] [CrossRef]
- Kuil, J.; van Wandelen, L.T.; de Mol, N.J.; Liskamp, R.M. Peptidomimetic ligands for the tandem SH2 domain of the Syk protein involved in signal transduction. Adv. Exp. Med. Biol. 2009, 611, 81–82. [Google Scholar]
- Plummer, M.S.; Lunney, E.A.; Para, K.S.; Shahripour, A.; Stankovic, C.J.; Humblet, C.; Fergus, J.H.; Marks, J.S.; Herrera, R.; Hubbell, S.; et al. Design of peptidomimetic ligands for the pp60src SH2 domain. Bioorg Med. Chem. 1997, 5, 41–47. [Google Scholar] [CrossRef]
- Mandal, P.K.; Liao, W.S.; McMurray, J.S. Synthesis of phosphatase-stable, cell-permeable peptidomimetic prodrugs that target the SH2 domain of Stat3. Org. Lett. 2009, 11, 3394–3397. [Google Scholar] [CrossRef]
- Mandal, P.K.; Morlacchi, P.; Knight, J.M.; Link, T.M.; Lee, G.R.t.; Nurieva, R.; Singh, D.; Dhanik, A.; Kavraki, L.; Corry, D.B.; et al. Targeting the Src Homology 2 (SH2) Domain of Signal Transducer and Activator of Transcription 6 (STAT6) with Cell-Permeable, Phosphatase-Stable Phosphopeptide Mimics Potently Inhibits Tyr641 Phosphorylation and Transcriptional Activity. J. Med. Chem. 2015, 58, 8970–8984. [Google Scholar] [CrossRef]
- McCusker, C.T.; Wang, Y.; Shan, J.; Kinyanjui, M.W.; Villeneuve, A.; Michael, H.; Fixman, E.D. Inhibition of experimental allergic airways disease by local application of a cell-penetrating dominant-negative STAT-6 peptide. J. Immunol. 2007, 179, 2556–2564. [Google Scholar] [CrossRef]
- Bobone, S.; Pannone, L.; Biondi, B.; Solman, M.; Flex, E.; Canale, V.C.; Calligari, P.; De Faveri, C.; Gandini, T.; Quercioli, A.; et al. Targeting Oncogenic Src Homology 2 Domain-Containing Phosphatase 2 (SHP2) by Inhibiting Its Protein-Protein Interactions. J. Med. Chem. 2021, 64, 15973–15990. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Yu, T.; Li, X.; Zhang, N.; Foster, L.J.; Peng, C.; Huang, W.; He, G. Recent advances in targeting the “undruggable” proteins: From drug discovery to clinical trials. Signal Transduct. Target. Ther. 2023, 8, 335. [Google Scholar] [CrossRef]
- Ramachandran, S.; Makukhin, N.; Haubrich, K.; Nagala, M.; Forrester, B.; Lynch, D.M.; Casement, R.; Testa, A.; Bruno, E.; Gitto, R.; et al. Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2. Nat. Commun. 2023, 14, 6345. [Google Scholar] [CrossRef] [PubMed]
- Bashore, F.M.; Katis, V.L.; Du, Y.; Sikdar, A.; Wang, D.; Bradshaw, W.J.; Rygiel, K.A.; Leisner, T.M.; Chalk, R.; Mishra, S.; et al. Characterization of covalent inhibitors that disrupt the interaction between the tandem SH2 domains of SYK and FCER1G phospho-ITAM. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Mo, J.; Zhang, Y.; Peng, K.; Li, H.; Ouyang, S.; Feng, Z.; Fang, W.; Wei, J.; Rong, D.; et al. Boronic Acid: A Novel Pharmacophore Targeting Src Homology 2 (SH2) Domain of STAT3. J. Med. Chem. 2022, 65, 13094–13111. [Google Scholar] [CrossRef]
- Zheng, M.; Liu, Y.; Wu, C.; Yang, K.; Wang, Q.; Zhou, Y.; Chen, L.; Li, H. Novel PROTACs for degradation of SHP2 protein. Bioorg Chem. 2021, 110, 104788. [Google Scholar] [CrossRef]
- Kaneshige, A.; Bai, L.; Wang, M.; McEachern, D.; Meagher, J.L.; Xu, R.; Kirchhoff, P.D.; Wen, B.; Sun, D.; Stuckey, J.A.; et al. Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia. J. Med. Chem. 2023, 66, 2717–2743. [Google Scholar] [CrossRef] [PubMed]
- A Phase 1, Multicenter, Open-Label, Dose-Escalation and Expansion Study to Evaluate Safety, Tolerability, Pharmacokinetics, Pharmacodynamics, and Clinical Activity of Intravenously Administered KT-333 in Adult Patients with Relapsed or Refractory Lymphomas, Large Granular Lymphocytic Leukemia, and Solid Tumors. In 2021.
- A Phase 1, Randomized, Placebo-Controlled, First-in-Human, Single and Multiple Ascending Dose Study Designed to Evaluate the Safety, Tolerability, Pharmacokinetics, and Pharmacodynamics of Orally Administered KT-621 in Healthy Adult Participants. In 2024.
- Rehman, A.U.; Zhao, C.; Wu, Y.; Zhu, Q.; Luo, R. Targeting SHP2 Cryptic Allosteric Sites for Effective Cancer Therapy. Int. J. Mol. Sci. 2024, 25, 6201. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Bharadwaj, U.; Eckols, T.K.; Kolosov, M.; Wu, H.; Cruz-Pavlovich, F.J.S.; Shaw, A.; Ifelayo, O.I.; Zhao, H.; Kasembeli, M.M.; et al. Novel STAT3 small-molecule inhibitors identified by structure-based virtual ligand screening incorporating SH2 domain flexibility. Pharmacol. Res. 2021, 169, 105637. [Google Scholar] [CrossRef] [PubMed]
- Vajda, S.; Beglov, D.; Wakefield, A.E.; Egbert, M.; Whitty, A. Cryptic binding sites on proteins: Definition, detection, and druggability. Curr. Opin. Chem. Biol. 2018, 44, 1–8. [Google Scholar] [CrossRef]
- Kang, Z.; Li, S.; Li, Y.; Song, J.; Peng, Y.; Chen, Y. Small molecular inhibitors and degraders targeting STAT3 for cancer therapy: An updated review (from 2022 to 2024). Chin. Chem. Lett. 2024, 36, 110447. [Google Scholar] [CrossRef]
- Greisman, J.B.; Willmore, L.; Yeh, C.Y.; Giordanetto, F.; Shahamadtar, S.; Nisonoff, H.; Maragakis, P.; Shaw, D.E. Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography. J. Chem. Inf. Model. 2023, 63, 2644–2650. [Google Scholar] [CrossRef]
- Samanta, P.; Doerksen, R.J. Identifying p56lck SH2 Domain Inhibitors Using Molecular Docking and In Silico Scaffold Hopping. Appl. Sci. 2024, 14, 4277. [Google Scholar] [CrossRef]
- Kraskouskaya, D.; Duodu, E.; Arpin, C.C.; Gunning, P.T. Progress towards the development of SH2 domain inhibitors. Chem. Soc. Rev. 2013, 42, 3337–3370. [Google Scholar] [CrossRef]
- Day, J.E.H.; Berdini, V.; Castro, J.; Chessari, G.; Davies, T.G.; Day, P.J.; St Denis, J.D.; Fujiwara, H.; Fukaya, S.; Hamlett, C.C.F.; et al. Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2). Correction in J. Med. Chem. 2024, 67, 4655–4675. [Google Scholar] [CrossRef]
- Potjewyd, F.M.; Katis, V.; Du, Y.; Bradshaw, W.; Sikdar, A.; Nwogbo, F.; Leisner, T.; Wang, D.; Hardy, B.P.; Kireev, D.B. Towards the development of a chemical probe targeting the disruption of SYK tandem SH2 domain and FCεR1γ interactions for Alzheimer’s disease. Alzheimer’s Dement. 2023, 19, e067392. [Google Scholar] [CrossRef]
- Liang, J.; Lambrecht, M.J.; Arenzana, T.L.; Aubert-Nicol, S.; Bao, L.; Broccatelli, F.; Cai, J.; Eidenschenk, C.; Everett, C.; Garner, T.; et al. Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding. ACS Med. Chem. Lett. 2024, 15, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, U.; Kasembeli, M.M.; Robinson, P.; Tweardy, D.J. Targeting Janus Kinases and Signal Transducer and Activator of Transcription 3 To Treat Inflammation, Fibrosis, and Cancer: Rationale, Progress, and Caution. Correction in Pharmacol. Rev. 2020, 72, 486–526. [Google Scholar] [CrossRef]
- Shao, H.; Xu, X.; Mastrangelo, M.-A.A.; Jing, N.; Cook, R.G.; Legge, G.B.; Tweardy, D.J. Structural Requirements for Signal Transducer and Activator of Transcription 3 Binding to Phosphotyrosine Ligands Containing the Y<em>XX</em>Q Motif *. J. Biol. Chem. 2004, 279, 18967–18973. [Google Scholar]
- Xu, X.; Kasembeli, M.M.; Jiang, X.; Tweardy, B.J.; Tweardy, D.J. Chemical probes that competitively and selectively inhibit Stat3 activation. PLoS ONE 2009, 4, e4783. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Vining, D.J.; Arora, S.P.; Achaval, S.d.; Larson, J.; Cartwright, C.; Avritscher, R.; Alibhai, I.; Kaseb, A.O. Phase 1 trial evaluating TTI-101, a first-in-class, orally bioavailable, small molecule, inhibitor of STAT3, in patients with advanced solid tumors. J. Clin. Oncol. 2023, 41, 3018. [Google Scholar] [CrossRef]
- REVERT-IPF: A Phase 2 Multicenter, Randomized, Double-blind, Placebo-controlled Study to Evaluate the Safety, Tolerability, Pharmacokinetics, and Efficacy of TTI-101 in Participants With Idiopathic Pulmonary Fibrosis. In 2023.
- Liao, Z.; Gu, L.; Vergalli, J.; Mariani, S.A.; De Dominici, M.; Lokareddy, R.K.; Dagvadorj, A.; Purushottamachar, P.; McCue, P.A.; Trabulsi, E.; et al. Structure-Based Screen Identifies a Potent Small Molecule Inhibitor of Stat5a/b with Therapeutic Potential for Prostate Cancer and Chronic Myeloid Leukemia. Mol. Cancer Ther. 2015, 14, 1777–1793. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
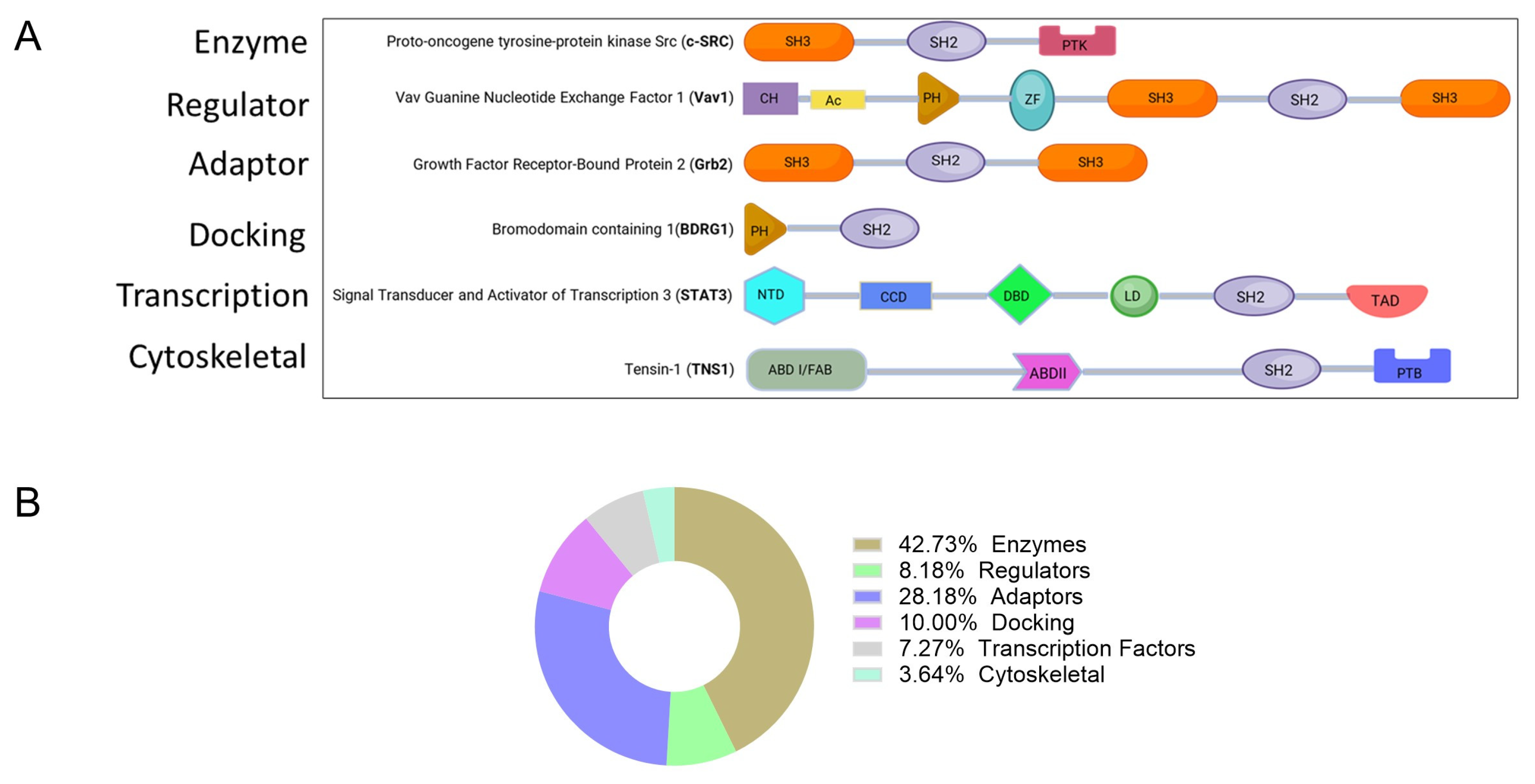
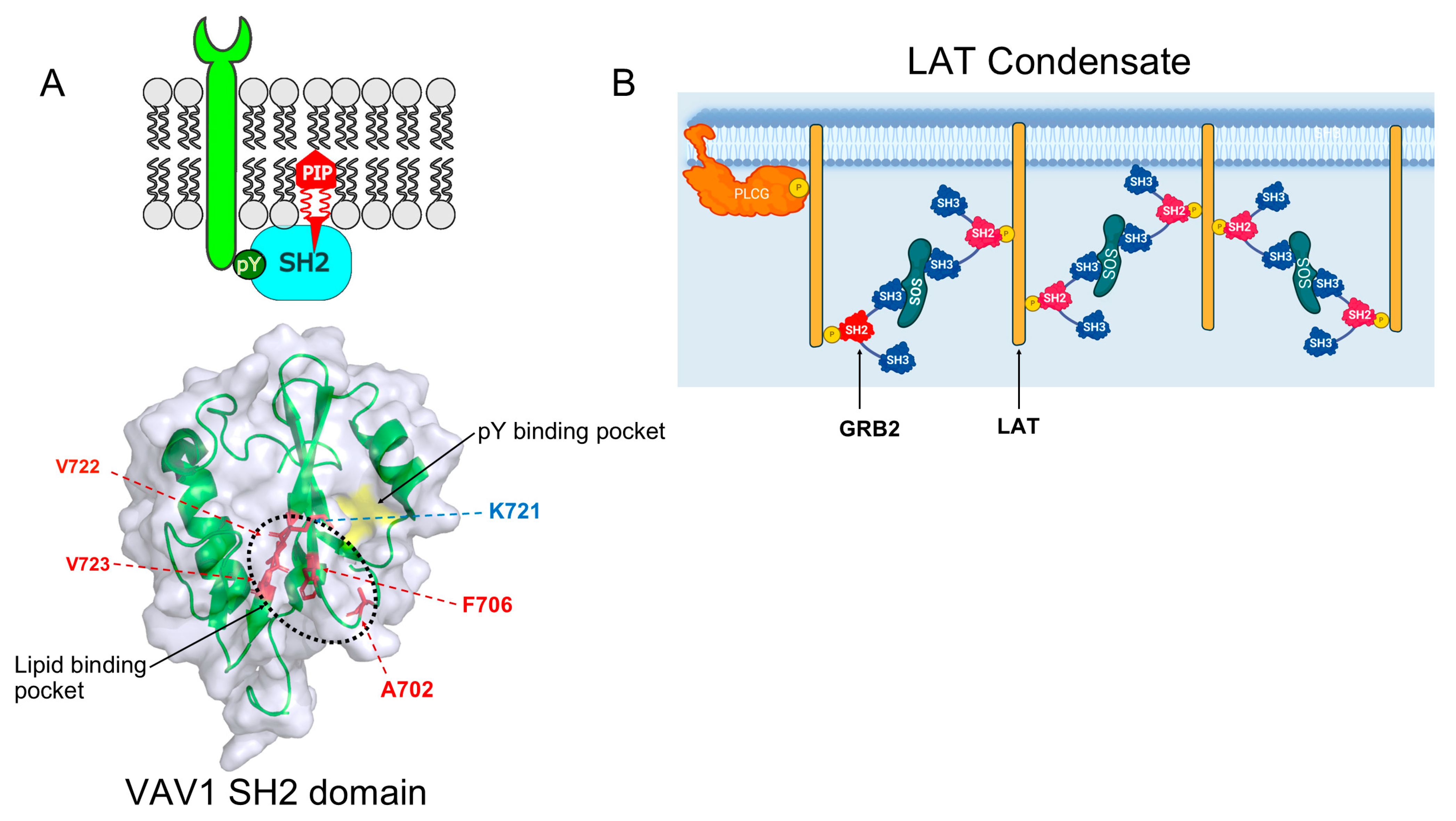
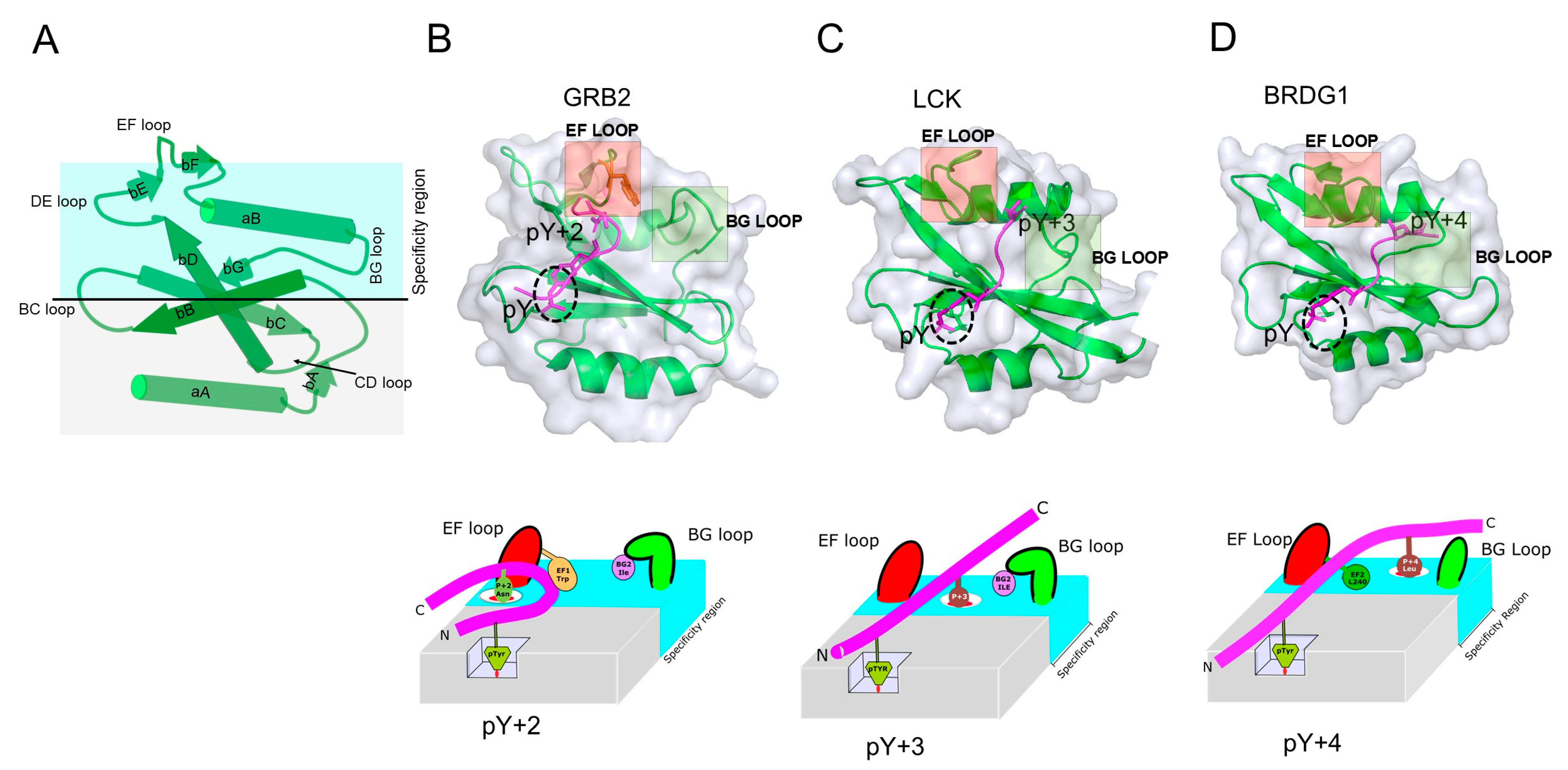
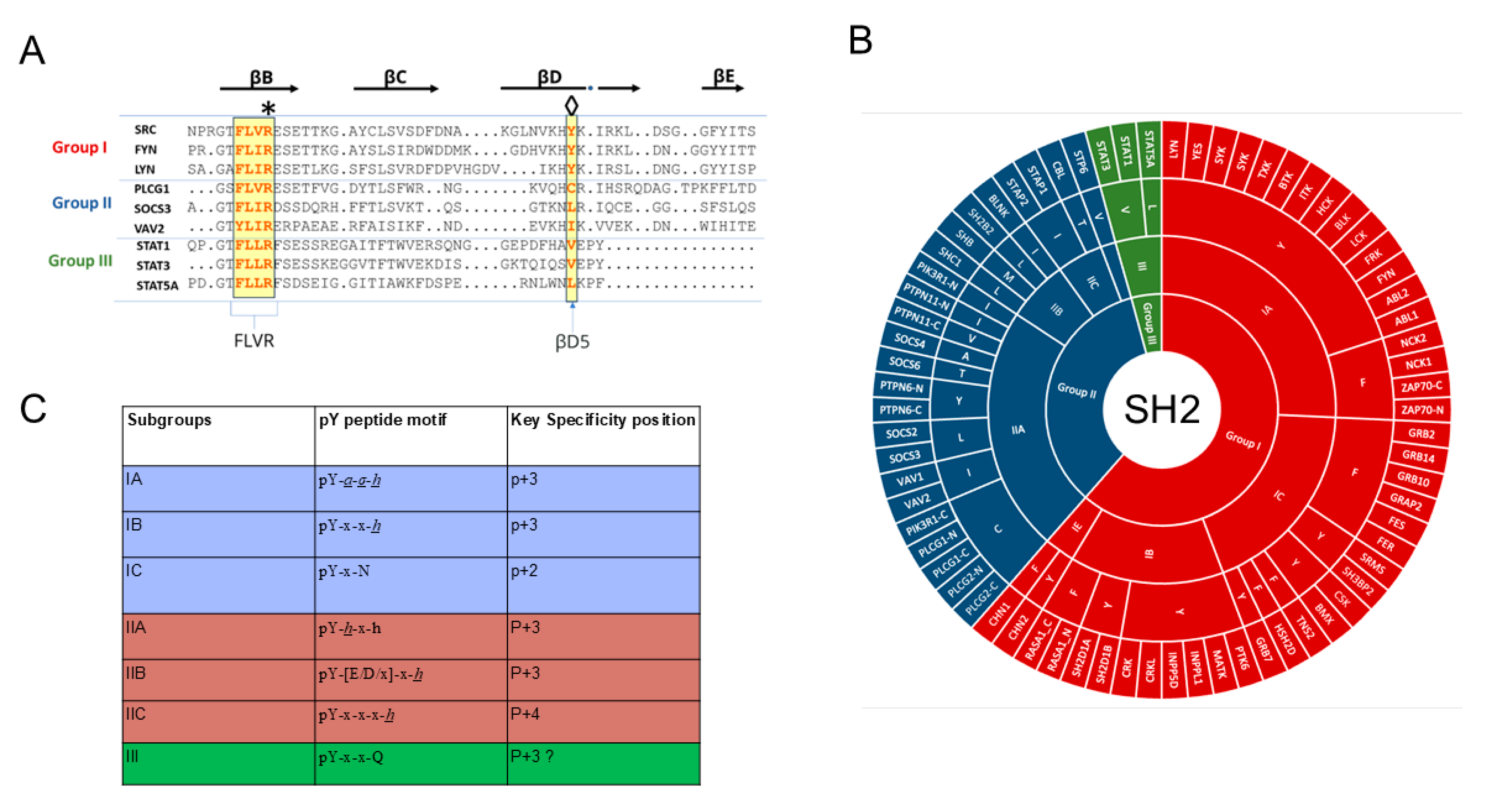

| Function | Protein |
|---|---|
| Enzymes | ABL1, ABL2, CBL, CSK, MATK, FER, JAK2, PIK3R2, PLCG1, PTPN6, PTPN11, SOCS2, SOCS4, SOCS6, SRC, FYN, LCK, HCK, ZAP70, SYK, BMX, BTK, TXK, FPS, FRK, BRK, SRMS, TYK2, JAK1, JAK3, PIK3R3, PLCG2, SHIP1, SHIP2, SOCS1, SOCS3, SOCS5, SOCS7, CISH, FGR, YES, BLK, TEC, ITK |
| Regulator (GTPase activity activator) | CHN1, CHN2, RASA1, VAV1, VAV2, RIN1, RIN2, RIN3, VAV3 |
| Adaptor proteins | CRK, CRKL, GRB2, GRB7, GRB10, GRB14, NCK1, NCK2, SH2D1A, HSH2D, SH2D1B, DAPP1, GADS GRAP, APS, LNK, SH2B, SH2D2A, SH2D7, SH2D3A SH2D3C, BCAR3, SH2D4A, SH2D4B, SH2D5, SLAP, SLAP2, BLNK, LCP2 CLNK, LCP2, CLNK |
| Docking proteins | BRDG1, SHC1, SH3BP2, SHB, SHD, SHE, SHF, SHC2, SHC3, SHC4, BKS |
| Transcription factor | STAT1, STAT2, STAT3, STAT4, STAT5, STAT5B, STAT6, SUPT6H |
| Cytoskeletal protein | TNS1, TENS2, TNS3, TNS4 |
| Condensate Complex | Role | SH2-Containing Proteins | Ref |
|---|---|---|---|
| FGFR2:SHP2:PLCγ1 | Increased activity of RTK Signaling | SHP2, PLCγ1 | [13] |
| LAT-GRB2-SOS1 | The ligand binding/T-cell activation and phosphorylation. | ZAP70, LCK, GRB2, PLCγ1 | [14] |
| N-WASP–NCK | T-cell signaling | NCK | [15] |
| SLP65, CIN85 | B-cell signaling | SLP65 | [16] |
| Target | Drug | Targeting Mechanism | Phase | Identifier |
|---|---|---|---|---|
| STAT3 | TTI-101 (C188-9) | STAT3 SH2 domain | I (Completed) II (Liver cancer) II (Lung fibrosis) | NCT03195699 NCT05440708 NCT05671835 |
| STAT3 | WP1066 | STAT3/JAK2 | I/II | NCT04334863 |
| STAT3 | Napabucasin (BBI608 or GB201) | STAT3 SH2 domain | II/III | NCT03721744 |
| STAT3 | DSP-0337 | STAT3 SH2 domain | I | NCT03416816 |
| STAT3 | SC-43 | STAT3/SHP-1 | I/II | NCT04733521 |
| STAT3 | Silibinin | STAT3 SH2 domain | Not applicable | NCT05689619 |
| STAT3 | YY201 (YY002) | STAT3 SH2 domain | I | NCT06225856 |
| STAT3 | KT-333 | STAT3 degrader | I | NCT05225584 |
| Shp2 | TNO155 | N-SH2/C-SH2/PTP interface | ||
| Shp | JAB-3312 | “latch” N-SH2/PTP | I/II/III | NCT05288205 NCT06416410 CTR20241931 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasembeli, M.M.; Rodas, J.; Tweardy, D.J. Update on Structure and Function of SH2 Domains: Mechanisms and Emerging Targeting Strategies. Int. J. Mol. Sci. 2025, 26, 9060. https://doi.org/10.3390/ijms26189060
Kasembeli MM, Rodas J, Tweardy DJ. Update on Structure and Function of SH2 Domains: Mechanisms and Emerging Targeting Strategies. International Journal of Molecular Sciences. 2025; 26(18):9060. https://doi.org/10.3390/ijms26189060
Chicago/Turabian StyleKasembeli, Moses M., Jorge Rodas, and David J. Tweardy. 2025. "Update on Structure and Function of SH2 Domains: Mechanisms and Emerging Targeting Strategies" International Journal of Molecular Sciences 26, no. 18: 9060. https://doi.org/10.3390/ijms26189060
APA StyleKasembeli, M. M., Rodas, J., & Tweardy, D. J. (2025). Update on Structure and Function of SH2 Domains: Mechanisms and Emerging Targeting Strategies. International Journal of Molecular Sciences, 26(18), 9060. https://doi.org/10.3390/ijms26189060





