SenolyticSynergy: An Attention-Based Network for Discovering Novel Senolytic Combinations via Human Aging Genomics
Abstract
1. Introduction
2. Results and Discussion
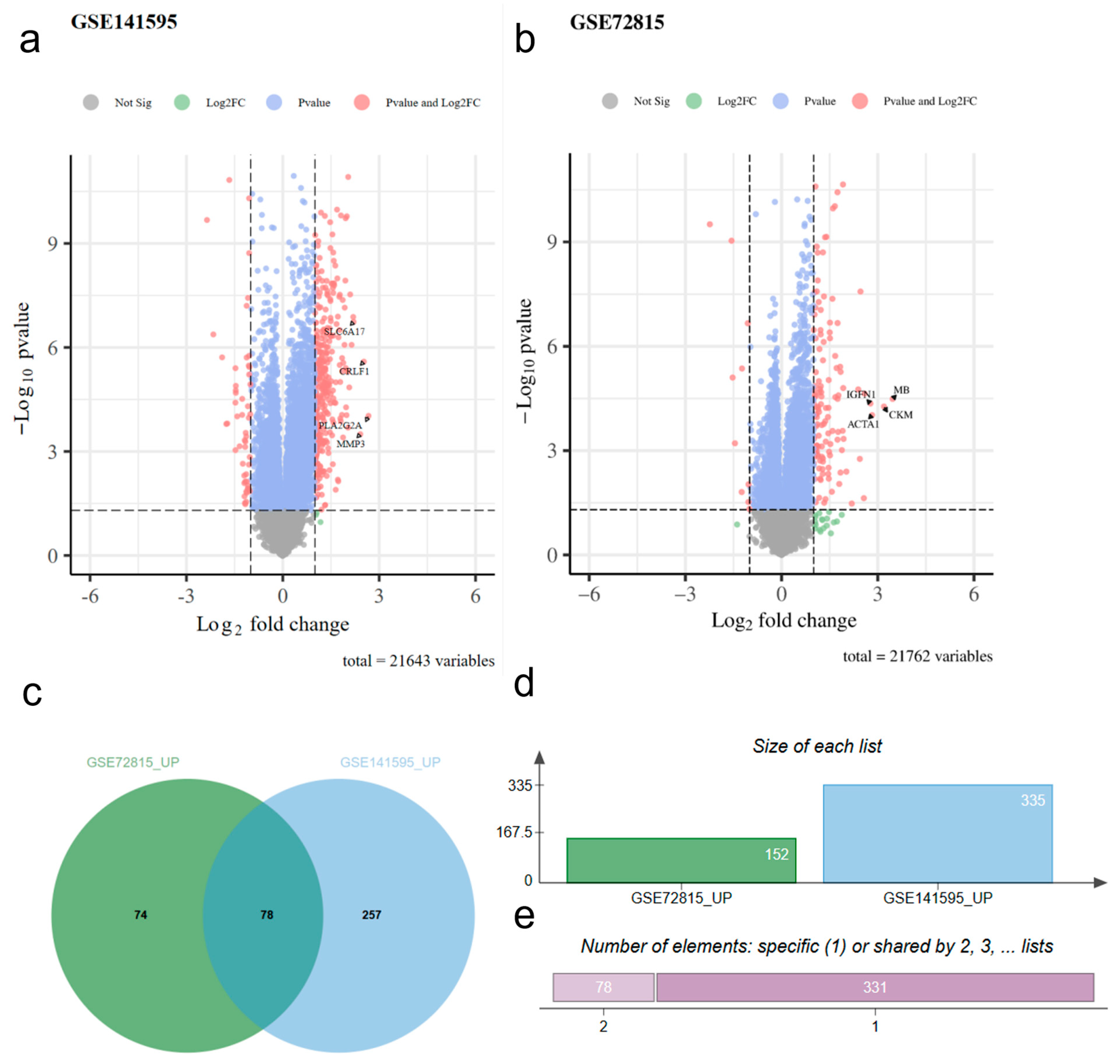
2.1. Differential Genetic Analysis
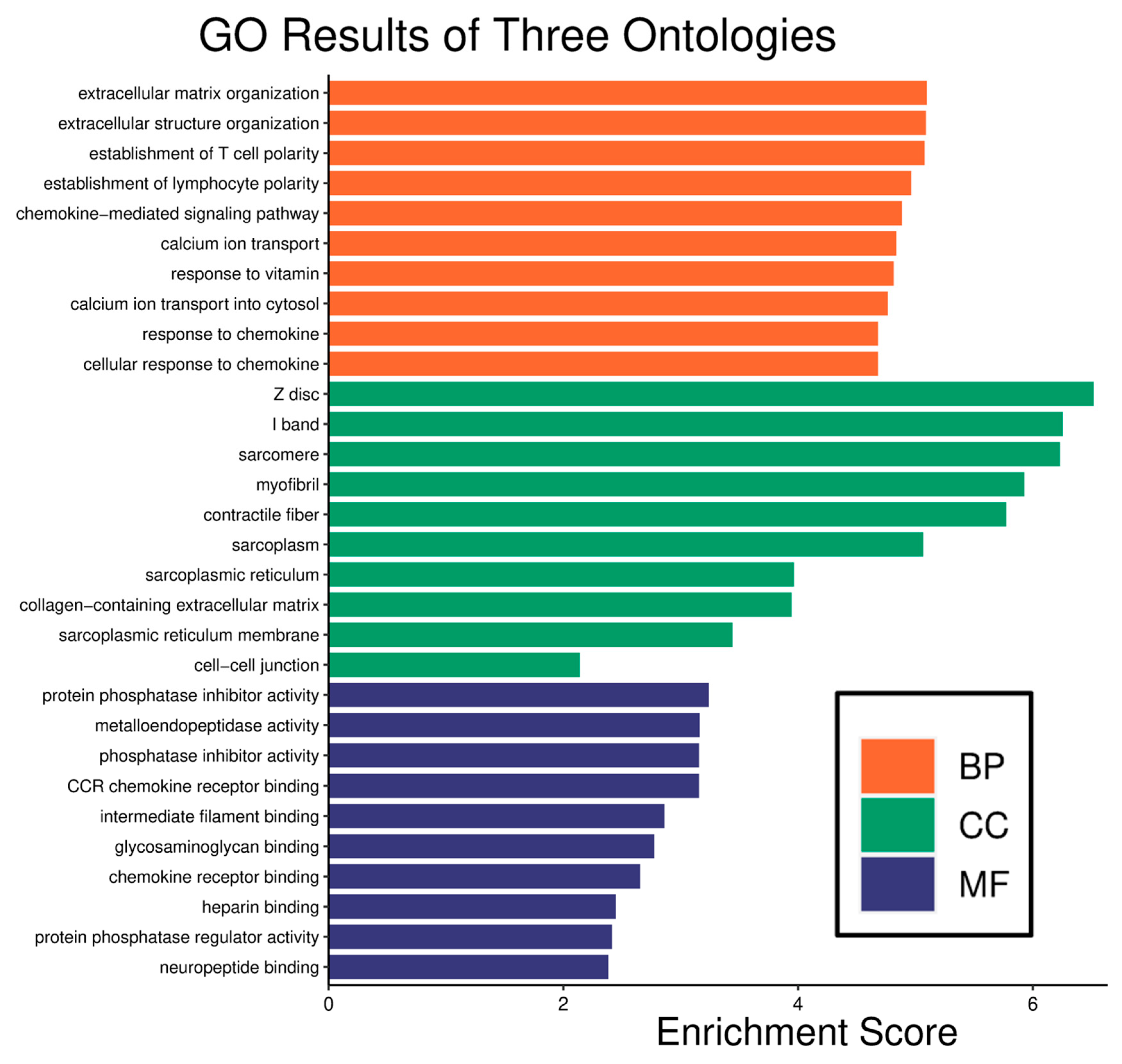
2.2. Enrichment Analysis
2.3. Pathway Analysis
2.4. Synergy Prediction and Interpretation
2.5. Verification
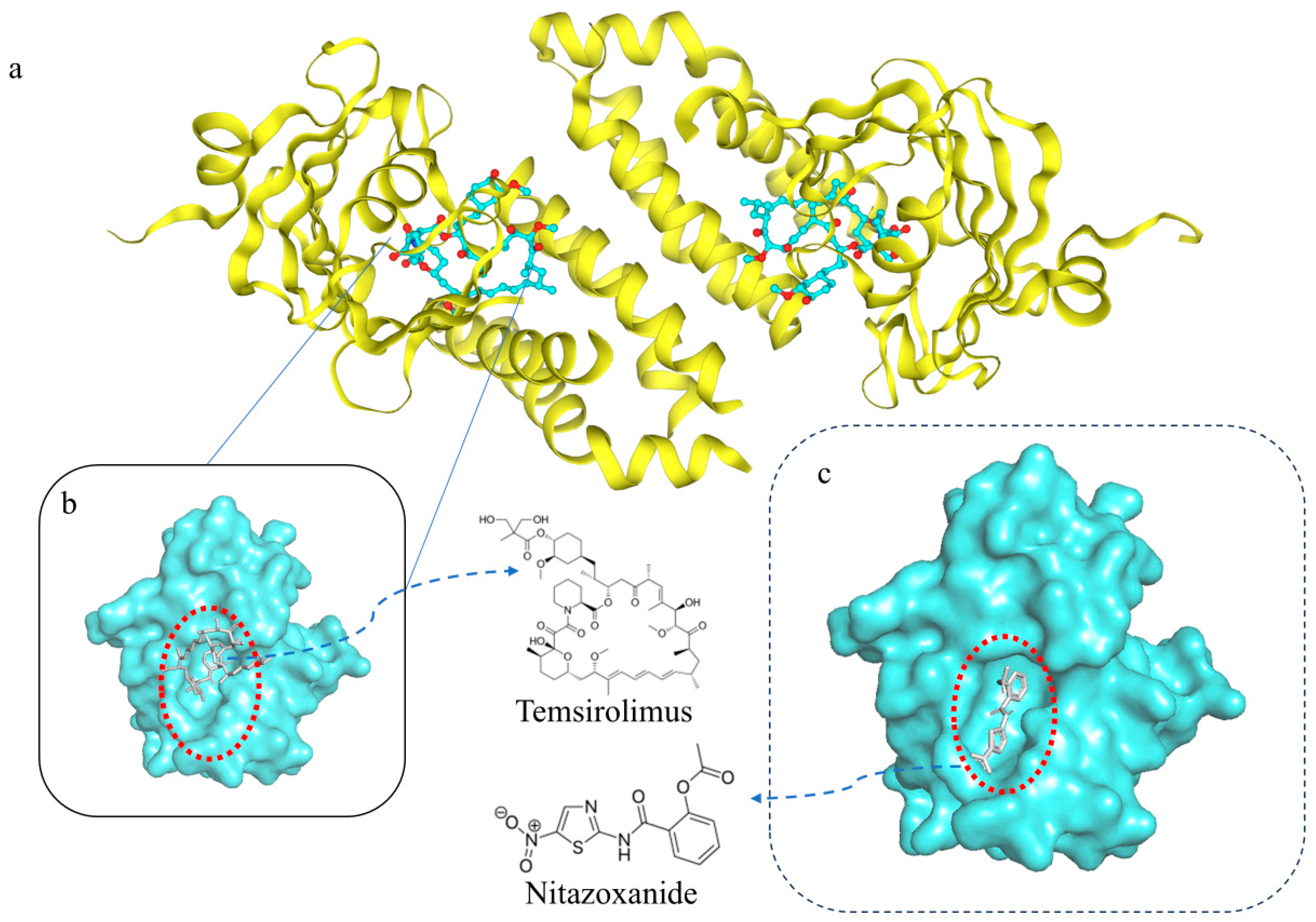
2.6. Molecular Docking Verification
2.7. Literature Verification
3. Materials and Methods
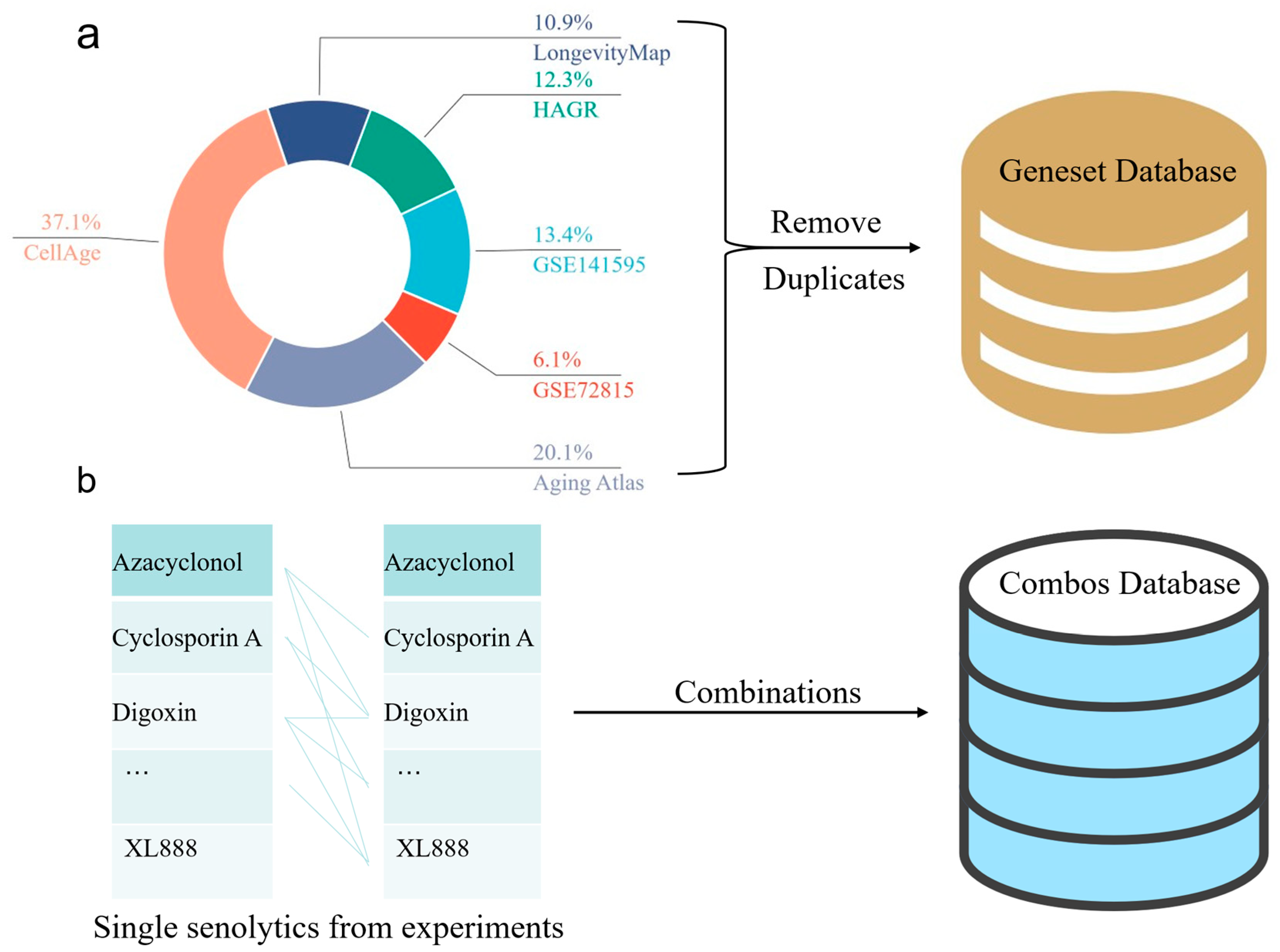
3.1. Youth-Old Age Differential Gene Expression Dataset
3.2. Senolytics Dataset
3.3. Aging-Related Target Gene Dataset
- (1)
- From the differential gene analysis of GSE72815, 152 entries with logFC > 1 and 11 entries with logFC < −1 were selected.
- (2)
- From the differential gene analysis of GSE141595, 335 entries with logFC > 1 and 90 with logFC < −1 were selected.
- (3)
- The Human Ageing Genomic Resources (HAGR) database [64], specifically the GenAge resource package (https://www.genomics.senescence.info/genes/index.html (accessed on 4 September 2024)), was used to download the latest stable version of human aging-related genes (https://www.genomics.senescence.info/genes/human_genes.zip (accessed on 4 September 2024)). A total of 307 gene entries were extracted from GenAge and designated as Source Three in the tables, named GenAge_human.
- (4)
- The latest stable version of the LongevityMap [65] (https://www.genomics.senescence.info/longevity/ (accessed on 4 September 2024)) was obtained from the LongevityMap (https://www.genomics.senescence.info/longevity/longevity_genes.zip (accessed on 4 September 2024)), reflecting the current understanding of human longevity genetics. However, this database includes records of negative results; therefore, only 273 gene entries with “significant” status in the Association column were selected and recorded in the longevityMap table.
- (5)
- A list of genes associated with cellular senescence was obtained from the CellAge database26 (https://genomics.senescence.info/cells/ (accessed on 4 September 2024)), which focuses on cellular senescence genes (https://genomics.senescence.info/cells/cellAge.zip (accessed on 4 September 2024)). Entries under the “Unclear” attribute of the Senescence Effect were excluded, and the remaining 927 records were extracted and recorded in the table.
- (6)
- From the Aging Atlas database [66], aging-related gene sets were selected for download within the Aging-related gene sets section, resulting in 503 entries recorded in the Aging Atlas table.
3.4. Senolytics Combination Efficacy Prediction Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of Aging: An Expanding Universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Xu, M.; Zhao, X.; Wang, J.; Feng, W.; Wen, N.; Wang, C.; Wang, J.; Liu, Y.; Zhao, L. DFFNDDS: Prediction of Synergistic Drug Combinations with Dual Feature Fusion Networks. J. Cheminform. 2023, 15, 33. [Google Scholar] [CrossRef]
- Wang, H.; Zhuang, L.; Ding, Y.; Tiwari, P.; Liang, C. EDDINet: Enhancing Drug–Drug Interaction Prediction via Information Flow and Consensus Constrained Multi-Graph Contrastive Learning. Artif. Intell. Med. 2025, 159, 103029. [Google Scholar] [CrossRef]
- Li, X.; Shen, B.; Feng, F.; Li, K.; Tang, Z.; Ma, L.; Li, H. Dual-View Jointly Learning Improves Personalized Drug Synergy Prediction. Bioinformatics 2024, 40, btae604. [Google Scholar] [CrossRef] [PubMed]
- Monem, S.; Hassanien, A.E.; Abdel-Hamid, A.H. A Multi-Task Graph Deep Learning Model to Predict Drugs Combination of Synergy and Sensitivity Scores. BMC Bioinform. 2024, 25, 327. [Google Scholar] [CrossRef]
- Guo, Y.; Hu, H.; Chen, W.; Yin, H.; Wu, J.; Hsieh, C.-Y.; He, Q.; Cao, J. SynergyX: A Multi-Modality Mutual Attention Network for Interpretable Drug Synergy Prediction. Brief. Bioinform. 2024, 25, bbae015. [Google Scholar] [CrossRef]
- Yan, S.; Yu, G.; Yang, J.; Chen, L. Predicting Synergistic Drug Combinations Based on Fusion of Cell and Drug Molecular Structures. Interdiscip. Sci. Comput. Life Sci. 2025, 17, 321–331. [Google Scholar] [CrossRef]
- Yan, S.; Zheng, D. A Deep Neural Network for Predicting Synergistic Drug Combinations on Cancer. Interdiscip. Sci. Comput. Life Sci. 2024, 16, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Du, H.; Hou, S.; Hu, Q.; Pang, X.; Wei, D.; Wang, X. Enhancing Drug Synergy Combination: Integrating Graph Transformers and BiLSTM for Accurate Drug Synergy Prediction. IEEE J. Biomed. Health Inform. 2025, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A Free Online Platform for Data Visualization and Graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Hickson, L.J.; Langhi Prata, L.G.P.; Bobart, S.A.; Evans, T.K.; Giorgadze, N.; Hashmi, S.K.; Herrmann, S.M.; Jensen, M.D.; Jia, Q.; Jordan, K.L.; et al. Senolytics Decrease Senescent Cells in Humans: Preliminary Report from a Clinical Trial of Dasatinib plus Quercetin in Individuals with Diabetic Kidney Disease. eBioMedicine 2019, 47, 446–456. [Google Scholar] [CrossRef]
- Farr, J.N.; Roforth, M.M.; Fujita, K.; Nicks, K.M.; Cunningham, J.M.; Atkinson, E.J.; Therneau, T.M.; McCready, L.K.; Peterson, J.M.; Drake, M.T.; et al. Effects of Age and Estrogen on Skeletal Gene Expression in Humans as Assessed by RNA Sequencing. PLoS ONE 2015, 10, e0138347. [Google Scholar] [CrossRef]
- Ren, J.; Li, G.; Zhao, W.; Lin, L.; Ye, T. Norcantharidin Combined with ABT-737 for Hepatocellular Carcinoma: Therapeutic Effects and Molecular Mechanisms. World J. Gastroenterol. 2016, 22, 3962–3968. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Xia, Y.; Pei, T.; Zhao, J.; Wang, Z.; Shen, Y.; Yang, Y.; Liang, J. Long Noncoding RNA H19: Functions and Mechanisms in Regulating Programmed Cell Death in Cancer. Cell Death Discov. 2024, 10, 76. [Google Scholar] [CrossRef]
- Jaskiewicz, M.; Moszynska, A.; Serocki, M.; Króliczewski, J.; Bartoszewska, S.; Collawn, J.F.; Bartoszewski, R. Hypoxia-Inducible Factor (HIF)-3a2 Serves as an Endothelial Cell Fate Executor during Chronic Hypoxia. EXCLI J. 2022, 21, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Jay, T.R.; von Saucken, V.E.; Landreth, G.E. TREM2 in Neurodegenerative Diseases. Mol. Neurodegener. 2017, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Navarro, A.; González-Soria, I.; Caldiño-Bohn, R.; Bobadilla, N.A. An Integrative View of Serpins in Health and Disease: The Contribution of SerpinA3. Am. J. Physiol. Cell Physiol. 2021, 320, C106–C118. [Google Scholar] [CrossRef] [PubMed]
- El Hour, M.; Moncada-Pazos, A.; Blacher, S.; Masset, A.; Cal, S.; Berndt, S.; Detilleux, J.; Host, L.; Obaya, A.J.; Maillard, C.; et al. Higher Sensitivity of Adamts12-Deficient Mice to Tumor Growth and Angiogenesis. Oncogene 2010, 29, 3025–3032. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Clevers, H. Wnt/β-Catenin Signaling in Development and Disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- Frezzato, F.; Visentin, A.; Davanzo, V.; Severin, F.; Pizzo, S.; Ruggeri, E.; Tonini, A.; Facco, M.; Piazza, F.; Semenzato, G.; et al. Strategies to Fight Ibrutinib-Resistance in Chronic Lymphocytic Leukemia. Haematologica 2020, 105, S97–S98. [Google Scholar]
- Shao, L.; Wang, N.; Yan, Y.; Tan, Y.; Wu, Q.; Lei, L.; Wang, M.; Liu, L. Quercetin of Huoxuehuayu Tongluo Decoction and Azithromycin Combination Therapy Effectively Improves Rat Tubal Factor Infertility by Inhibiting Inflammation. Iran. J. Basic Med. Sci. 2024, 27, 685–694. [Google Scholar] [CrossRef]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef]
- Leiphrakpam, P.D.; Are, C. PI3K/Akt/mTOR Signaling Pathway as a Target for Colorectal Cancer Treatment. Int. J. Mol. Sci. 2024, 25, 3178. [Google Scholar] [CrossRef]
- Dancey, J.E. Temsirolimus. In Encyclopedia of Cancer; Schwab, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 2933–2935. ISBN 978-3-540-47648-1. [Google Scholar]
- Khan, S.A.; Lee, T.K.W. Investigations of Nitazoxanide Molecular Targets and Pathways for the Treatment of Hepatocellular Carcinoma Using Network Pharmacology and Molecular Docking. Front. Pharmacol. 2022, 13, 968148. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Qiu, X.; Zhu, Z.; Lv, J.; Lu, J.; Mao, F.; Zhu, J.; Wang, J.; Guan, X.; Chen, J.; et al. Nitazoxanide, an Anti-Parasitic Drug, Efficiently Ameliorates Learning and Memory Impairments in AD Model Mice. Acta Pharmacol. Sin. 2019, 40, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Liu, J.; Xie, C.; Wang, T.; Sun, P.; Wang, J.; Li, J.; Li, G.; Qiu, J.; Zhang, Y.; et al. High-Throughput Screening Unveils Nitazoxanide as a Potent PRRSV Inhibitor by Targeting NMRAL1. Nat. Commun. 2024, 15, 4813. [Google Scholar] [CrossRef]
- Sun, Y.; Qin, H.; Zhang, H.; Feng, X.; Yang, L.; Hou, D.-X.; Chen, J. Fisetin inhibits inflammation and induces autophagy by mediating PI3K/AKT/mTOR signaling in LPS-induced RAW264.7 cells. Food Nutr. Res. 2021, 65, 6355. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.; Garg, V.K.; Tuli, H.S.; Yerer, M.B.; Sak, K.; Sharma, A.K.; Kumar, M.; Aggarwal, V.; Sandhu, S.S. Fisetin and Quercetin: Promising Flavonoids with Chemopreventive Potential. Biomolecules 2019, 9, 174. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, W.; Feng, X.; Yang, F.; Qin, H.; Wu, S.; Hou, D.-X.; Chen, J. Nrf2–ARE Signaling Acts as Master Pathway for the Cellular Antioxidant Activity of Fisetin. Molecules 2019, 24, 708. [Google Scholar] [CrossRef]
- Xu, S.; Xing, J.; Zheng, L.; Su, H.; Zou, Y.; Niu, Y.; Di, H. Azithromycin Regulates Mettl3-Mediated NF-κB Pathway to Enhance M2 Polarization of RAW264.7 Macrophages and Attenuate LPS-Triggered Cytotoxicity of MLE-12 Alveolar Cells. Int. Immunopharmacol. 2024, 137, 112426. [Google Scholar] [CrossRef]
- Zaccolo, M.; Zerio, A.; Lobo, M.J. Subcellular Organization of the cAMP Signaling Pathway. Pharmacol. Rev. 2021, 73, 278. [Google Scholar] [CrossRef]
- Jia, X.; Zheng, Y.; Guo, Y.; Chen, K. Sodium butyrate and panobinostat induce apoptosis of chronic myeloid leukemia cells via multiple pathways. Mol. Genet. Genom. Med. 2019, 7, e613. [Google Scholar] [CrossRef]
- Prystowsky, M.; Feeney, K.; Kawachi, N.; Montagna, C.; Willmott, M.; Wasson, C.; Antkowiak, M.; Loudig, O.; Parish, J. Inhibition of Plk1 and Cyclin B1 Expression Results in Panobinostat-Induced G2 Delay and Mitotic Defects. Sci. Rep. 2013, 3, 2640. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qin, G.; Li, Y.; Xu, X.; Wang, X.; Zhang, K.; Tang, Y.; Qiu, H.; Shi, D.; Zhang, C.; Long, Q.; et al. Panobinostat (LBH589) Inhibits Wnt/β-Catenin Signaling Pathway via Upregulating APCL Expression in Breast Cancer. Cell Signal. 2019, 59, 62–75. [Google Scholar] [CrossRef]
- Perrone, G.; Calabrese, E.; Hideshima, T.; Gorgun, G.; Hiroshi, I.; Cristea, D.; Santo, L.; Yiguo, H.; Anderson, K.C. Panobinostat Inhibits JAK2/STAT3 Pathway in Multiple Myeloma. Blood 2009, 114, 2849. [Google Scholar] [CrossRef]
- Liu, C.; Ye, Y.; Zhou, Q.; Zhang, R.; Zhang, H.; Liu, W.; Xu, C.; Liu, L.; Huang, S.; Chen, L. Crosstalk between Ca2+ Signaling and Mitochondrial H2O2 Is Required for Rotenone Inhibition of mTOR Signaling Pathway Leading to Neuronal Apoptosis. Oncotarget 2016, 7, 7534–7549. [Google Scholar] [CrossRef]
- Li, N.; Ragheb, K.; Lawler, G.; Sturgis, J.; Rajwa, B.; Melendez, J.A.; Robinson, J.P. Mitochondrial Complex I Inhibitor Rotenone Induces Apoptosis through Enhancing Mitochondrial Reactive Oxygen Species Production. J. Biol. Chem. 2003, 278, 8516–8525. [Google Scholar] [CrossRef]
- Chen, B.; Ning, K.; Sun, M.-L.; Zhang, X.-A. Regulation and Therapy, the Role of JAK2/STAT3 Signaling Pathway in OA: A Systematic Review. Cell Commun. Signal. 2023, 21, 67. [Google Scholar] [CrossRef]
- Oh, Y.; Park, J.H.; Djunadi, T.A.; Shah, Z.; Chung, L.I.-Y.; Chae, Y.K. Deep Response to a Combination of mTOR Inhibitor Temsirolimus and Dual Immunotherapy of Nivolumab/Ipilimumab in Poorly Differentiated Thyroid Carcinoma with PTEN Mutation: A Case Report and Literature Review. Front. Endocrinol. 2024, 15, 1304188. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Li, B.; Ming, D. A Multilayer Dynamic Perturbation Analysis Method for Predicting Ligand-Protein Interactions. BMC Bioinform. 2022, 23, 456. [Google Scholar] [CrossRef]
- Na, W.; Ju-xin, Z.; Guang-zhi, L.I.U. Inducing Apoptosis of Norcantharidin in Combination with ABT-737 on Cervical Cancer Cells. J. Int. Reprod. Health/Fam. Plan. 2014, 33, 261. [Google Scholar]
- Smer-Barreto, V.; Quintanilla, A.; Elliott, R.J.R.; Dawson, J.C.; Sun, J.; Campa, V.M.; Lorente-Macías, Á.; Unciti-Broceta, A.; Carragher, N.O.; Acosta, J.C.; et al. Discovery of Senolytics Using Machine Learning. Nat. Commun. 2023, 14, 3445. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Guerrero, A.; Harranz, N. Senolytic Compounds. U.S. Patent 20200121620, 23 April 2020. [Google Scholar]
- Triana-Martínez, F.; Picallos-Rabina, P.; Da Silva-Álvarez, S.; Pietrocola, F.; Llanos, S.; Rodilla, V.; Soprano, E.; Pedrosa, P.; Ferreirós, A.; Barradas, M.; et al. Identification and Characterization of Cardiac Glycosides as Senolytic Compounds. Nat. Commun. 2019, 10, 4731. [Google Scholar] [CrossRef]
- Ozsvari, B.; Nuttall, J.R.; Sotgia, F.; Lisanti, M.P. Azithromycin and Roxithromycin Define a New Family of “Senolytic” Drugs That Target Senescent Human Fibroblasts. Aging 2018, 10, 3294–3307. [Google Scholar] [CrossRef] [PubMed]
- Yousefzadeh, M.J.; Zhu, Y.; McGowan, S.J.; Angelini, L.; Fuhrmann-Stroissnigg, H.; Xu, M.; Ling, Y.Y.; Melos, K.I.; Pirtskhalava, T.; Inman, C.L.; et al. Fisetin Is a Senotherapeutic That Extends Health and Lifespan. eBioMedicine 2018, 36, 18–28. [Google Scholar] [CrossRef]
- Guerrero, A.; Herranz, N.; Sun, B.; Wagner, V.; Gallage, S.; Guiho, R.; Wolter, K.; Pombo, J.; Irvine, E.E.; Innes, A.J.; et al. Cardiac Glycosides Are Broad-Spectrum Senolytics. Nat. Metab. 2019, 1, 1074–1088. [Google Scholar] [CrossRef]
- Wang, Y.; Chang, J.; Liu, X.; Zhang, X.; Zhang, S.; Zhang, X.; Zhou, D.; Zheng, G. Discovery of Piperlongumine as a Potential Novel Lead for the Development of Senolytic Agents. Aging 2016, 8, 2915–2926. [Google Scholar] [CrossRef]
- Zhu, Y.; Tchkonia, T.; Pirtskhalava, T.; Gower, A.C.; Ding, H.; Giorgadze, N.; Palmer, A.K.; Ikeno, Y.; Hubbard, G.B.; Lenburg, M.; et al. The Achilles’ Heel of Senescent Cells: From Transcriptome to Senolytic Drugs. Aging Cell 2015, 14, 644–658. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann-Stroissnigg, H.; Ling, Y.Y.; Zhao, J.; McGowan, S.J.; Zhu, Y.; Brooks, R.W.; Grassi, D.; Gregg, S.Q.; Stripay, J.L.; Dorronsoro, A.; et al. Identification of HSP90 Inhibitors as a Novel Class of Senolytics. Nat. Commun. 2017, 8, 422. [Google Scholar] [CrossRef]
- Guerrero, A.; Guiho, R.; Herranz, N.; Uren, A.; Withers, D.J.; Martínez-Barbera, J.P.; Tietze, L.F.; Gil, J. Galactose-Modified Duocarmycin Prodrugs as Senolytics. Aging Cell 2020, 19, e13133. [Google Scholar] [CrossRef]
- Lafontaine, J.; Cardin, G.B.; Malaquin, N.; Boisvert, J.-S.; Rodier, F.; Wong, P. Senolytic Targeting of Bcl-2 Anti-Apoptotic Family Increases Cell Death in Irradiated Sarcoma Cells. Cancers 2021, 13, 386. [Google Scholar] [CrossRef]
- Li, W.; He, Y.; Zhang, R.; Zheng, G.; Zhou, D. The Curcumin Analog EF24 Is a Novel Senolytic Agent. Aging 2019, 11, 771–782. [Google Scholar] [CrossRef]
- Samaraweera, L.; Adomako, A.; Rodriguez-Gabin, A.; McDaid, H.M. A Novel Indication for Panobinostat as a Senolytic Drug in NSCLC and HNSCC. Sci. Rep. 2017, 7, 1900. [Google Scholar] [CrossRef] [PubMed]
- Yosef, R.; Pilpel, N.; Tokarsky-Amiel, R.; Biran, A.; Ovadya, Y.; Cohen, S.; Vadai, E.; Dassa, L.; Shahar, E.; Condiotti, R.; et al. Directed Elimination of Senescent Cells by Inhibition of BCL-W and BCL-XL. Nat. Commun. 2016, 7, 11190. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tchkonia, T.; Fuhrmann-Stroissnigg, H.; Dai, H.M.; Ling, Y.Y.; Stout, M.B.; Pirtskhalava, T.; Giorgadze, N.; Johnson, K.O.; Giles, C.B.; et al. Identification of a Novel Senolytic Agent, Navitoclax, Targeting the Bcl-2 Family of Anti-Apoptotic Factors. Aging Cell 2016, 15, 428–435. [Google Scholar] [CrossRef]
- Wong, F.; Omori, S.; Donghia, N.M.; Zheng, E.J.; Collins, J.J. Discovering Small-Molecule Senolytics with Deep Neural Networks. Nat. Aging 2023, 3, 734–750. [Google Scholar] [CrossRef]
- de Magalhães, J.P.; Abidi, Z.; dos Santos, G.A.; Avelar, R.A.; Barardo, D.; Chatsirisupachai, K.; Clark, P.; De-Souza, E.A.; Johnson, E.J.; Lopes, I.; et al. Human Ageing Genomic Resources: Updates on Key Databases in Ageing Research. Nucleic Acids Res. 2024, 52, D900–D908. [Google Scholar] [CrossRef]
- Budovsky, A.; Craig, T.; Wang, J.; Tacutu, R.; Csordas, A.; Lourenço, J.; Fraifeld, V.E.; de Magalhães, J.P. LongevityMap: A Database of Human Genetic Variants Associated with Longevity. Trends Genet. 2013, 29, 559–560. [Google Scholar] [CrossRef] [PubMed]
- Aging Atlas Consortium. Aging Atlas: A Multi-Omics Database for Aging Biology. Nucleic Acids Res. 2021, 49, D825–D830. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Auckland, New Zealand, 2–6 December 2024; Curran Associates Inc.: Red Hook, NY, USA, 4 December 2017; pp. 6000–6010. [Google Scholar]
- Malyutina, A.; Majumder, M.M.; Wang, W.; Pessia, A.; Heckman, C.A.; Tang, J. Drug Combination Sensitivity Scoring Facilitates the Discovery of Synergistic and Efficacious Drug Combinations in Cancer. PLoS Comput. Biol. 2019, 15, e1006752. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Wang, Y.; Zhang, L.; Chu, Y.; Liu, Y.; Fang, Y.; Jiang, M.; Wang, Q.; Zhao, B.; Xiong, Y.; et al. MDF-SA-DDI: Predicting Drug-Drug Interaction Events Based on Multi-Source Drug Fusion, Multi-Source Feature Fusion and Transformer Self-Attention Mechanism. Brief. Bioinform. 2022, 23, bbab421. [Google Scholar] [CrossRef]

| Co-Expressed Differential Genes | Gene Name |
|---|---|
| Upregulated genes | IGFN1, PTCHD4, PKP1, NRAP, CMA1, MIR675, AQP7B, ADCYAP1R1, ANGPTL7, MYOZ1, HOTS, LOC112267876, JPH2, LOC102724852, PLN, H19, LINC01436, MUC3A, MYH1, LMO3, PLCXD3, HIF3A, ADAMTS15, SRL, CASQ2, HOXD9, REM1, HSPB6, MEOX1, NTM, SLC52A3, CCL21, PTN, CYP26B1, INSRR, IGHV7-4-1, ADAMTSL1, COX4I2, LAMA3, GPR15, SERPINA3, FLNC, PERM1, TLL1, TREM2, STXBP6, NES, CCDC85A, LOC105375249, RERGL, CLIC5, SH3RF2, SYPL2, CCL19, RASD2, TCF15, CACNA1H, SLCO2A1, ALDH1A2, SSTR1, C1QTNF7, GPR17, KRT222, POSTN, FAM107A, PLPPR4, L1CAM, ANKRD29, TRIM63, IRX6, STC1, LOC105377979, MET, SHISAL1, TCEAL2, EBF2, ADAMTS12, SLIT3 |
| Downregulated genes | PTPRQ, CRH |
| ID | Description | Gene ID | Count |
|---|---|---|---|
| hsa04020 | Calcium signaling pathway | PLN/CASQ2/CACNA1H/MET | 4 |
| hsa04360 | Axon guidance | L1CAM/MET/SLIT3 | 3 |
| hsa04510 | Focal adhesion | LAMA3/FLNC/MET | 3 |
| hsa05415 | Diabetic cardiomyopathy | CMA1/PLN/COX4I2 | 3 |
| hsa04024 | cAMP signaling pathway | ADCYAP1R1/PLN/SSTR1 | 3 |
| hsa04010 | MAPK signaling pathway | FLNC/CACNA1H/MET | 3 |
| hsa00830 | Retinol metabolism | CYP26B1/ALDH1A2 | 2 |
| hsa04260 | Cardiac muscle contraction | CASQ2/COX4I2 | 2 |
| hsa04713 | Circadian entrainment | ADCYAP1R1/CACNA1H | 2 |
| hsa04061 | Viral protein interaction with cytokine and cytokine receptor | CCL21/CCL19 | 2 |
| hsa04064 | NF-kappa B signaling pathway | CCL21/CCL19 | 2 |
| hsa04062 | Chemokine signaling pathway | CCL21/CCL19 | 2 |
| hsa05205 | Proteoglycans in cancer | FLNC/MET | 2 |
| hsa05208 | Chemical carcinogenesis—reactive oxygen species | COX4I2/MET | 2 |
| hsa04060 | Cytokine-cytokine receptor interaction | CCL21/CCL19 | 2 |
| hsa04151 | PI3K-Akt signaling pathway | LAMA3/MET | 2 |
| hsa04080 | Neuroactive ligand–receptor interaction | ADCYAP1R1/SSTR1 | 2 |
| drugA_Name | drug_B_Name | Predicted Synergy Score | Available Reference |
|---|---|---|---|
| Temsirolimus | Nitazoxanide | 13.28 | / |
| Temsirolimus | Fisetin | 11.86 | / |
| * Cantharidin | * Fisetin | 10.35 | Frezzato et al. [24] |
| Fisetin | Enoxacin | 9.89 | / |
| * Cantharidin | * ABT-737 | 9.70 | Ren et al. [14] |
| * Fisetin | * Azithromycin | 9.64 | Shao et al. [25] |
| Panobinostat | Cantharidin | 9.62 | / |
| Temsirolimus | Rotenone | 9.60 | / |
| Temsirolimus | Azithromycin | 9.14 | / |
| Cantharidin | Enoxacin | 8.32 | / |
| Compounds & Signaling Pathway | Description | Reference |
|---|---|---|
| Temsirolimus | ||
| PI3K-Akt signaling pathway | Inhibits the proliferation and survival of cancer cells by blocking the PI3K/Akt/mTOR signaling pathway through mTOR inhibition, associated with the core differential genes LAMA3/MET. | Are et al. [27] |
| mTOR signaling pathway | Directly acts on the mTOR signaling pathway, inhibiting the activity of mTORC1 and mTORC2, thereby inhibiting tumor cell growth and proliferation. | Dancey et al. [28] |
| Nitazoxanide | ||
| MAPK signaling pathway | κ receptor-induced p38 MAPK phosphorylation mediates restlessness and anxiety in animals, unrelated to analgesic effects, and is mediated by the β-arrestin2 pathway [29]. | Khan et al. [29] |
| PI3K-Akt signaling pathway | Inhibition of mTOR pathway activation can eliminate κ receptor-induced conditioned place aversion (CPA), distinguishing varying degrees of restlessness and anxiety caused by these agonists. | Fan et al. [30] |
| Neuroactive ligand–receptor interaction | As an opioid receptor agonist-antagonist, involves the interaction of neuroactive ligands with opioid receptors in its analgesic effect. | Cui et al. [31] |
| Fisetin | ||
| PI3K-Akt signaling pathway | Inhibit the PI3K/AKT signaling pathway, thereby inhibiting mTOR and inducing cell death. | Sun et al. [32] |
| MAPK signaling pathway | Upregulates HO-1 expression via the p38 MAPK pathway, inhibiting doxorubicin-induced senescence of pulmonary artery endothelial cells. | Kashyap et al. [33] |
| Nrf2/HO-1 signaling pathway | Inhibits doxorubicin-induced senescence of pulmonary artery endothelial cells and inhibits the proliferation of pulmonary artery smooth muscle cells, thereby preventing pulmonary artery remodeling. | Zhang et al. [34] |
| Azithromycin | ||
| NF-kappa B signaling pathway | Mitigates inflammatory responses by suppressing the NF-κB signaling pathway. | Xu et al. [35] |
| Panobinostat | ||
| cAMP signaling pathway | May indirectly affect the cAMP signaling pathway by inhibiting HDAC activity, as HDAC inhibitors can affect multiple cellular signaling pathways, including the cAMP signaling pathway. | Zaccolo et al. [36] |
| Apoptosis signaling pathway | Increases the acetylation of histones and tubulins, leading to cell cycle arrest and apoptosis by inhibiting HDACs. | Jia et al. [37] |
| Cell cycle signaling pathway | Induces cell cycle arrest by increasing the level of p21 cell cycle protein. | Prystowsky et al. [38] |
| Wnt/β-catenin signaling pathway | Inhibits the Wnt/β-catenin signaling pathway by upregulating the expression of APCL. | Qin et al. [39] |
| JAK2/STAT3 signaling pathway | Inhibits the JAK2/STAT3 signaling pathway in multiple myeloma. | Perrone et al. [40] |
| Rotenone | ||
| Calcium signaling pathway | Elevates intracellular free calcium ion levels ([Ca2+]i) and activates CaMKII, leading to the inhibition of mTOR signaling and the induction of neuronal apoptosis. | Liu et al. [41] |
| Apoptosis signaling pathway | Induces the production of reactive oxygen species (ROS) in neuronal cells and leads to neuronal apoptosis by inhibiting the mTOR-mediated S6K1 and 4E-BP1 pathways. | Li et al. [42] |
| mTOR signaling pathway | Induces ROS/H2O2 to inhibit the mTOR signaling pathway, leading to neuronal apoptosis. | Liu et al. [41] |
| JAK/STAT3 signaling pathway | Influences the proliferation and apoptosis of oral squamous cell carcinoma cells by regulating the JAK/STAT3 pathway. | Chen et al. [43] |
| Chemical carcinogenesis—reactive oxygen species | Causes mitochondrial dysfunction, increases the generation of ROS, and results in oxidative damage to proteins, lipids, and nucleic acids. | Li et al. [42] |
| Mode | Affinity (kcal/mol) | RMSD l.b. | RMSD u.b. |
|---|---|---|---|
| 1 | −7.259 | 0 | 0 |
| 2 | −6.705 | 22.38 | 23.67 |
| 3 | −6.652 | 22.23 | 23.29 |
| 4 | −6.494 | 3.092 | 8.357 |
| 5 | −6.484 | 15.39 | 17.38 |
| 6 | −6.461 | 15.81 | 18.06 |
| 7 | −6.417 | 15.16 | 17.87 |
| 8 | −6.395 | 14.52 | 16.83 |
| 9 | −6.386 | 18.74 | 20.63 |
| 10 | −6.386 | 16.06 | 18.75 |
| Drug_Name | Proposed/Known Target(s) | Source |
|---|---|---|
| Azacyclonol | Histamine | Patent US 2020/0121620 [49] |
| Cyclosporin A | Calcineurin, NFAT | Patent US 2020/0121620 [49] |
| Digoxin | Na+/K+-ATPase | Triana et al., 2019 [50] |
| Nitrofural | ROS generation | Patent US 2020/0121620 [49] |
| Roxithromycin | Protein homeostasis | Ozsvari et al., 2018 [51] |
| Luteolin | PI3K/Akt, Nrf2, NF-κB | Yousefzadeh et al., 2018 [52] |
| Enoxacin | TRBP | Patent US 2020/0121620 [49] |
| Atorvastatin | HMG-CoA, Rho/ROCK | Patent US 2020/0121620 [49] |
| Azithromycin | Mitochondrial translation | Ozsvari et al., 2018 [51] |
| Nitazoxanide | phosphorylation, | Patent US 2020/0121620 [49] |
| Adapalene | RAR/RXR nuclear receptors | Patent US 2020/0121620 [49] |
| Amiloride hydrochloride | NHE1, ENaC | Triana et al., 2019 [50] |
| Cantharidin | PP2A | Patent US 2020/0121620 [49] |
| Calmidazolium chloride | Calmodulin | Guerrero et al., 2019 [53] |
| Dequalinium chloride hydrate | Mitochondria membrane potential | Patent US 2020/0121620 [49] |
| Diphenyleneiodonium chloride | NADPH oxidase, flavoproteins | Patent US 2020/0121620 [49] |
| 2,3-Dimethoxy-1,4-naphthoquinone | Redox cycling | Patent US 2020/0121620 [49] |
| Idarubicin | Topoisomerase II | Patent US 2020/0121620 [49] |
| JFD00244 | SIRT6 | Guerrero et al., 2019 [53] |
| Mibefradil dihydrochloride | T-type calcium channels | Guerrero et al., 2019 [53] |
| Piperlongumine | TrxR/GPx | Wang et al., 2016 [54] |
| Ouabain | Na+/K+-ATPase | Guerrero et al., 2019 [53] |
| Quercetin dihydrate | PI3K, HSP90, AMPK, Nrf2 | Zhu et al., 2015 [55] |
| Rottlerin | PKCδ | Guerrero et al., 2019 [53] |
| Rotenone | Complex I (ETC) | Guerrero et al., 2019 [53] |
| BIX 01294 trihydrochloride hydrate | G9a/GLP (EHMT2/1) | Guerrero et al., 2019 [53] |
| Tyrphostin AG 879 | ErbB2, TrkA | Patent US 2020/0121620 [49] |
| Vincristine sulfate | Tubulin | Patent US 2020/0121620 [49] |
| Tanespimycin | HSP90 | Fuhrmann-Stroissnigg et al., 2017 [56] |
| Geldanamycin | HSP90 | Fuhrmann-Stroissnigg et al., 2017 [56] |
| Alvespimycin | HSP90 | Fuhrmann-Stroissnigg et al., 2017 [56] |
| ProDrug A | unknown | Guerrero et al., 2020 [57] |
| JHB76B | KRAS/ERK pathway | Guerrero et al., 2020 [57] |
| CGP-74514A | CDK1/2 | Guerrero et al., 2019 [53] |
| Ouabagenin | Na+/K+-ATPase | Guerrero et al., 2019 [53] |
| K-Strophanthin | Na+/K+-ATPase | Guerrero et al., 2019 [53] |
| Strophanthidin | Na+/K+-ATPase | Guerrero et al., 2019 [53] |
| PF-573228 | FAK | Patent US 2020/0121620 [49] |
| LY-367265 | 5-HT1B/1D | Patent US 2020/0121620 [49] |
| Temsirolimus | mTORC1 | Patent US 2020/0121620 [49] |
| Eltrombopag | MPL (TPO -R) | Patent US 2020/0121620 [49] |
| Raltegravir | HIV integrase | Patent US 2020/0121620 [49] |
| Venetoclax | BCL-2 | Lafontaine et al., 2021 [58] |
| EF24 | NF-κB/IκB-α | Li et al., 2019 [59] |
| Panobinostat | HDAC | Samaraweera et al., 2017 [60] |
| Bufalin | Na+/K+-ATPase | Triana et al., 2019 [50] |
| Proscillaridin A | Na+/K+-ATPase | Triana et al., 2019 [50] |
| Cinobufagin | Na+/K+-ATPase | Triana et al., 2019 [50] |
| Peruvoside | Na+/K+-ATPase | Triana et al., 2019 [50] |
| Digitoxin | Na+/K+-ATPase | Triana et al., 2019 [50] |
| Convallotoxin | Na+/K+-ATPase | Triana et al., 2019 [50] |
| ABT-737 | BCL-2, BCL-xL, BCL-w | Yosef et al., 2016 [61] |
| Fisetin | PI3K, NF-κB, HIF-1α, Nrf2 | Yousefzadeh et al., 2018 [52] |
| Curcumin | NF-κB, Nrf2, HAT/HDAC | Yousefzadeh et al., 2018 [52] |
| Dasatinib | SRC/ABL kinases | Zhu et al., 2015 [55] |
| Navitoclax | BCL-2, BCL-xL | Zhu et al., 2016 [62] |
| A1331852 | BCL-xL | Zhu et al., 2017 [56] |
| A1155463 | BCL-xL | Zhu et al., 2017 [56] |
| ginkgetin | JAK/STAT, NF-κB | Smer-Barreto et al., 2023 [48] |
| oleandrin | Na+/K+-ATPase | Smer-Barreto et al., 2023 [48] |
| periplocin | Na+/K+-ATPase | Smer-Barreto et al., 2023 [48] |
| BRD-K56819078 | HSP90 | Wong et al., 2023 [63] |
| XL888 | HSP90 | Wong et al., 2023 [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Y.; Su, T.; Gao, J.; Ming, D. SenolyticSynergy: An Attention-Based Network for Discovering Novel Senolytic Combinations via Human Aging Genomics. Int. J. Mol. Sci. 2025, 26, 9004. https://doi.org/10.3390/ijms26189004
Ye Y, Su T, Gao J, Ming D. SenolyticSynergy: An Attention-Based Network for Discovering Novel Senolytic Combinations via Human Aging Genomics. International Journal of Molecular Sciences. 2025; 26(18):9004. https://doi.org/10.3390/ijms26189004
Chicago/Turabian StyleYe, Yaowen, Ting Su, Jiayi Gao, and Dengming Ming. 2025. "SenolyticSynergy: An Attention-Based Network for Discovering Novel Senolytic Combinations via Human Aging Genomics" International Journal of Molecular Sciences 26, no. 18: 9004. https://doi.org/10.3390/ijms26189004
APA StyleYe, Y., Su, T., Gao, J., & Ming, D. (2025). SenolyticSynergy: An Attention-Based Network for Discovering Novel Senolytic Combinations via Human Aging Genomics. International Journal of Molecular Sciences, 26(18), 9004. https://doi.org/10.3390/ijms26189004








