Research Progress on the Regulation of Plant Floral Organ Development by the MADS-box Gene Family
Abstract
1. Introduction
2. Classification and Structure of MADS-box Genes
3. Regulation of Floral Organ Development by the MADS-box Gene Family
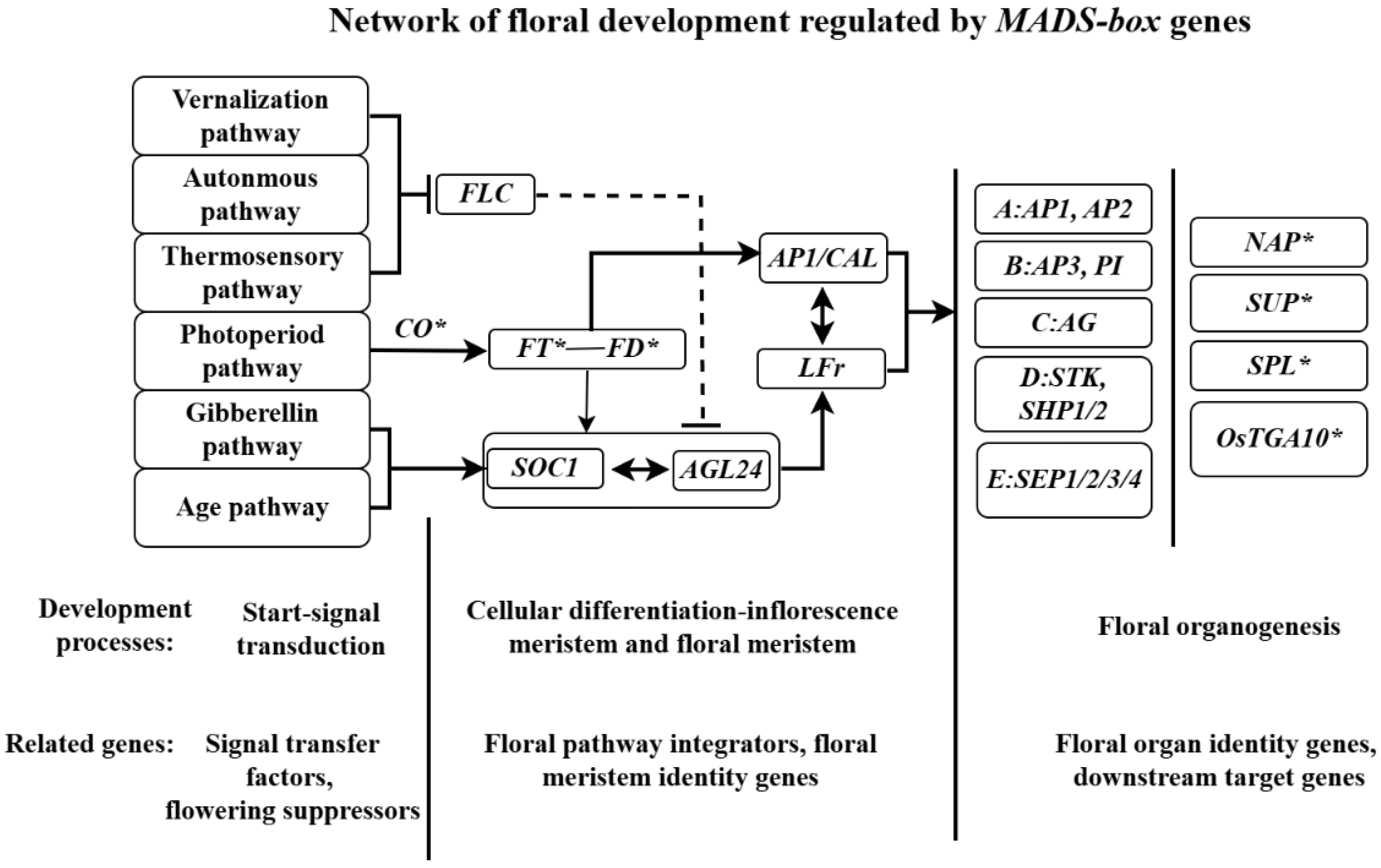
3.1. Role of MADS-box Genes in Floral Initiation
3.2. Regulation of Cell Differentiation by MADS-box Genes
3.3. Regulation of Floral Organ Formation by MADS-box Genes
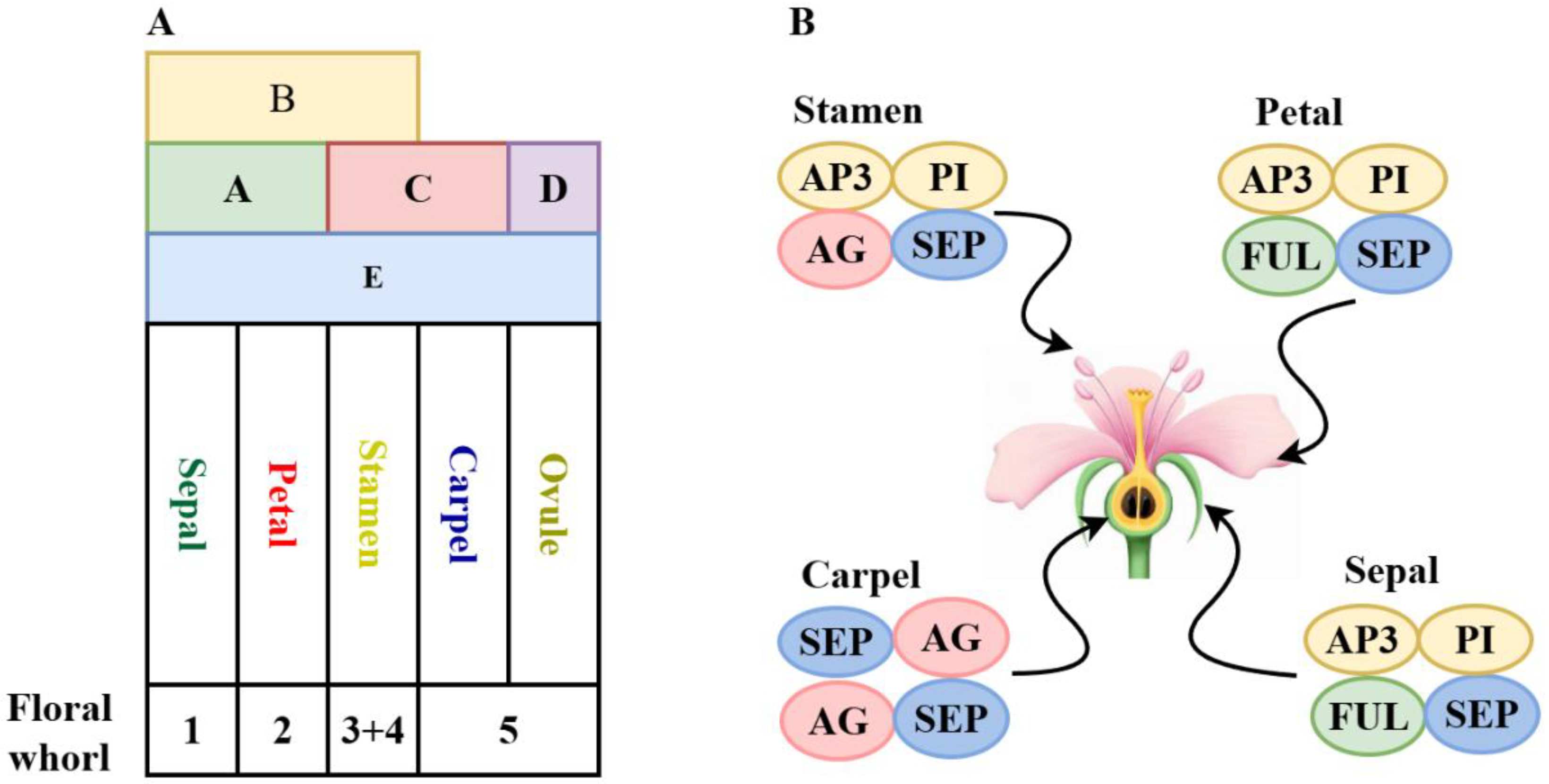
3.4. Research Progress on ABCDE-Class Genes
3.4.1. A-Class Genes
3.4.2. B-Class Genes
3.4.3. C- and D-Class Genes
3.4.4. E-Class Genes
3.5. Regulation of Downstream Target Genes by MADS-box Genes
4. Conclusions and Prospectives
Author Contributions
Funding
Conflicts of Interest
References
- Xu, H.X.; Meng, D.; Yang, Q.; Chen, T.; Qi, M.; Li, X.Y.; Ge, H.; Chen, J.W. Sorbitol induces flower bud formation via the MADS-box transcription factor EjCAL in loquat. J. Integr. Plant Biol. 2023, 65, 1241–1261. [Google Scholar] [CrossRef]
- Adhikari, P.B.; Kasahara, R.D. An overview on MADS-box members in plants: A meta-review. Int. J. Mol. Sci. 2024, 25, 8233. [Google Scholar] [CrossRef] [PubMed]
- Chopy, M.; Cavallini-Speisser, Q.; Chambrier, P.; Morel, P.; Just, J.; Hugouvieux, V.; Rodrigues Bento, S.; Zubieta, C.; Vandenbussche, M.; Monniaux, M. Cell layer–specific expression of the homeotic MADS-box transcription factor PhDEF contributes to modular petal morphogenesis in petunia. Plant Cell 2024, 36, 324–345. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.B.; Jian, H.M.; Wang, Y.; Wang, G.Y.; Zhou, C.C.; Jia, H.J.; Gao, Z.S. Genome-wide identification and analysis of the MADS-box gene family and its potential role in fruit development and ripening in red bayberry (Morella rubra). Gene 2019, 717, 144045. [Google Scholar] [CrossRef] [PubMed]
- Schwarz-Sommer, Z.; Huijser, P.; Nacken, W.; Saedler, H.; Sommer, H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef]
- Heijmans, K.; Morel, P.; Vandenbussche, M. MADS-box genes and floral development: The dark side. J. Exp. Bot. 2012, 63, 5397–5404. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Ribas de Pouplana, L.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Lai, X.; Daher, H.; Galien, A.; Hugouvieux, V.; Zubieta, C. Structural basis for plant MADS transcription factor oligomerization. Comput. Struct. Biotechnol. J. 2019, 17, 946–953. [Google Scholar] [CrossRef]
- Bemer, M.; Heijmans, K.; Airoldi, C.; Davies, B.; Angenent, G.C. An atlas of type I MADS-box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 2010, 154, 287–300. [Google Scholar] [CrossRef]
- Masiero, S.; Colombo, L.; Grini, P.E.; Schnittger, A.; Kater, M.M. The emerging importance of type I MADS-box transcription factors for plant reproduction. Plant Cell 2011, 23, 865–872. [Google Scholar] [CrossRef]
- Kang, I.H.; Steffen, J.G.; Portereiko, M.F.; Lloyd, A.; Drews, G.N. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 2008, 20, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, D.; Lin, X.; Ding, M.; Tong, Z. Genome-wide identification of MADS-box family genes in moso bamboo (Phyllostachys edulis) and a functional analysis of PeMADS5 in flowering. BMC Plant Biol. 2018, 18, 176. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Meyerowitz, E.M. MADS domain proteins in plant development. Biol. Chem. 1997, 378, 1079–1101. [Google Scholar] [PubMed]
- Lai, X.; Vega-Léon, R.; Hugouvieux, V.; Blanc-Mathieu, R.; van der Wal, F.; Lucas, J.; Silva, C.S.; Jourdain, A.; Muino, J.M.; Nanao, M.H.; et al. The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors. Nat. Commun. 2021, 12, 4760. [Google Scholar] [CrossRef]
- Käppel, S.; Rümpler, F.; Theißen, G. Cracking the Floral Quartet Code: How do multimers of MIKCC-type MADS-domain transcription factors recognize their target genes? Int. J. Mol. Sci. 2023, 24, 8253. [Google Scholar] [CrossRef]
- Yoon, J.; Baek, G.; Pasriga, R.; Tun, W.; Min, C.W.; Kim, S.T.; Cho, L.H.; An, G. Homeobox transcription factors OsZHD1 and OsZHD2 induce inflorescence meristem activity at floral transition in rice. Plant Cell Environ. 2023, 46, 1327–1339. [Google Scholar] [CrossRef]
- Wang, C.; Yang, X.; Li, G. Molecular insights into inflorescence meristem specification for yield potential in cereal crops. Int. J. Mol. Sci. 2021, 22, 3508. [Google Scholar] [CrossRef]
- Sharma, N.; Ruelens, P.; D’hauw, M.; Maggen, T.; Dochy, N.; Torfs, S.; Kaufmann, K.; Rohde, A.; Geuten, K. A flowering locus C homolog is a vernalization-regulated repressor in Brachypodium and is cold regulated in wheat. Plant Physiol. 2017, 173, 1301–1315. [Google Scholar] [CrossRef]
- Sharma, N.; Geuten, K.; Giri, B.S.; Varma, A. The molecular mechanism of vernalization in Arabidopsis and cereals: Role of Flowering Locus C and its homologs. Physiol. Plant. 2020, 170, 373–383. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, D.; He, Y. FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat. Plants 2018, 4, 836–846. [Google Scholar] [CrossRef]
- Li, Z.; Ou, Y.; Zhang, Z.; Li, J.; He, Y. Brassinosteroid signaling recruits histone 3 lysine-27 demethylation activity to FLOWERING LOCUS C chromatin to inhibit the floral transition in Arabidopsis. Mol. Plant 2018, 11, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.K.; Park, S.R.; Suh, E.J.; Park, J. Effects of overexpression of Brassica rapa SHORT VEGETATIVE PHASE gene on flowering time. Korean J. Breed. Sci. 2020, 52, 244–251. [Google Scholar] [CrossRef]
- Liu, X.; Sun, Z.; Dong, W.; Wang, Z.; Zhang, L. Expansion and functional divergence of the SHORT VEGETATIVE PHASE (SVP) genes in eudicots. Genome Biol. Evol. 2018, 10, 3026–3037. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Zhang, Y.; Wang, K.; Luo, X.; Xu, D.; Tian, X.; Li, L.; Ye, X.; Xia, X.; Li, W.; et al. TaVrt2, an SVP-like gene, cooperates with TaVrn1 to regulate vernalization-induced flowering in wheat. New Phytol. 2021, 231, 834–848. [Google Scholar] [CrossRef]
- Liu, X.; Sun, Z.; Dong, W.; Wang, Z.; Zhang, L. Transcriptome profiling reveals phase-specific gene expression in the developing barley inflorescence. Crop J. 2020, 8, 71–86. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, W.; Leung, C.C.; Tarté, D.A.; Gendron, J.M. Plants distinguish different photoperiods to independently control seasonal flowering and growth. Science 2024, 383, eadg9196. [Google Scholar] [CrossRef]
- Rehman, S.; Bahadur, S.; Xia, W. An overview of floral regulatory genes in annual and perennial plants. Gene 2023, 885, 147699. [Google Scholar] [CrossRef]
- Wang, P.; Su, L.; Cao, L.; Hu, H.; Wan, H.; Wu, C.; Zheng, Y.; Bao, C.; Liu, X. AtSRT1 regulates flowering by regulating flowering integrators and energy signals in Arabidopsis. Plant Physiol. Biochem. 2024, 213, 108841. [Google Scholar] [CrossRef]
- Cai, F.; Jin, X.; Han, L.; Chen, H.; Shao, C.; Shi, G.; Bao, M.; Sun, Y.; Zhang, J. AINTEGUMENTA-LIKE genes regulate reproductive growth and bud dormancy in Platanus acerifolia. Plant Cell Rep. 2024, 43, 261. [Google Scholar] [CrossRef]
- Lee, J.; Oh, M.; Park, H.; Lee, I. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J. 2008, 55, 832–843. [Google Scholar] [CrossRef]
- Torti, S.; Fornara, F. AGL24 acts in concert with SOC1 and FUL during Arabidopsis floral transition. Plant Signal. Behav. 2012, 7, 1251–1254. [Google Scholar] [CrossRef]
- Goslin, K.; Zheng, B.; Serrano-Mislata, A.; Rae, L.; Ryan, P.T.; Kwaśniewska, K.; Thomson, B.; Ó’Maoiléidigh, D.S.; Madueño, F.; Wellmer, F.; et al. Transcription factor interplay between LEAFY and APETALA1/CAULIFLOWER during floral initiation. Plant Physiol. 2017, 174, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Pabón-Mora, N.; Ambrose, B.A.; Litt, A. Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 2012, 158, 1685–1704. [Google Scholar] [CrossRef]
- Sri, T.; Gupta, B.; Tyagi, S.; Singh, A. Homeologs of Brassica SOC1, a central regulator of flowering time, are differentially regulated due to partitioning of evolutionarily conserved transcription factor binding sites in promoters. Mol. Phylogenetics Evol. 2020, 147, 106777. [Google Scholar] [CrossRef] [PubMed]
- Hill, K.; Wang, H.; Perry, S.E. A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 2008, 53, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Agliassa, C.; Narayana, R.; Bertea, C.M.; Rodgers, C.T.; Maffei, M.E. Reduction of the geomagnetic field delays Arabidopsis thaliana flowering time through downregulation of flowering-related genes. Bioelectromagnetics 2018, 39, 361–374. [Google Scholar] [CrossRef]
- Tan, X.M.; Li, Y.R.; Song, M.R.; Yuan, L.N.; Zhao, Z.X.; Liu, Y.; Meng, Q.; Huang, X.; Ma, Y.Y.; Xu, Z.Q. The Molecular mechanism of interaction between SEPALLATA3 and APETALA1 in Arabidopsis thaliana. Plant Direct 2025, 9, e70052. [Google Scholar] [CrossRef]
- Xu, Y.; Prunet, N.; Gan, E.S.; Wang, Y.; Stewart, D.; Wellmer, F.; Huang, J.; Yamaguchi, N.; Tatsumi, Y.; Kojima, M.; et al. SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 2018, 37, e97499. [Google Scholar] [CrossRef]
- Kong, L.; Duan, Y.; Ye, Y.; Cai, Z.; Wang, F.; Qu, X.; Qiu, R.; Wu, C.; Wu, W. Screening and analysis of proteins interacting with OsMADS16 in rice (Oryza sativa L.). PLoS ONE 2019, 14, e0221473. [Google Scholar] [CrossRef]
- Otani, M.; Aoyagi, K.; Nakano, M. Suppression of B function by chimeric repressor gene-silencing technology (CRES-T) reduces the petaloid tepal identity in transgenic Lilium sp. PLoS ONE 2020, 15, e0237176. [Google Scholar] [CrossRef]
- Angent, G.C.; Colombo, L. Molecular control of ovule development. Am. Ind. Hyg. Assoc. Q. 1996, 1, 228–232. [Google Scholar]
- Colombo, L.; Franken, J.; Koetje, E.; van Went, J.; Dons, H.J.M.; Angenent, G.C.; van Tunen, A.J. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 1995, 7, 1859–1868. [Google Scholar]
- Immink, R.G.; Tonaco, I.A.; de Folter, S.; Shchennikova, A.; van Dijk, A.D.; Busscher-Lange, J.; Borst, J.W.; Angenent, G.C. SEPALLATA3: The ‘glue’ for MADS-box transcription factor complex formation. Genome Biol. 2009, 10, R24. [Google Scholar] [CrossRef]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Gioppato, H.A.; Dornelas, M.C. When Bs are better than As: The relationship between B-class MADS-box gene duplications and the diversification of perianth morphology. Trop. Plant Biol. 2019, 12, 1–11. [Google Scholar] [CrossRef]
- Xiang, L.; Chen, Y.; Chen, L.; Fu, X.; Zhao, K.; Zhang, J.; Sun, C. B and E MADS-box genes determine the perianth formation in Cymbidium goeringii Rchb.f. Physiol. Plant. 2018, 162, 353–369. [Google Scholar] [CrossRef]
- Mandel, M.A.; Gustafson-Brown, C.; Savidge, B.; Yanofsky, M.F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992, 360, 273–277. [Google Scholar] [CrossRef]
- Gustafson-Brown, C.; Savidge, B.; Yanofsky, M.F. Regulation of the arabidopsis floral homeotic gene APETALA1. Cell 1994, 76, 131–143. [Google Scholar] [CrossRef]
- Liu, Z.; Fei, Y.; Zhang, K.; Fang, Z. Ectopic Expression of a Fagopyrum esculentum APETALA1 Ortholog only Rescues Sepal Development in Arabidopsis ap1 Mutant. Int. J. Mol. Sci. 2019, 20, 2021. [Google Scholar] [CrossRef]
- Yang, Q.; Luo, L.; Jiao, X.; Chen, X.; Liu, Y.; Liu, Z. APETALA2-like Floral Homeotic Protein Up-Regulating FaesAP1_2 Gene Involved in Floral Development in Long-Homostyle Common Buckwheat. Int. J. Mol. Sci. 2024, 25, 7193. [Google Scholar] [CrossRef]
- Bowman, J.L.; Alvarez, J.; Weigel, D.; Meyerowitz, E.M.; Smyth, D.R. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 1993, 119, 721–743. [Google Scholar] [CrossRef]
- Kater, M.M.; Dreni, L.; Colombo, A.L. Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 2006, 57, 3433–3444. [Google Scholar] [CrossRef]
- Ma, Y.Q.; Pu, Z.Q.; Tan, X.M.; Meng, Q.; Zhang, K.L.; Yang, L.; Ma, Y.Y.; Huang, X.; Xu, Z.Q. SEPALLATA-like genes of Isatis indigotica can affect the architecture of the inflorescences and the development of the floral organs. PeerJ 2022, 10, e13034. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; Sablowski, R.W.; Meyerowitz, E.M. Transcriptional activation of APETALA1 by LEAFY. Science 1999, 285, 582–584. [Google Scholar] [CrossRef]
- Zhang, S.; Lu, S.; Yi, S.; Han, H.; Liu, L.; Zhang, J.; Bao, M.; Liu, G. Functional conservation and divergence of five SEPALLATA-like genes from a basal eudicot tree, Platanus acerifolia. Planta 2017, 245, 439–457. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Bari, A.; Li, H.; Chen, L.L. Identification and analysis of micro-exons in AP2/ERF and MADS gene families. FEBS Open Bio 2020, 10, 2564–2577. [Google Scholar] [CrossRef] [PubMed]
- Dipp-Álvarez, M.; Cruz-Ramírez, A. A Phylogenetic Study of the ANT Family Points to a preANT Gene as the ancestor of basal and euANT transcription factors in land plants. Front. Plant Sci. 2019, 10, 17. [Google Scholar] [CrossRef]
- Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular events of rice AP2/ERF transcription factors. Int. J. Mol. Sci. 2022, 23, 12013. [Google Scholar] [CrossRef]
- Shannon, S.; Meeks-Wagner, D.R. Genetic interactions that regulate inflorescence development in Arabidopsis. Plant Cell 1993, 5, 639–655. [Google Scholar] [CrossRef]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef]
- Schmid, M.; Uhlenhaut, N.H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J.U. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.S.; Amasino, R.; Scheres, B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Klucher, K.M.; Chow, H.; Reiser, L.; Fischer, R.L. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 1996, 8, 137–153. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Elliott, R.C.; Betzner, A.S.; Huttner, E.; Oakes, M.P.; Tucker, W.Q.; Gerentes, D.; Perez, P.; Smyth, D.R. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 1996, 8, 155–168. [Google Scholar][Green Version]
- Zhao, L.M.; Kong, H.Z.; Leebens-Mack, J.H.; Kim, S.; Soltis, P.S.; Landherr, L.L.; Soltis, D.E.; de Pamphilis, C.W.; Ma, H. The evolution of the SEPALLATA subfamily of MADS-box genes: A preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 2005, 169, 2209–2223. [Google Scholar][Green Version]
- Goto, K.; Meyerowitz, E.M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994, 8, 1548–1560. [Google Scholar] [CrossRef]
- Fukui, M.; Futamura, N.; Mukai, Y.; Wang, Y.Q.; Nagao, A.; Shinohara, K. Ancestral MADS-box genes in Sugi, Cryptomeria japonica D. Don (Taxodiaceae), homologous to the B function genes in Angiosperms. Plant Cell Physiol. 2001, 42, 566–575. [Google Scholar] [CrossRef]
- Zhou, L.; Iqbal, A.; Yang, M.; Yang, Y. Research progress on gene regulation of plant floral organogenesis. Genes 2025, 16, 79. [Google Scholar] [CrossRef]
- Takeda, S.; Hamamura, Y.; Sakamoto, T.; Kimura, S.; Aida, M.; Higashiyama, T. Non-cell-autonomous regulation of petal initiation in Arabidopsis thaliana. Development 2022, 149, dev200684. [Google Scholar] [CrossRef]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Rodríguez-Cazorla, E.; Ripoll, J.J.; Ortuño-Miquel, S.; Martínez-Laborda, A.; Vera, A. Dissection of the Arabidopsis HUA-PEP gene activity reveals that ovule fate specification requires restriction of the floral A-function. New Phytol. 2020, 227, 1222–1234. [Google Scholar] [CrossRef]
- Hussin, S.H.; Wang, H.; Tang, S.; Zhi, H.; Tang, C.; Zhang, W.; Jia, G.; Diao, X. SiMADS34, an E-class MADS-box transcription factor, regulates inflorescence architecture and grain yield in Setaria italica. Plant Mol. Biol. 2021, 105, 419–434. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, M.; Zhang, R.; Xie, J.; Duan, X.; Shan, H.; Xu, G.; Kong, H. Identification of the target genes of AqAPETALA3-3(AqAP3-3) in Aquilegia coerulea (Ranunculaceae) helps understand the molecular bases of the conserved and nonconserved features of petals. New Phytol. 2020, 227, 1235–1248. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zhang, Y.; Hou, X. BcAP3, a MADS-box gene, controls stamen development and male sterility in Pak-choi (Brassica rapa ssp. chinensis). Gene 2020, 747, 144698. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Liang, B.; Yang, L.; Hu, W.; Kuang, L.; Song, J.; Xie, J.; Huang, Y.; Liu, D.; Liu, Y. The MADS-box family gene PtrANR1 encodes a transcription activator promoting root growth and enhancing plant tolerance to drought stress. Plant Cell Rep. 2023, 43, 16. [Google Scholar] [CrossRef] [PubMed]
- Mao, T.; Wang, X.; Gao, H.; Gong, Z.; Liu, R.; Jiang, N.; Zhang, Y.; Zhang, H.; Guo, X.; Yu, C. Ectopic Expression of MADS-box transcription factor VvAGL12 from grape promotes early flowering, plant growth, and production by regulating cell-wall architecture in Arabidopsis. Genes 2023, 14, 2078. [Google Scholar] [CrossRef]
- Prunet, N. My favourite flowering image: An Arabidopsis inflorescence expressing fluorescent reporters for the APETALA3 and SUPERMAN genes. J. Exp. Bot. 2019, 70, e6499–e6501. [Google Scholar] [CrossRef]
- Zhang, S.; Yao, J.; Wang, L.; Wu, N.; van Nocker, S.; Li, Z.; Gao, M.; Wang, X. Role of grapevine SEPALLATA-related MADS-box gene VvMADS39 in flower and ovule development. Plant J. 2022, 111, 1565–1579. [Google Scholar] [CrossRef]
- Uemura, A.; Yamaguchi, N.; Xu, Y.; Wee, W.; Ichihashi, Y.; Suzuki, T.; Shibata, A.; Shirasu, K.; Ito, T. Regulation of floral meri-stem activity through the interaction of AGAMOUS, SUPERMAN, and CLAVATA3 in Arabidopsis. Plant Reprod. 2018, 31, 89–105. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Cozzolino, S.; Widmer, A. Orchid diversity: An evolutionary consequence of deception? Trends Ecol. Evol. 2005, 20, 487–494. [Google Scholar] [CrossRef]
- Sun, J.; Liu, Y.; Zheng, Y.; Xue, Y.; Fan, Y.; Ma, X.; Ji, Y.; Liu, G.; Zhang, X.; Li, Y.; et al. The MADS-box transcription factor GmFULc promotes GmZTL4 gene transcription to modulate maturity in soybean. J. Integr. Plant Biol. 2024, 66, 1603–1619. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.S.; Liu, X.F.; Wang, D.H.; Chen, R.; Zhang, X.L.; Xu, Z.H.; Bai, S.N. Transcription factor OsTGA10 is a target of the MADS protein OsMADS8 and is required for tapetum development. Plant Physiol. 2018, 176, 819–835. [Google Scholar] [CrossRef] [PubMed]
- Shulga, O.A.; Mitiouchkina, T.Y.; Shchennikova, A.V.; Skryabin, K.G.; Dolgov, S.V. Chrysanthemum modification via ectopic expression of sunflower MADS-box gene HAM59. Acta Hortic. 2015, 1087, 105–111. [Google Scholar] [CrossRef]
- Mouradov, A.; Glassick, T.V.; Hamdorf, B.A.; Murphy, L.C.; Marla, S.S.; Yang, Y.; Teasdale, R.D. Family of MADS-box genes expressed early in male and female reproductive structures of Monterey pine. Plant Physiol. 1998, 117, 55–62. [Google Scholar] [CrossRef][Green Version]
- Winter, K.U.; Becker, A.; Münster, T.; Kim, J.T.; Saedler, H.; Theissen, G. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc. Natl. Acad. Sci. USA 1999, 96, 7342–7347. [Google Scholar] [CrossRef]
- Thangavel, G.; Nayar, S. A survey of MIKC type MADS-box genes in non-seed plants: Algae, bryophytes, lycophytes and ferns. Front. Plant Sci. 2018, 9, 510. [Google Scholar] [CrossRef]
- Mao, L.; Begum, D.; Chuang, H.W.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R.A. JOINTLESS is a MADS-box gene controlling tomato flower abscissionzone development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef]
- Gan, Y.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef]
- Xing, M.; Li, H.; Liu, G.; Zhu, B.; Zhu, H.; Grierson, D.; Luo, Y.; Fu, D. A MADS-box transcription factor, SlMADS1, interacts with SlMACROCALYX to regulate tomato sepal growth. Plant Sci. 2022, 322, 111366. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Yu, L.H.; Xiang, C.B. ARABIDOPSIS NITRATE REGULATED 1 acts as a negative modulator of seed germination by activating ABI3 expression. New Phytol. 2020, 225, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Srivastava, H.; Das, A.; Tribhuvan, K.U.; Durgesh, K.; Joshi, R.; Sevanthi, A.M.; Jain, P.K.; Singh, N.K.; Gaikwad, K. Identification and characterization of MADS-box gene family in pigeonpea for their role during floral transition. 3 Biotech 2021, 11, 108. [Google Scholar] [CrossRef]
- Zhang, C.; Shen, J.; Wang, C.; Wang, Z.; Guo, L.; Hou, X. Characterization of PsmiR319 during flower development in early- and late-flowering tree peonies cultivars. Plant Signal. Behav. 2022, 17, 2120303. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, T.; Saito, M.; Yamada, E.; Fujita, K.; Yamagishi, N.; Yoshikawa, N.; Nishihara, M. Isolation and characterization of the C-class MADS-box gene involved in the formation of double flowers in Japanese gentian. BMC Plant Biol. 2015, 15, 182. [Google Scholar] [CrossRef]
- Zhao, J.; Gong, P.; Liu, H.; Zhang, M.; He, C. Multiple and integrated functions of floral C-class MADS-box genes in flower and fruit development of Physalis floridana. Plant Mol. Biol. 2021, 107, 101–116. [Google Scholar] [CrossRef]
- Garceau, D.C.; Batson, M.K.; Pan, I.L. Variations on a theme in fruit development: The PLE lineage of MADS-box genes in tomato (TAGL1) and other species. Planta 2017, 246, 313–321. [Google Scholar] [CrossRef]
- Zhong, S.; Yang, H.; Guan, J.; Shen, J.; Ren, T.; Li, Z.; Tan, F.; Li, Q.; Luo, P. Characterization of the MADS-box gene family in Akebia trifoliata and their evolutionary events in Angiosperms. Genes 2022, 13, 1777. [Google Scholar] [CrossRef]
- Zhao, D.; Chen, Z.; Xu, L.; Zhang, L.; Zou, Q. Genome-wide analysis of the MADS-box gene family in Maize: Gene structure, evolution, and relationships. Genes 2021, 12, 1956. [Google Scholar] [CrossRef]
- Preston, J.C.; Fjellheim, S. Flowering time runs hot and cold. Plant Physiol. 2022, 190, 5–18. [Google Scholar] [CrossRef]
- Feng, Y.Y.; Du, H.; Huang, K.Y.; Ran, J.H.; Wang, X.Q. Reciprocal expression of MADS-box genes and DNA methylation reconfiguration initiate bisexual cones in spruce. Commun. Biol. 2024, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; Wu, Y.; Zhang, X.H.; Feng, X.; Wu, H.L.; Zhou, B.J.; Zhang, Y.Q.; Cao, M.; Hou, Z.X. Characterization of the MIKCC-type MADS-box gene family in blueberry and its possible mechanism for regulating flowering in response to the chilling requirement. Planta 2024, 259, 77. [Google Scholar] [CrossRef]
- Li, N.; Huang, B.; Tang, N.; Jian, W.; Zou, J.; Chen, J.; Cao, H.; Habib, S.; Dong, X.; Wei, W.; et al. The MADS-box gene SlMBP21 regulates sepal size mediated by ethylene and auxin in tomato. Plant Cell Physiol. 2017, 58, 2241–2256. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, S.; Guan, J.; Wang, S.; Zhang, H.; Li, G.; Sun, R.; Li, F.; Zhang, S. Single-cell transcriptomic analysis of flowering regulation and vernalization in Chinese cabbage shoot apex. Hortic. Res. 2024, 11, uhae214. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Yan, X.; Ding, L.; Shen, L.; Yu, H. Characterization of C- and D-class MADS-box genes in Orchids. Plant Physiol. 2020, 184, 1469–1481. [Google Scholar] [CrossRef]
- Zuo, Z.W.; Zhang, Z.H.; Huang, D.R.; Fan, Y.Y.; Yu, S.B.; Zhuang, J.Y.; Zhu, Y.J. Control of thousand-grain weight by OsMADS56 in rice. Int. J. Mol. Sci. 2021, 23, 125. [Google Scholar] [CrossRef]
- Zhou, P.; Qu, Y.; Wang, Z.; Huang, B.; Wen, Q.; Xin, Y.; Ni, Z.; Xu, L. Gene structural specificity and expression of MADS-box gene family in Camellia chekiangoleosa. Int. J. Mol. Sci. 2023, 24, 3434. [Google Scholar] [CrossRef]
- Yu, Y.; Chu, X.; Ma, X.; Hu, Z.; Wang, M.; Li, J.; Yin, H. Genome-wide analysis of MADS-box gene family reveals CjSTK as a key regulator of seed abortion in Camellia japonica. Int. J. Mol. Sci. 2024, 25, 5770. [Google Scholar] [CrossRef]


| Gene | Function | Expression Pattern | Homologous Gene |
|---|---|---|---|
| FLC | Control of flowering time, flowering suppressor | Except shoot apex, widely expressed before flowering, download expression caused flowering | Cereal plants have no FLC homologous genes |
| SVP | Control of flowering time, flowering suppressor | Apical meristem of inflorescence, buds, leaves before flowering | Barley: BM1, BM9, HvVRT2; Wheat: TaVRT2 |
| SOC1 | Control of flowering time | Shoot apical meristem, leaves, flower buds | Brassica: LF, MF1, MF2 |
| CAL | Floral meristem identity | Floral meristem | Paralogs of AP1 |
| FUL | Control of flowering time, floral meri stemidentity, fruit development | Inflorescence meristem, ovules, cauline leaves | Paralogs of AP1 |
| AGL24 | Control of flowering time, flowering activator | Floral meristem | Paralogs of SVP |
| AP1 | Floral meristem identity, A-class homeotic gene, regulated sepals and petals | Throughout floral meristem, whorls 1 and whorls 2 of floral organs | Snapdragon: SQUA, DEFH28; Rice: OsMADS14, OsMADS15, OsMADS18 |
| AP3 | B-class homeotic gene, regulated petals and stamens | Whorls 2 and whorls 3 of floral organs | Snapdragon: DEF, Petunia: PhGLO1/2, Rice: OsMADS16, Maize: Si1 |
| PI | B-class homeotic gene, regulated petals and stamens | Whorls 2 and whorls 3 of floral organs | Snapdragon: GLO; Petunia: pMADS1,GP; Rice: OsMADS2, OsMADS4 |
| AG | C-class homeotic gene, regulated stamens and carpels | Whorls 3 and whorls 4 of floral organs | Snapdragon: FAR, Petunia: pMADS3, Rice: OsMADS3, Maize: ZAG1 |
| SHP1/2 | D-class homeotic gene, fruit development and dehiscence | Ovules, valve margin, fruit dehiscence zone | Snapdragon: PLE; Petunia: FBP6 |
| STK | D-class homeotic gene, regulated ovules development | Ovules | Petunia: FBP7, FBP11, Rice: OsMADS13, OsMADS21 |
| SEP1/2/3/4 | E-class homeotic gene, co-regulated floral development, activated B-and C class genes | SEP1/2: all whorls of floral organs; SEP3: whorls 2 and whorls 3; SEP4: whorls 1 | Petunia: FBP2, Tomato: TM5, Rice: OsMADS1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Wu, Y.; Li, R.; Cao, H.; Li, Z.; Li, Q.; Zhou, L. Research Progress on the Regulation of Plant Floral Organ Development by the MADS-box Gene Family. Int. J. Mol. Sci. 2025, 26, 8946. https://doi.org/10.3390/ijms26188946
Wu Q, Wu Y, Li R, Cao H, Li Z, Li Q, Zhou L. Research Progress on the Regulation of Plant Floral Organ Development by the MADS-box Gene Family. International Journal of Molecular Sciences. 2025; 26(18):8946. https://doi.org/10.3390/ijms26188946
Chicago/Turabian StyleWu, Qiufei, Yi Wu, Rui Li, Hongxing Cao, Zongming Li, Qihong Li, and Lixia Zhou. 2025. "Research Progress on the Regulation of Plant Floral Organ Development by the MADS-box Gene Family" International Journal of Molecular Sciences 26, no. 18: 8946. https://doi.org/10.3390/ijms26188946
APA StyleWu, Q., Wu, Y., Li, R., Cao, H., Li, Z., Li, Q., & Zhou, L. (2025). Research Progress on the Regulation of Plant Floral Organ Development by the MADS-box Gene Family. International Journal of Molecular Sciences, 26(18), 8946. https://doi.org/10.3390/ijms26188946






