Exploratory Analysis of Circulating Serum miR-197-3p, miR-1236, and miR-1271 Expression in Early Breast Cancer
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Cohort
4.2. Quality Control Measurements of Biological Material
4.3. Determination of miRNA Expressions
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOCOBAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Borri, F.; Granaglia, A. Pathology of triple negative breast cancer. Semin. Cancer Biol. 2021, 72, 136–145. [Google Scholar] [CrossRef]
- Brinton, L.A.; Gaudet, M.M.; Gierach, G.L. Breast Cancer; Thun, M., Linet, M., Cerhan, J.R., Haiman, C., Schottenfeld, D., Eds.; Oxford University Press: Oxford, UK, 2018; pp. 861–888. [Google Scholar]
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar] [CrossRef]
- Hanker, A.B.; Sudhan, D.R.; Arteaga, C.L. Overcoming Endocrine Resistance in Breast Cancer. Cancer Cell 2020, 37, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Lewis, B.; Harris, O.B.; Watson, C.J.; Davis, F.M. Mammary stem cells: Premise, properties, and perspectives. Trends Cell Biol. 2017, 27, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Bao, T.; Davidson, N.E. Gene expression profiling of breast cancer. Adv. Surg. 2008, 42, 249–260. [Google Scholar] [CrossRef][Green Version]
- Lo, P.K.; Wolfson, B.; Zhou, X.; Duru, N.; Gernapudi, N.; Zhou, Q. Noncoding RNAs in breast cancer. Brief. Funct. Genom. 2016, 15, 200–221. [Google Scholar] [CrossRef] [PubMed]
- Shademan, B.; Avci, C.B.; Karamad, V.; Sogutlu, F.; Nourazarian, A. MicroRNAs as a new target for Alzheimer’s disease treatment. MicroRNA 2023, 12, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Tomasini, S.; Vigo, P.; Margiotta, F.; Scheele, U.S.; Panella, R.; Kauppinen, S. The Role of microRNA-22 in metabolism. Int. J. Mol. Sci. 2025, 26, 782. [Google Scholar] [CrossRef]
- Xie, W.; Shui, C.; Fang, X.; Peng, Y.; Qin, L. miR-197-3p reduces ephitelial-mesenchymal transition by targeting ABCA7 in ovarian cancer cells. 3 Biotech 2020, 10, 375. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cao, L.; Lin, J.-T.; Yuan, Y.; Cao, Z.-L.; Jia, J.-D. Upregulated miRNA-1236-3p in osteosarcoma inhibits cell proliferation and induces apoptosis via targeting KLF8. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6053–6061. [Google Scholar] [PubMed]
- Sun, J.; Zhang, X.; Chen, Z.; Ye, X.; Zhang, C. MiR-1271-5p promotes the growth and migration of neuroblastoma cells by regulating ACY-1. Transl. Cancer Res. 2024, 13, 3397–3406. [Google Scholar] [CrossRef]
- Yu, T.; Yu, H.-R.; Sun, J.-Y. miR-1271 inhibits ERα expression and confers letrozole resistance in breast cancer. Oncotarget 2017, 8, 107134–107148. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Liu, J.; Jia, W.; Jiang, P.; Cheng, Z.; Zhang, Y.; Li, J.; Liu, X.; Tian, C. MiR-197-3p affects angiogenesis and inflammation of endothelial cells by targeting CXCR2/COX2 axis. Am. J. Transl. Res. 2022, 14, 4666–4677. [Google Scholar]
- Wang, W.-T.; Huang, Z.-P.; Sui, S.; Liu, J.-H.; Yu, D.-M.; Wang, W.-B. microRNA-1236 promotes chondrocyte apoptosis in osteoarthritis via direct suppression of PIK3R3. Life Sci. 2020, 253, 117694. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.; Maher, M.; Nassar, Y.; Morcos, G.; Gad, Z. Role of miRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancers females. Gene. 2015, 560, 77–82. [Google Scholar] [CrossRef]
- Liang, T.-C.; Fu, W.-G.; Zhong, Y.-S. MicroRNA-1236-3p inhibits proliferation and invasion of breast cancer cells by targeting ZEB1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9988–9995. [Google Scholar] [PubMed]
- Babaei, Z.; Shahrestanaki, M.K.; Aghaei, M. MiR-1236: Key controller of tumor development and progression: Focus on the biological functions and molecular mechanisms. Pathol. Res. Pract. 2023, 248, 154671. [Google Scholar] [CrossRef]
- Du, H.-Y.; Liu, B. MiR-1271 as a tumor suppressor in breast cancer proliferation and progression via targeting SPIN1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2697–2706. [Google Scholar] [PubMed]
- Fu, Y.; Yang, Q.; Xu, N.; Zhang, X. MiRNA affects the advancement of breast cancer by modulating the immune system’s response. Biochim. Biopys. Acta Mol. Basis Dis. 2025, 1871, 167759. [Google Scholar] [CrossRef]
- de Miranda, F.S.; Slaibi-Filho, J.; Dos Santos, G.C.; Carmo, N.T.; Kenato, C.M.; Borin, T.F.; Luiz, W.B.; Campos, L.C.G. MicroRNA as a promising molecular biomarker in the diagnosis of breast cancer. Front. Mol. Biosci. 2024, 11, 1337706. [Google Scholar] [CrossRef]
- Zablon, F.; Desai, P.; Dellinger, K.; Aravamudhan, S. Cellular and Exosomal microRNAs: Emerging clinical relevance as targets for breast cancer diagnosis and prognosis. Adv. Biol. 2024, 8, e2300532. [Google Scholar] [CrossRef]
- Wang, L.; Wang, G.; Song, J.; Yao, D.; Wang, Y.; Chen, T. A comprehensive analysis of the prognostic characteristics of microRNAs in breast cancer. Front. Genet. 2024, 15, 1293824. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, D.; Wang, H.; Hu, S.; Xie, X.; Zhang, L.; Jia, H.; Qi, Q. MicroRNA-197-3p mediates damage to human coronary artery endothelial cells via targeting TIMP3 in Kawasaki disease. Mol. Cell. Biochem. 2021, 476, 4245–4263. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, Y.; Hu, X.; Zhang, X.; Xu, L.; Yang, Y.; Wu, R.; Wang, E.; Lv, T. Identification of ceRNA (lncRNA-miRNA-mRNA) Regulatory Network in Myocardial Fibrosis After Acute Myocardial Infarction. Int. J. Gen. Med. 2021, 14, 9977–9990. [Google Scholar] [CrossRef]
- Akkaya-Ulum, Y.Z.; Akbaba, T.H.; Tavukcuoglu, Z.; Chae, J.J.; Yilmaz, E.; Ozen, S.; Balcı-Peynircioglu, B. Familial Mediterranean fever-related miR-197-3p targets IL1R1 gene and modulates inflammation in monocytes and synovial fibroblasts. Sci. Rep. 2021, 11, 685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, S.; Bai, Y.; Ali, A.M.; Deng, J.; Chen, Y.; Fu, Y.; He, M. MiR-197-3p Promotes Osteosarcoma Stemness and Chemoresistance by Inhibiting SPOPL. J. Clin. Med. 2023, 12, 1177. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.-M.; Fu, Y.; Zeng, J.; Zhu, X.-Y.; Gao, Y. Cancer-derived exosomal miR-197-3p confers angiogenesis via targeting TIMP2/3 in lung adenocarcinoma metastasis. Cell Death Dis. 2022, 13, 1032. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Saikia, J.; Sharawat, S.K.; Malik, P.S.; Kumar, S.; Mohan, A. Analysis of miR-375-3p, miR-197-3p, and miR-15a-5p Expression and Their Clinical Relevance as Biomarkers in Lung Cancer. Technol. Cancer Res. Treat. 2022, 21, 15330338221080981. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Liu, Z.; Liu, J.; Fan, R. Circ_0060055 Promotes the Growth, Invasion, and Radioresistance of Glioblastoma by Targeting MiR-197-3p/API5 Axis. Neurotox. Res. 2022, 40, 1292–1303. [Google Scholar] [CrossRef]
- Huang, Q.; Ma, B.; Su, Y.; Chan, K.; Qu, H.; Huang, J.; Wang, D.; Qiu, J.; Liu, H.; Yang, X.; et al. miR-197-3p Represses the Proliferation of Prostate Cancer by Regulating the VDAC1/AKT/β-catenin Signaling Axis. Int. J. Biol. Sci. 2020, 16, 1417–1426. [Google Scholar] [CrossRef]
- Li, H.; Li, Q.; He, S. Hsa_circ_0025202 suppresses cell tumorigenesis and tamoxifen resistance via miR-197-3p/HIPK3 axis in breast cancer. World J. Surg. Oncol. 2021, 19, 39. [Google Scholar] [CrossRef]
- Ye, F.; Gao, G.; Zou, Y.; Zheng, S.; Zhang, L.; Ou, X.; Xie, X.; Tang, H. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Mol. Ther. Nucleic Acids 2019, 18, 88–98. [Google Scholar] [CrossRef]
- Yu, Z.; Zhu, X.; Li, Y.; Liang, M.; Liu, M.; Liu, Z.; Qin, L.; Wu, X.; Du, K.; Liu, L.; et al. Circ-HMGA2 (hsa_circ_0027446) promotes the metastasis and epithelial-mesenchymal transition of lung adenocarcinoma cells through the miR-1236-3p/ZEB1 axis. Cell Death Dis. 2021, 12, 313. [Google Scholar] [CrossRef]
- Zhong, S.; Li, Y.; Zhu, X.; Li, Z.; Jin, C.; Zhao, Y. Knockdown of circ_0006225 overcomes resistance to cisplatin and suppresses growth in lung cancer by miR-1236-3p/ANKRD22 axis. J. Biochem. Mol. Toxicol. 2024, 38, e23830. [Google Scholar] [CrossRef] [PubMed]
- Song, T.-F.; Xu, A.-L.; Chen, X.-H.; Gao, J.-Y.; Gao, F.; Kong, X.-C. Circular RNA circRNA_101996 promoted cervical cancer development by regulating miR-1236-3p/TRIM37 axis. Kaohsiung J. Med. Sci. 2021, 37, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Cui, X.; Li, X.; Zang, W.; Chang, M.; Sun, Z.; Liu, Z.; Sun, Y.; Jia, J.; Li, W. CircMAN1A2 is upregulated by Helicobacter pylori and promotes development of gastric cancer. Cell Death Dis. 2022, 13, 409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, P.; Zhang, C. Hepatitis B virus X protein upregulates alpha-fetoprotein to promote hepatocellular carcinoma by targeting miR-1236 and miR-329. J. Cell. Biochem. 2020, 121, 2489–2499. [Google Scholar] [CrossRef]
- Yu, H.; Luo, H.; Liu, X. Knockdown of circ_0102273 inhibits the proliferation, metastasis and glycolysis of breast cancer through miR-1236-3p/PFKFB3 axis. Anti-Cancer Drugs 2022, 33, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Zhang, Y.; Sun, P.; Lin, J.; Wang, C. Knockdown of circADAM9 inhibits cell progression and glycolysis by targeting the miR-1236-3p/FGF7 axis in breast cancer. Thorac. Cancer 2023, 14, 2350–2360. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, J.; Wang, H.; Li, J. Interference of the Circular RNA Sperm Antigen with Calponin Homology and Coiled-Coil Domains 1 Suppresses Growth and Promotes Apoptosis of Breast Cancer Cells Partially Through Targeting miR-1236-3p/Chromobox 8 Pathway. Clin. Breast Cancer. 2024, 24, e138–e151. [Google Scholar] [CrossRef]
- Hao, J.; Du, X.; Lv, F.; Shi, Q. Knockdown of circ_0006528 Suppresses Cell Proliferation, Migration, Invasion, and Adriamycin Chemoresistance via Regulating the miR-1236-3p/CHD4 Axis in Breast Cancer. J. Surg. Res. 2021, 260, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Huang, P.; Wu, H.; Wang, S.; Liu, G. LINC02381 aggravates breast cancer through the miR-1271-5p/FN1 axis to activate PI3K/AKT pathway. Mol. Carcinog. 2022, 61, 346–358. [Google Scholar] [CrossRef]
- Fontana, A.; Barbano, R.; Dama, E.; Pasculli, B.; Rendina, M.; Morritti, M.G.; Melocchi, V.; Castelvetere, M.; Valori, V.M.; Ravaioli, S.; et al. Combined analysis of miR-200 family and its significance for breast cancer. Sci. Rep. 2021, 11, 2980. [Google Scholar] [CrossRef]
- Garrido-Placios, A.; Carvajal, A.M.R.; Nunez-Negrillo, A.M.; Cortes-Martin, J.; Sanchez-Garcia, J.C.; Aguilar-Cordero, M.J. MicroRNA Dysregulation in Early Breast Cancer Diagnosis: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2023, 24, 8270. [Google Scholar] [CrossRef] [PubMed]
- de Santana Silva, I.T.S.; Fehlberg, H.F.; Ferreira, F.B.; da Silvai, M.F.; dos Santos, P.R.; Porto, V.M.; Dias, J.C.T.; Alboquerque, G.R.; Mariano, A.P.M.; Gadelha, S.R.; et al. Identification of SnRNA U6 as an endogenous reference gene for normalization of MiRNA expression data in COVİD-19 patients. Sci. Rep. 2025, 15, 16636. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
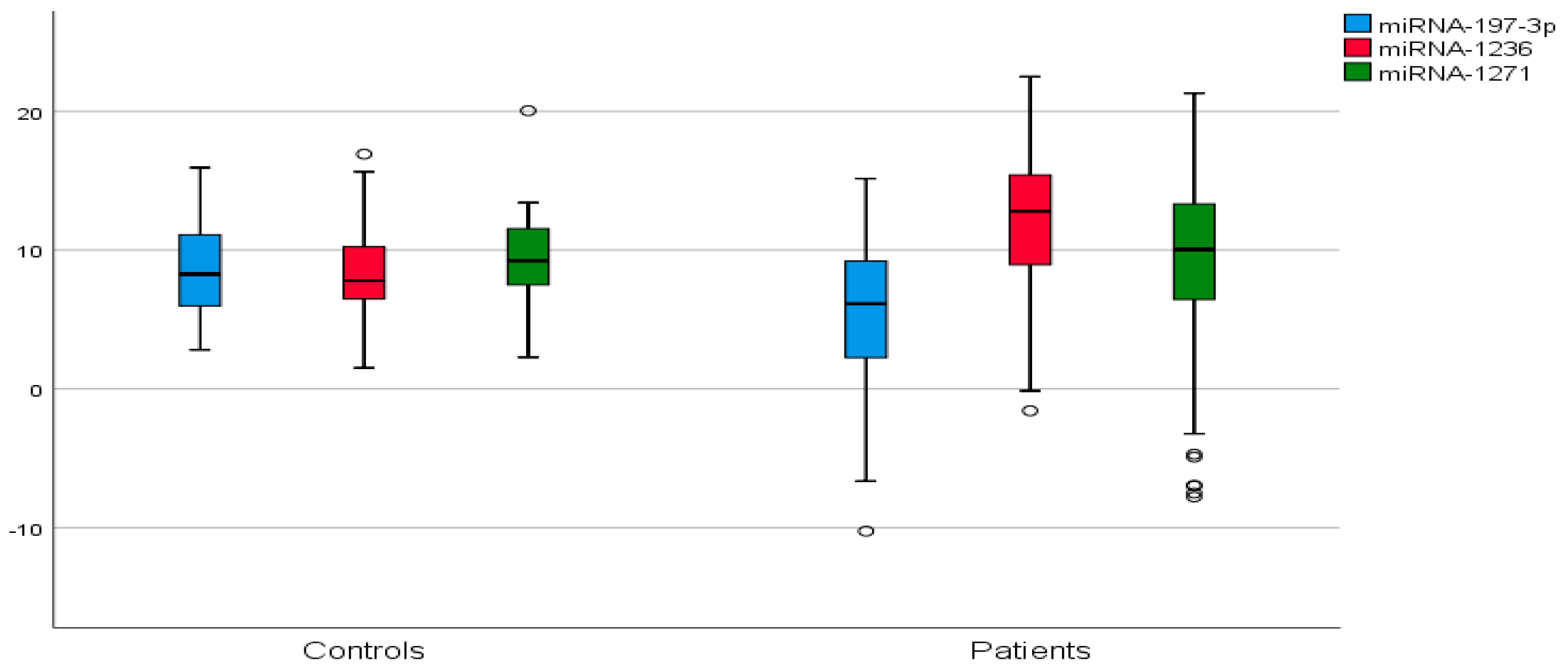
| miRNA | ΔCt | STD | 2−ΔΔCt | Fold Change | p | |||
|---|---|---|---|---|---|---|---|---|
| Patients | Controls | Patients | Controls | Patients | Controls | |||
| miR-197-3p | 5.32 | 8.48 | 5.31 | 3.47 | 0.02503 | 0.00280 | 8.939 | 0.0048 |
| miR-1236 | 11.66 | 8.49 | 5.08 | 3.23 | 0.00031 | 0.00278 | 0.112 | 0.0029 |
| miR-1271 | 8.69 | 9.20 | 6.29 | 3.63 | 0.00242 | 0.001710 | 1.42 | 0.6712 |
| Univariate Model | Regression Model | ||||||
|---|---|---|---|---|---|---|---|
| Patients | Controls | p | OR | 95%CI Exp(B) | p | ||
| miR-197-3p | Low | 29 (31.5%) | 18 (58.1%) | 0.008 | 0.33 | 0.14–0.77 | 0.010 |
| High | 63 (68.5%) | 13 (41.9%) | |||||
| miR-1236 | Low | 65 (70.7%) | 10 (32.3%) | <0.001 | 5.06 | 2.10–12.15 | <0.001 |
| High | 27 (29.3%) | 21 (67.7%) | |||||
| miR-1271 | Low | 36 (39.1%) | 13 (41.9%) | 0.472 | 1.12 | 0.49–2.59 | 0.783 |
| High | 56 (60.9%) | 18 (58.1%) | |||||
| Age | <50 | 43 (46.7%) | 19 (61.3%) | 0.161 | 1.80 | 0.77–4.14 | 0.164 |
| >50 | 49 (53.3%) | 12 (38.7%) | |||||
| Menopause | Pre | 42 (45.7%) | 20 (64.5%) | 0.069 | 2.17 | 0.93–5.03 | 0.072 |
| Post | 50 (54.3%) | 11 (35.5%) | |||||
| Birth history | No | 13 (14.1%) | 15 (48.4%) | <0.001 | 5.70 | 2.28–14.25 | <0.001 |
| Yes | 79 (85.9%) | 16 (51.6%) | |||||
| Bad habits | No | 84 (91.3%) | 19 (61.3%) | <0.001 | 0.15 | 0.05–0.42 | <0.001 |
| Yes | 8 (8.7%) | 12 (38.7%) | |||||
| Oral contraceptive | No | 86 (93.5%) | 18 (58.1%) | <0.001 | 0.09 | 0.03–0.29 | <0.001 |
| Yes | 6 (6.5%) | 13 (41.9%) | |||||
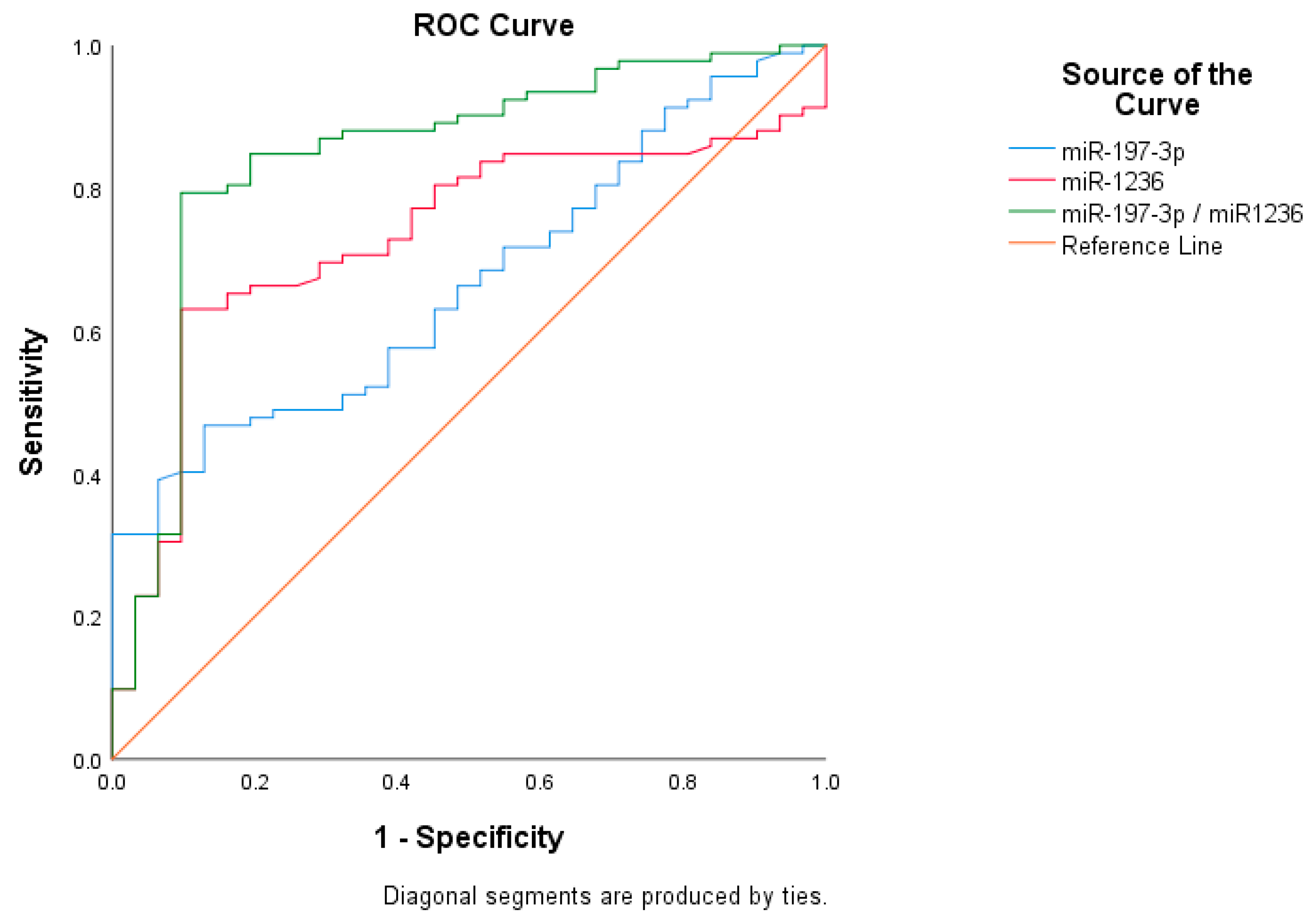
| Assay | Cut-Off Value | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | AUC (CI) | p |
|---|---|---|---|---|---|---|---|
| miR-197-3p | 7.32 | 67.0% | 45.6% | 79.6% | 30.9% | 0.667 (0.567–0.786) | 0.0056 |
| miR-1236 | 9.87 | 80.7% | 51.3% | 89.3% | 53.7% | 0.731 (0.636–0.827) | 0.0001 |
| Asymptotic 95% Confidence Interval | ||||
|---|---|---|---|---|
| Test Variable | AUC | Asymptotic Sig | Lower Bound | Upper Bound |
| miR-197-3p–miR-1236 | 0.842 | <0.001 | 0.764 | 0.936 |
| miR-197-3p | miR-1236 | |||||
|---|---|---|---|---|---|---|
| Variables | n (%) | Mean ± STD | p | Mean ± STD | p | |
| Age | <50 | 35 (38.0%) | 5.68 ± 4.39 | 0.053 | 12.32 ± 3.59 | 0.326 |
| ≥50 | 57 (62.0%) | 7.62 ± 4.48 | 11.25 ± 5.80 | |||
| Tumor size | <2 cm | 36 (39.1%) | 4.86 ± 5.34 | 0.510 | 11.17 ± 3.58 | 0.463 |
| ≥2 cm | 56 (60.9%) | 5.61 ± 5.32 | 11.97 ± 4.75 | |||
| Breast cancer subtype | IDC | 75 (81.5%) | 5.59 ± 5.13 | 0.303 | 11.57 ± 4.95 | 0.733 |
| Other BC subtypes * | 17 (18.5%) | 4.11 ± 6.04 | 12.04 ± 5.76 | |||
| Histologic grade | 1–2 | 59 (64.1%) | 5.58 ± 5.07 | 0.531 | 11.84 ± 5.15 | 0.647 |
| 3 | 33 (35.9%) | 4.85 ± 5.77 | 11.33 ± 5.01 | |||
| cT | 1–2 | 65 (70.7%) | 5.24 ± 5.02 | 0.842 | 11.53 ± 5.13 | 0.716 |
| 3 | 27 (29.3%) | 5.50 ± 5.25 | 11.85 ± 5.02 | |||
| cN | Negative | 52 (56.5%) | 5.86 ± 5.07 | 0.265 | 11.82 ± 5.27 | 0.723 |
| Positive | 40 (43.5%) | 4.81 ± 5.59 | 11.44 ± 4.88 | |||
| ER | Negative | 15 (16.3%) | 5.33 ± 5.14 | 0.891 | 11.95 ± 4.91 | 0.806 |
| Positive | 77 (83.7%) | 5.31 ± 5.37 | 11.60 ± 5.14 | |||
| PR | Negative | 20 (21.7%) | 4.96 ± 4.97 | 0.737 | 11.06 ± 4.91 | 0.554 |
| Positive | 72 (78.3%) | 5.41 ± 5.43 | 11.82 ± 5.15 | |||
| HER2 | Negative | 72 (78.3%) | 5.48 ± 5.16 | 0.569 | 11.97 ± 4.84 | 0.261 |
| Positive | 20 (21.7%) | 4.71 ± 5.91 | 10.52 ± 5.84 | |||
| Receptor status | Luminal | 76 (82.6%) | 5.38 ± 5.37 | 0.807 | 11.66 ± 5.13 | 0.956 |
| Non-luminal | 16 (17.4%) | 5.02 ± 5.12 | 11.59 ± 4.96 | |||
| Tumor stage | 1–2 | 79 (85.9%) | 5.14 ± 5.41 | 0.439 | 11.53 ± 5.13 | 0.718 |
| 3 | 13 (14.1%) | 6.38 ± 4.72 | 11.95 ± 5.02 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
İlhan, B.; Kuraş, S.; Kılıç, B.; Tilgen Yasasever, C.; Oğuz Soydinç, H.; Alsaadoni, H.; Öztan, G.; Adamnejad Ghafour, A.; Ucuncu, M.; Kunduz, E.; et al. Exploratory Analysis of Circulating Serum miR-197-3p, miR-1236, and miR-1271 Expression in Early Breast Cancer. Int. J. Mol. Sci. 2025, 26, 8944. https://doi.org/10.3390/ijms26188944
İlhan B, Kuraş S, Kılıç B, Tilgen Yasasever C, Oğuz Soydinç H, Alsaadoni H, Öztan G, Adamnejad Ghafour A, Ucuncu M, Kunduz E, et al. Exploratory Analysis of Circulating Serum miR-197-3p, miR-1236, and miR-1271 Expression in Early Breast Cancer. International Journal of Molecular Sciences. 2025; 26(18):8944. https://doi.org/10.3390/ijms26188944
Chicago/Turabian Styleİlhan, Burak, Sibel Kuraş, Berkay Kılıç, Ceren Tilgen Yasasever, Hilal Oğuz Soydinç, Hani Alsaadoni, Gözde Öztan, Arash Adamnejad Ghafour, Muhammed Ucuncu, Enver Kunduz, and et al. 2025. "Exploratory Analysis of Circulating Serum miR-197-3p, miR-1236, and miR-1271 Expression in Early Breast Cancer" International Journal of Molecular Sciences 26, no. 18: 8944. https://doi.org/10.3390/ijms26188944
APA Styleİlhan, B., Kuraş, S., Kılıç, B., Tilgen Yasasever, C., Oğuz Soydinç, H., Alsaadoni, H., Öztan, G., Adamnejad Ghafour, A., Ucuncu, M., Kunduz, E., & Bademler, S. (2025). Exploratory Analysis of Circulating Serum miR-197-3p, miR-1236, and miR-1271 Expression in Early Breast Cancer. International Journal of Molecular Sciences, 26(18), 8944. https://doi.org/10.3390/ijms26188944






