Exploring Genetic Diversity and Inter-/Intraspecific Polymorphism in Rheum sp. (Polygonaceae) Using the iPBS Retrotransposon Marker System
Abstract
1. Introduction
2. Results
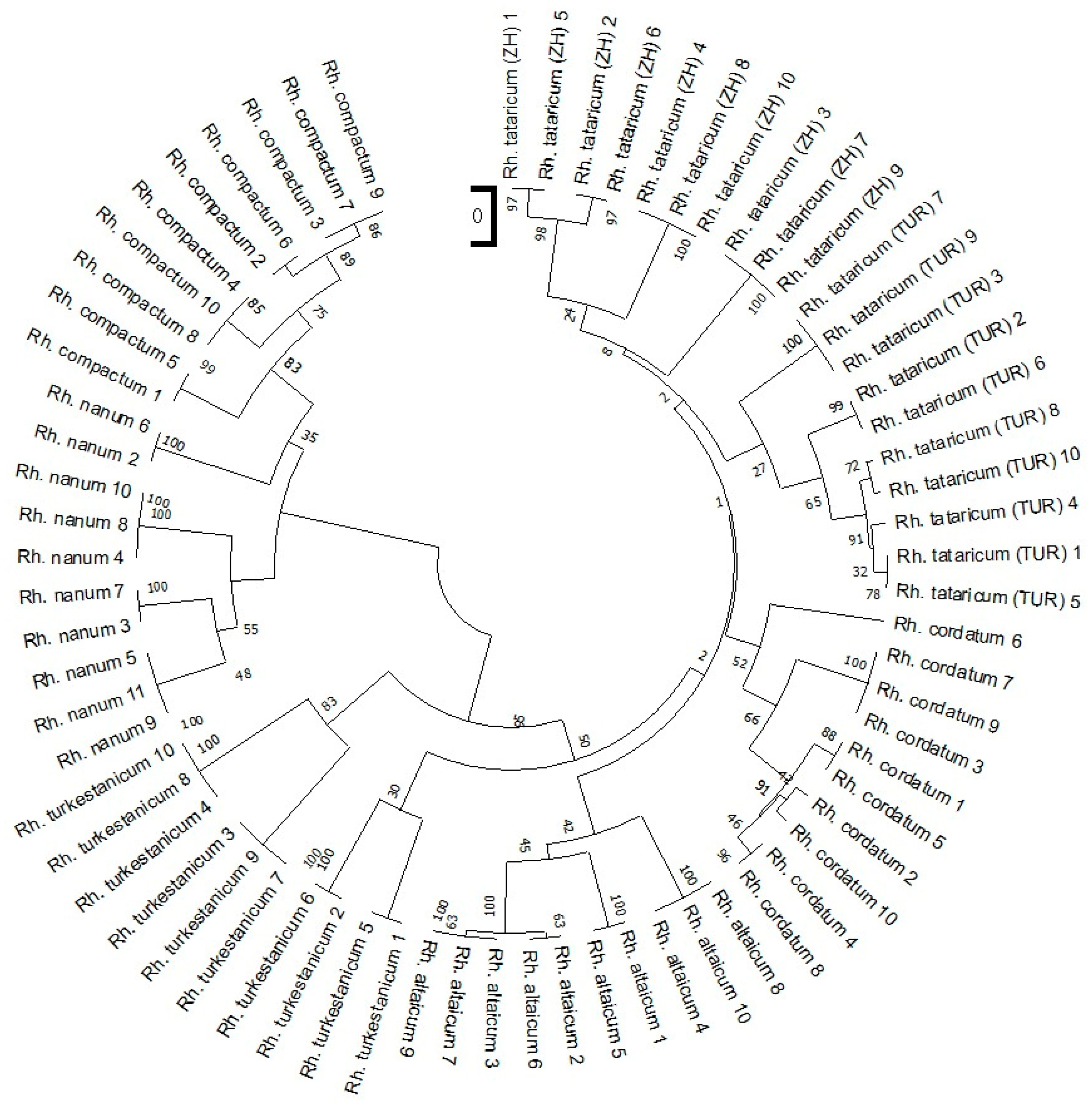
2.1. iPBS Profiling of Rheum Species and Ecopopulations
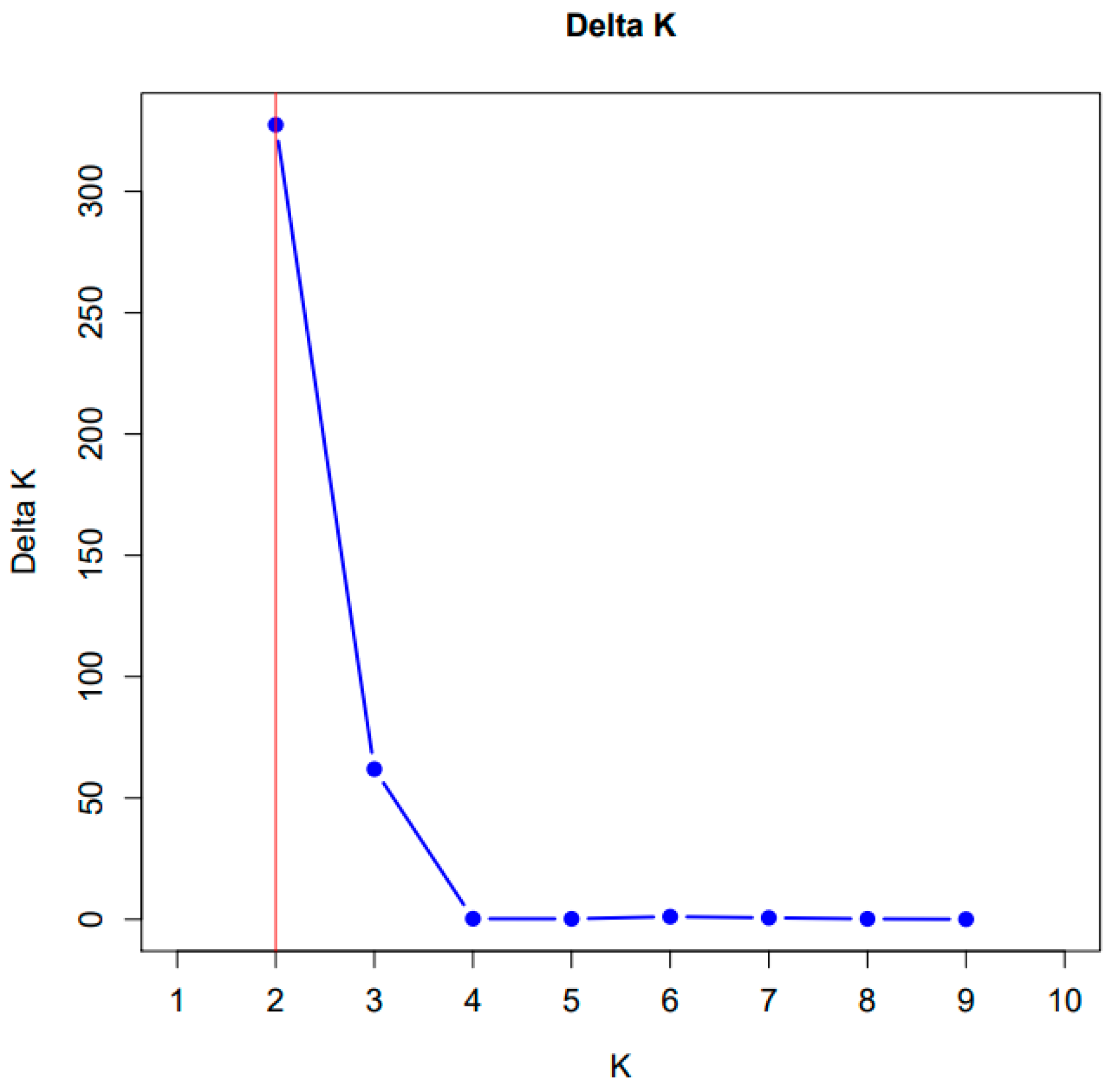
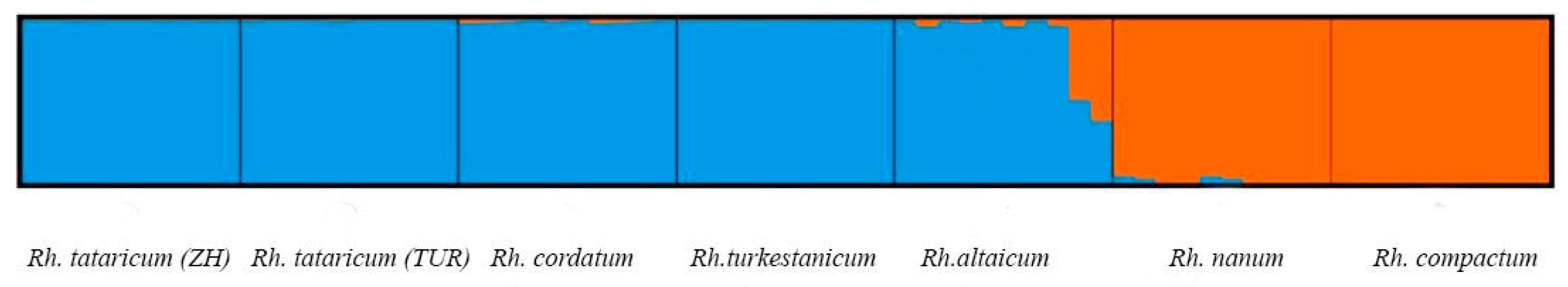
2.2. Statistical Analysis of iPBS Profiling Data of Rheum Species and Ecopopulations
3. Discussion
4. Materials and Methods
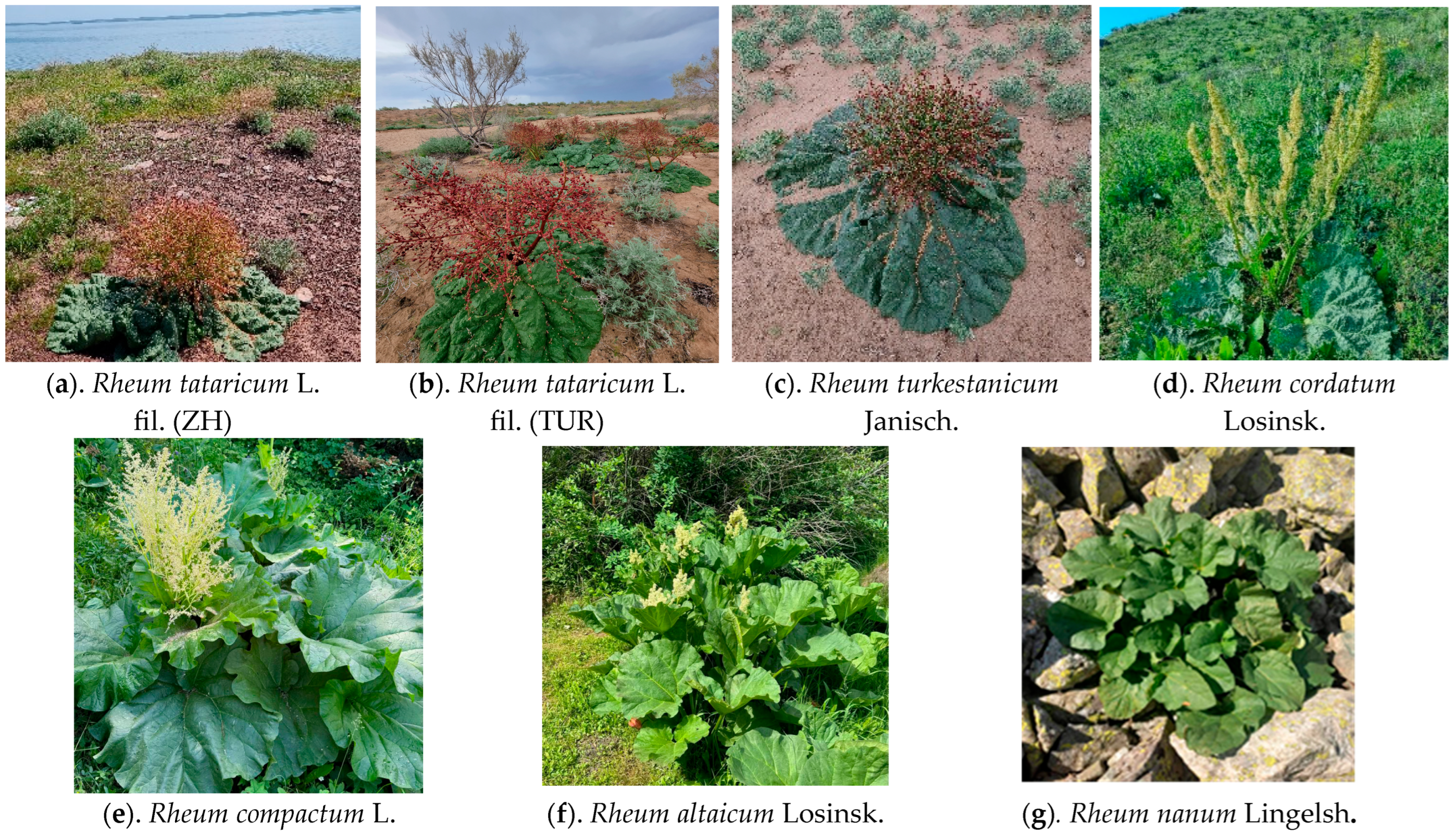
4.1. Research Materials
4.1.1. Rheum tataricum L. fil.
4.1.2. Rheum turkestanicum Janisch.
4.1.3. Rheum cordatum Losinsk.
4.1.4. Rheum compactum L.
4.1.5. Rheum altaicum Losinsk.
4.1.6. Rheum nanum Lingelsh.
4.2. Research Methods
4.2.1. DNA Extraction
4.2.2. iPBS Profiling of Rheum Species
4.2.3. Data Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMOVA | Analysis of Molecular Variance |
| a.s.l. | Above sea level |
| iPBS | Inter-Primer Binding Site |
| SSR | Simple Sequence Repeats |
| ISSR | Inter-Simple Sequence Repeats |
| RAPD | Random Amplified Polymorphic DNA |
| DNA | Deoxyribonucleic Acid |
| LTR | Long Terminal Repeat |
| PCR | Polymerase Chain Reaction |
| PCoA | Principal Coordinate Analysis |
| UPGMA | Unweighted Pair Group Method with Arithmetic Mean |
| ROS | Reactive Oxygen Species |
| PVP | Polyvinylpyrrolidone |
| CTAB | Cetyltrimethylammonium Bromide |
References
- Chen, F.; Wang, A.; Chen, K.; Wan, D.; Liu, J. Genetic Diversity and Population Structure of the Endangered and Medically Important Rheum tanguticum (Polygonaceae) Revealed by SSR Markers. Biochem. Syst. Ecol. 2009, 37, 613–621. [Google Scholar] [CrossRef]
- Sanchez, A.; Schuster, T.M.; Burke, J.M.; Kron, K.A. Taxonomy of Polygonoideae (Polygonaceae): A New Tribal Classification. TAXON 2011, 60, 151–160. [Google Scholar] [CrossRef]
- Wani, I.A.; Verma, S.; Ahmad, P.; El-Serehy, H.A.; Hashim, M.J. Reproductive Biology of Rheum webbianum Royle, a Vulnerable Medicinal Herb From Alpines of North-Western Himalaya. Front. Plant Sci. 2022, 13, 699645. [Google Scholar] [CrossRef]
- Grudzinskaya, L.M.; Gemedzhieva, N.G.; Nelina, N.V.; Karzhaubekova, Z.Z. Annotated List of Medicinal Plants of Kazakhstan: Reference Edition; Institute of Botany and Phytointroduction: Almaty, Kazakhstan, 2014; p. 114. [Google Scholar]
- Uniyal, A.; Kumar, A.; Upadhyay, S.; Kumar, V.; Gupta, S. Assessment of Genetic Diversity of Rheum Species (Endangered Medicinal Herb of Indian Himalayan Region) Using Molecular Markers. Res. J. Biotechnol. 2021, 16, 147–154. [Google Scholar] [CrossRef]
- Aitzhan, M.; Aimenova, Z.; Sumbembayev, A. Studying the Species Distribution of Rhubarbs (Rheum L.) in Kazakhstan. BIO Web Conf. 2024, 100, 04034. [Google Scholar] [CrossRef]
- Gemedzhieva, N.G.; Sayakova, G.M.; Zhumashova, G.T. Review of the Current State of Knowledge of Kazakh Species of Rheum L. (Polygonaceae Juss.). Pharm. Kazakhstan 2015, 12, 22–28. [Google Scholar]
- Vysochina, G.I. Anthraquinones and Biological Activity of Species of The Genus rheum L. (Polygonaceae). Khimiya Rastit. Syr’ya 2018, 29–41. [Google Scholar] [CrossRef]
- Xu, Q.Q.Y.; Su, X.J.; Luo, W.S. Chemical Constituents in Rheum palmatum. Chin. Tradit. Herb. Drugs 2009, 40, 533–536. [Google Scholar]
- He, J.; Wang, L.; Guo, H.; Zhao, H.; Sun, J. Chemistry, Pharmacology and Processing Method of Rhubarb (Rheum Species): A Review. J. Food Bioact. 2019, 8, 42–50. [Google Scholar] [CrossRef]
- Matsuda, H.; Tomohiro, N.; Hiraba, K.; Harima, S.; Ko, S.; Matsuo, K.; Yoshikawa, M.; Kubo, M. Study on Anti-Oketsu Activity of Rhubarb II. Anti-Allergic Effects of Stilbene Components from Rhei Undulati Rhizoma (Dried Rhizome of Rheum undulatum Cultivated in Korea). Biol. Pharm. Bull. 2001, 24, 264–267. [Google Scholar] [CrossRef]
- Zheng, Q.; Wu, H.; Guo, J.; Nan, H.; Chen, S.; Yang, J.; Xu, X. Review of Rhubarbs: Chemistry and Pharmacology. Chin. Herb. Med. 2013, 5, 9–32. [Google Scholar] [CrossRef]
- Cole, C.T. Genetic Variation in Rare and Common Plants. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 213–237. [Google Scholar] [CrossRef]
- Kim, S.-C.; Lee, C.; Santos-Guerra, A. Genetic Analysis and Conservation of the Endangered Canary Island Woody Sow-Thistle, Sonchus Gandogeri (Asteraceae). J. Plant Res. 2005, 118, 147–153. [Google Scholar] [CrossRef]
- Ayala, F.J.; Kiger, J.A. Modern Genetics, 2nd ed.; Benjamin/Cummings Publishing Company: San Francisco, CA, USA, 1984; ISBN 978-0-8053-0316-2. [Google Scholar]
- Zhivotovsky, L.A. Population Structure of Species: Eco-Geographic Units and Genetic Differentiation between Populations. Russ. J. Mar. Biol. 2016, 42, 373–382. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of Genetic Diversity Assessment in Crop Plants and Its Recent Advances: An Overview of Its Analytical Perspectives. Genet. Res. Int. 2015, 2015, 431487. [Google Scholar] [CrossRef]
- Gitzendanner, M.A.; Soltis, P.S. Patterns of Genetic Variation in Rare and Widespread Plant Congeners. Am. J. Bot. 2000, 87, 783–792. [Google Scholar] [CrossRef]
- Bornet, B.; Branchard, M. Nonanchored Inter Simple Sequence Repeat (ISSR) Markers: Reproducible and Specific Tools for Genome Fingerprinting. Plant Mol. Biol. Rep. 2001, 19, 209–215. [Google Scholar] [CrossRef]
- Liu, S.; Gao, J.; Xiao, B.; Guo, W.; Yu, Q.; Wang, A.; Li, W. Genetic Differentiation and Historical Dynamics of the Endemic Species Rheum pumilum on the Qinghai-Tibetan Plateau Inferred from Phylogeography Implications. BMC Plant Biol. 2025, 25, 162. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, N.; Chen, L.; Zhang, Z.; Lei, H.; Wei, S.; Liu, C. Genetic Diversity and Genetic Differentiation of Rheum palmatum by Chloroplast matK Sequences. Nat. Prod. J. 2020, 10, 96–103. [Google Scholar] [CrossRef]
- Wang, X.; Yang, R.; Feng, S.; Hou, X.; Zhang, Y.; Li, Y.; Ren, Y. Genetic Variation in Rheum palmatum and Rheum tanguticum (Polygonaceae), Two Medicinally and Endemic Species in China Using ISSR Markers. PLoS ONE 2012, 7, e51667. [Google Scholar] [CrossRef] [PubMed]
- Tabin, S.; Kamili, A.N.; Ganie, S.A.; Zargar, O.; Sharma, V.; Gupta, R.C. Genetic Diversity and Population Structure of Rheum Species in Kashmir Himalaya Based on ISSR Markers. Flora 2016, 223, 121–128. [Google Scholar] [CrossRef]
- Kumar, A.; Bisht, Y.; Rautela, K.; Jugran, A.K.; Bhatt, I.D.; Bargali, S.S. Morphological and Genetic Diversity Assessment of Rheum Australe D. Don–A High Value Medicinal Herb from the Himalaya, and Implications for Conservation Strategies. S. Afr. J. Bot. 2023, 163, 620–629. [Google Scholar] [CrossRef]
- Ekincialp, A.; Erdinç, Ç.; Turan, S.; Cakmakci, O.; Nadeem, M.A.; Baloch, F.S.; Sensoy, S. Genetic Characterization of Rheum ribes (Wild Rhubarb) Genotypes in Lake Van Basin of Turkey through ISSR and SSR Markers. Int. J. Agric. Biol. 2019, 21, 795–802. [Google Scholar] [CrossRef]
- Persson, H.A.; Rumpunen, K.; Möllerstedt, L.K. Identification of Culinary Rhubarb (Rheum Spp.) Cultivars Using Morphological Characterization and RAPD Markers. J. Hortic. Sci. Biotechnol. 2000, 75, 684–689. [Google Scholar] [CrossRef]
- Zhai, Y.; Zhang, T.; Guo, Y.; Gao, C.; Zhou, L.; Feng, L.; Zhou, T.; Xumei, W. Phylogenomics, Phylogeography and Germplasms Authentication of the Rheum palmatum Complex Based on Complete Chloroplast Genomes. J. Plant Res. 2023, 136, 291–304. [Google Scholar] [CrossRef]
- Schulman, A.H.; Flavell, A.J.; Ellis, T.H.N. The Application of LTR Retrotransposons as Molecular Markers in Plants. In Mobile Genetic Elements: Protocols and Genomic Applications; Miller, W.J., Capy, P., Eds.; Humana Press: Totowa, NJ, USA, 2004; pp. 145–173. ISBN 978-1-59259-755-0. [Google Scholar]
- Kalendar, R.; Antonius, K.; Smýkal, P.; Schulman, A.H. iPBS: A Universal Method for DNA Fingerprinting and Retrotransposon Isolation. Theor. Appl. Genet. 2010, 121, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Ivashuta, S.; Naumkina, M.; Gau, M.; Uchiyama, K.; Isobe, S.; Mizukami, Y.; Shimamoto, Y. Genotype-Dependent Transcriptional Activation of Novel Repetitive Elements during Cold Acclimation of Alfalfa (Medicago sativa). Plant J. 2002, 31, 615–627. [Google Scholar] [CrossRef]
- Naito, K.; Zhang, F.; Tsukiyama, T.; Saito, H.; Hancock, C.N.; Richardson, A.O.; Okumoto, Y.; Tanisaka, T.; Wessler, S.R. Unexpected Consequences of a Sudden and Massive Transposon Amplification on Rice Gene Expression. Nature 2009, 461, 1130–1134. [Google Scholar] [CrossRef]
- Borges, F.; Martienssen, R.A. The Expanding World of Small RNAs in Plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef]
- Ito, H.; Kim, J.-M.; Matsunaga, W.; Saze, H.; Matsui, A.; Endo, T.A.; Harukawa, Y.; Takagi, H.; Yaegashi, H.; Masuta, Y.; et al. A Stress-Activated Transposon in Arabidopsis Induces Transgenerational Abscisic Acid Insensitivity. Sci. Rep. 2016, 6, 23181. [Google Scholar] [CrossRef]
- Lanciano, S.; Mirouze, M. Transposable Elements: All Mobile, All Different, Some Stress Responsive, Some Adaptive? Curr. Opin. Genet. Dev. 2018, 49, 106–114. [Google Scholar] [CrossRef]
- Porquier, A.; Tisserant, C.; Salinas, F.; Glassl, C.; Wange, L.; Enard, W.; Hauser, A.; Hahn, M.; Weiberg, A. Retrotransposons as Pathogenicity Factors of the Plant Pathogenic Fungus Botrytis cinerea. Genome Biol. 2021, 22, 225. [Google Scholar] [CrossRef]
- Galindo-González, L.; Mhiri, C.; Deyholos, M.K.; Grandbastien, M.-A. LTR-Retrotransposons in Plants: Engines of Evolution. Gene 2017, 626, 14–25. [Google Scholar] [CrossRef]
- Papolu, P.K.; Ramakrishnan, M.; Mullasseri, S.; Kalendar, R.; Wei, Q.; Zou, L.-H.; Ahmad, Z.; Vinod, K.K.; Yang, P.; Zhou, M. Retrotransposons: How the Continuous Evolutionary Front Shapes Plant Genomes for Response to Heat Stress. Front. Plant Sci. 2022, 13, 1064847. [Google Scholar] [CrossRef]
- Kalendar, R. The Use of Retrotransposon-Based Molecular Markers to Analyze Genetic Diversity. Ratar. I Povrt. 2011, 48, 261–274. [Google Scholar] [CrossRef]
- Khapilina, O.; Turzhanova, A.; Zhumagul, M.; Tagimanova, D.; Raiser, O.; Kubentayev, S.; Shevtsov, V.; Hohn, M. Retrotransposon-Based Genetic Diversity of Rhodiola rosea L. (Crassulaceae) from Kazakhstan Altai. Diversity 2025, 17, 45. [Google Scholar] [CrossRef]
- Terletskaya, N.V.; Turzhanova, A.S.; Khapilina, O.N.; Zhumagul, M.Z.; Meduntseva, N.D.; Kudrina, N.O.; Korbozova, N.K.; Kubentayev, S.A.; Kalendar, R. Genetic Diversity in Natural Populations of Rhodiola Species of Different Adaptation Strategies. Genes 2023, 14, 794. [Google Scholar] [CrossRef] [PubMed]
- Uras, M.; Filiz, E.; Sen, U.; ÖZYİĞİT, İ. Genetic Diversity and Phylogenetic Analysis of Robinia pseudoacacia L. Populations Using ISSR Markers, ITS1 and trnL-F Intergenic Spacer Sequences. J. For. Sci. 2024, 70, 1–13. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, W.; Yang, Z.; Luo, C.; Zhang, W.; Xu, F.; Ye, J.; Liao, Y. Genetic Diversity Analysis and DNA Fingerprint Construction of Zanthoxylum Species Based on SSR and iPBS Markers. BMC Plant Biol. 2024, 24, 843. [Google Scholar] [CrossRef]
- Borna, F.; Luo, S.; Ahmad, N.M.; Nazeri, V.; Shokrpour, M.; Trethowan, R. Genetic Diversity in Populations of the Medicinal Plant Leonurus cardiaca L. Revealed by Inter-Primer Binding Site (iPBS) Markers. Genet. Resour. Crop Evol. 2017, 64, 479–492. [Google Scholar] [CrossRef]
- Sadık, G.; Yıldız, M.; Taşkın, B.; Koçak, M.; Cavagnaro, P.F.; Baloch, F.S. Application of iPBS-Retrotransposons Markers for the Assessment of Genetic Diversity and Population Structure among Sugar Beet (Beta vulgaris) Germplasm from Different Regions of the World. Genet. Resour. Crop Evol. 2025, 72, 3039–3049. [Google Scholar] [CrossRef]
- Yin, W.-Y.; Cui, X.-Q.; Zhao, Q.-J.; Deng, J.-L.; Huang, C.-Y.; Zhang, Z.-B.; Li, J.-W. Genetic Relationship Analysis and Fingerprint Construction of Some Species in the Genus Vanda (Orchidaceae) by Interprimer Binding Site (iPBS) Markers. Genet. Resour. Crop Evol. 2025, 72, 935–946. [Google Scholar] [CrossRef]
- Milovanov, A.; Zvyagin, A.; Daniyarov, A.; Kalendar, R.; Troshin, L. Genetic Analysis of the Grapevine Genotypes of the Russian Vitis Ampelographic Collection Using iPBS Markers. Genetica 2019, 147, 91–101. [Google Scholar] [CrossRef]
- Ali, F.; Yılmaz, A.; Nadeem, M.A.; Habyarimana, E.; Subaşı, I.; Nawaz, M.A.; Chaudhary, H.J.; Shahid, M.Q.; Ercişli, S.; Zia, M.A.B.; et al. Mobile Genomic Element Diversity in World Collection of Safflower (Carthamus tinctorius L.) Panel Using iPBS-Retrotransposon Markers. PLoS ONE 2019, 14, e0211985. [Google Scholar] [CrossRef]
- Kalendar, R.; Ivanov, K.I.; Akhmetollayev, I.; Kairov, U.; Samuilova, O.; Burster, T.; Zamyatnin, A.A., Jr. An Improved Method and Device for Nucleic Acid Isolation Using a High-Salt Gel Electroelution Trap. Anal. Chem. 2024, 96, 15526–15530. [Google Scholar] [CrossRef]
- Kalendar, R.; Ivanov, K.I.; Samuilova, O.; Kairov, U.; Zamyatnin, A.A., Jr. Isolation of High-Molecular-Weight DNA for Long-Read Sequencing Using a High-Salt Gel Electroelution Trap. Anal. Chem. 2023, 95, 17818–17825. [Google Scholar] [CrossRef]
- Bonchev, G.N.; Vassilevska-Ivanova, R. Fingerprinting the Genetic Variation and Intergeneric Hybrid Dynamics in the Family Asteraceae (Genera Helianthus, Echinaceae, Tagetes and Verbesina) Using iPBS Markers. Biologia 2020, 75, 457–464. [Google Scholar] [CrossRef]
- Khapilina, O.; Raiser, O.; Danilova, A.; Shevtsov, V.; Turzhanova, A.; Kalendar, R. DNA Profiling and Assessment of Genetic Diversity of Relict Species Allium altaicum Pall. on the Territory of Altai. PeerJ 2021, 9, e10674. [Google Scholar] [CrossRef] [PubMed]
- Naeem, H.; Awan, F.S.; Dracatos, P.M.; Sajid, M.W.; Saleem, S.; Yousafi, Q.; Khan, M.S.; Mehmood, A.; Zulfiqar, B. Population Structure and Phylogenetic Relationship of Peach [Prunus persica (L.) Batsch] and Nectarine [Prunus persica Var. Nucipersica (L.) C.K. Schneid.] Based on Retrotransposon Markers. Genet. Resour. Crop Evol. 2021, 68, 3011–3023. [Google Scholar] [CrossRef]
- Terletskaya, N.V.; Khapilina, O.N.; Turzhanova, A.S.; Erbay, M.; Magzumova, S.; Mamirova, A. Genetic Polymorphism in the Amaranthaceae Species in the Context of Stress Tolerance. Plants 2023, 12, 3470. [Google Scholar] [CrossRef]
- Sameeullah, M.; Kayaçetin, F.; Khavar, K.M.; Perkasa, A.Y.; Maesaroh, S.; Waheed, M.T.; Çiftçi, V. Decoding Genetic Diversity and Population Structure of Brassica Species by Inter Primer Binding Site (iPBS) Retrotransposon Markers. Genet. Resour. Crop Evol. 2025, 72, 417–427. [Google Scholar] [CrossRef]
- Backlund, P. Effects of Climate Change on Agriculture, Land Resources, Water Resources and Biodiversity in the United States; DIANE Publishing: Darby, PA, USA, 2009; 240p. [Google Scholar]
- Kurmanbayeva, M.S.; Terletskaya, N.V.; Gemejiyeva, N.G.; Karzhaubekova, Z.Z.; Kudrina, N.O.; Yerbay, M.; Kusmangazinov, A.B.; Karabalayeva, D. Comparative Analysis of the Anatomical and Morphological Features of Rheum tataricum L. Plants from Different Ecopopulations. Eurasian J. Ecol. 2024, 79, 127–134. [Google Scholar] [CrossRef]
- Tabin, S.; Kamili, A.N.; Gupta, R.C. Morphological Studies and Development of Ex-Situ Protocol for Rehabilitation of Threatened Rheum Species under Nursery Conditions. Curr. Bot. 2016, 7, 17–26. [Google Scholar] [CrossRef][Green Version]
- Bhardwaj, R.; Sood, M. Morphological, Phenological and Seed Germination Studies in Rheum australe D Don: An Endangered Medicinal Plant. J. Pharmacogn. Phytochem. 2020, 9, 2003–2006. [Google Scholar][Green Version]
- Sumbembayev, A.A.; Lagus, O.A.; Nowak, S. Seed Morphometry of Rheum L. (Polygonaceae) Species from Kazakhstan and Its Implications in Taxonomy and Species Identification. Biodiversitas J. Biol. Divers. 2023, 24, 4677–4692. [Google Scholar] [CrossRef]
- Vysochina, G.I. Phenolic Compounds in Systematics and Phylogeny of the Family Polygonaceae Juss. VI. Genus Knorringia (Chukav.) Tzvel. Turczaninowia 2014, 17, 33–41. [Google Scholar] [CrossRef]
- Tan, L.; Ji, T.; Jiang, G.; Hu, F. Simultaneous Identification and Quantification of Five Flavonoids in the Seeds of Rheum palmatum L. by Using Accelerated Solvent Extraction and HPLC–PDA–ESI/MSn. Arab. J. Chem. 2019, 12, 1345–1352. [Google Scholar] [CrossRef]
- Ruirui, L.; Wang, A.; Tian, X.; Wang, D.; Liu, J. Uniformity of Karyotypes in Rheum (Polygonaceae), a Species-Rich Genus in the Qinghai-Tibetan Plateau and Adjacent Regions. Caryologia 2010, 63, 82–90. [Google Scholar] [CrossRef]
- Mala, D.; Awasthi, S.; Sharma, N.K.; Swarnkar, M.K.; Shankar, R.; Kumar, S. Comparative Transcriptome Analysis of Rheum australe, an Endangered Medicinal Herb, Growing in Its Natural Habitat and Those Grown in Controlled Growth Chambers. Sci. Rep. 2021, 11, 3702. [Google Scholar] [CrossRef]
- Khan, M.I.; Bashir, N.; Pandith, S.A.; Patil, S.S.; Pable, A.A.; Shah, M.A.; Barvkar, V.T.; Shahzad, A. Low Temperature Stress Modulates the Biochemical, Metabolic, and Molecular Behavior of the Trans-Himalayan Medicinal Herb Rheum spiciforme Royle (Spiked Rhubarb). Ind. Crops Prod. 2023, 193, 116154. [Google Scholar] [CrossRef]
- Kahlon, P.S.; Stam, R. Polymorphisms in Plants to Restrict Losses to Pathogens: From Gene Family Expansions to Complex Network Evolution. Curr. Opin. Plant Biol. 2021, 62, 102040. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Nie, X.; Singh, M.; Coffin, R.; Duplessis, P. Sodium Sulphite Inhibition of Potato and Cherry Polyphenolics in Nucleic Acid Extraction for Virus Detection by RT-PCR. J. Virol. Methods 2002, 99, 123–131. [Google Scholar] [CrossRef]
- Kim, C.S.; Lee, C.H.; Shin, J.S.; Chung, Y.S.; Hyung, N.I. A Simple and Rapid Method for Isolation of High Quality Genomic DNA from Fruit Trees and Conifers Using PVP. Nucleic Acids Res. 1997, 25, 1085–1086. [Google Scholar] [CrossRef]
- Yerbay, M.; Khapilina, O.N.; Turzhanova, A.S.; Otradnykh, I.G.; Sedina, I.A.; Mamirova, A.; Korbozova, N.K.; Magzumova, S.; Ashimuly, K.; Kudrina, N.O.; et al. Metabolomic Adaptations and Genetic Polymorphism in Ecopopulations of Rhodiola linearifolia Boriss. Front. Plant Sci. 2025, 16, 1570411. [Google Scholar] [CrossRef] [PubMed]
- Karron, J.D. A Comparison of Levels of Genetic Polymorphism and Self-Compatibility in Geographically Restricted and Widespread Plant Congeners. Evol. Ecol. 1987, 1, 47–58. [Google Scholar] [CrossRef]
- Kent Wildlife Trust Habitat Fragmentation: Why It’s an Issue for Nature & Climate. Available online: https://www.kentwildlifetrust.org.uk/blog/habitat-fragmentation-impacts (accessed on 15 May 2025).
- Aralbayev, N.K.; Zhubanova, A.E.; Zhaparova, N.K.; Kasenova, B.T.; Aralbayeva, A.N.; Shadrina, N.V.; Lysenko, V.V. Red Book of Kazakhstan. Vol. 2: Plants; Baitulin, I.O., Sitpayeva, G.T., Eds.; 2nd, revised and expanded ed.; Art Print XXI: Astana, Kazakhstan, 2014. [Google Scholar]
- Botanical Salon “Robinzon”. Available online: http://plantarum.ru/ (accessed on 15 June 2025).
- Plants of the World Online|Kew Science. Available online: https://powo.science.kew.org/ (accessed on 15 June 2025).
- Pavlov, N.V. Flora of Kazakhstan; Academy of Sciences of the Kazakh SSR: Almaty, Kazakhstan, 1956; Volumes 1–9. [Google Scholar]
- Muchitdinov, N.M.; Muzychkina, R.A.; Korulkin, D.Y.; Gemedzhieva, N.G.; Ametov, A.A.; Kurbatova, N.V.; Abidkulova, K.T. State and prospects of studying of some Kazakhstan types of Polygonaceae Juss family. Exp. Biol. 2015, 63, 253–260. [Google Scholar]
- Sun, Y.; Wang, A.; Wan, D.; Wang, Q.; Liu, J. Rapid Radiation of Rheum (Polygonaceae) and Parallel Evolution of Morphological Traits. Mol. Phylogenetics Evol. 2012, 63, 150–158. [Google Scholar] [CrossRef]
- Zhang, H.-J.; Zhang, X.; Landis, J.B.; Sun, Y.-X.; Sun, J.; Kuang, T.-H.; Li, L.-J.; Tiamiyu, B.B.; Deng, T.; Sun, H.; et al. Phylogenomic and Comparative Analyses of Rheum (Polygonaceae, Polygonoideae). J. Syst. Evol. 2022, 60, 1229–1240. [Google Scholar] [CrossRef]
- Clark, A.G. Neutral Behavior of Shared Polymorphism. Proc. Natl. Acad. Sci. USA 1997, 94, 7730–7734. [Google Scholar] [CrossRef]
- Lozina-Lozinskaya, A.S. Systematic Review of Wild Species of the Genus rheum L. Flora Syst. High. Plants Proc. Bot. Inst. USSR Acad. Sci. 1936, 5, 67. [Google Scholar]
- Engelhardt, B.E.; Stephens, M. Analysis of Population Structure: A Unifying Framework and Novel Methods Based on Sparse Factor Analysis. PLOS Genet. 2010, 6, e1001117. [Google Scholar] [CrossRef]
- Suguiyama, V.F.; Vasconcelos, L.A.B.; Rossi, M.M.; Biondo, C.; Setta, N. de The Population Genetic Structure Approach Adds New Insights into the Evolution of Plant LTR Retrotransposon Lineages. PLoS ONE 2019, 14, e0214542. [Google Scholar] [CrossRef] [PubMed]
- Borredá, C.; Pérez-Román, E.; Ibanez, V.; Terol, J.; Talon, M. Reprogramming of Retrotransposon Activity during Speciation of the Genus Citrus. Genome Biol. Evol. 2019, 11, 3478–3495. [Google Scholar] [CrossRef]
- POWO Rheum compactum L. Available online: https://powo.science.kew.org/results?q=Rheum%20compactum (accessed on 25 April 2025).
- Grudzinskaya, L.M.; Yesimbekova, M.A.; Gemedzhieva, N.G.; Mukin, K.B. Wild Useful Plants of Kazakhstan (Catalog); Urazaliev, R.A., Ed.; Gylym: Almaty, Kazakhtan, 2008. [Google Scholar]
- Fedorov, A.A. Plant Resources of the USSR: Flowering Plants, Their Chemical Composition, and Use. Families Magnoliaceae-Limoniaceae; V.L. Komarov Botanical Institute, Academy of Sciences of the USSR; Nauka: Leningrad, Russia, 1985. [Google Scholar]
- Baitenov, M.S. Flora of Kazakhstan: The Generic Complex of Flora; Gylym: Almaty, Kazakhstan, 2001; Volume 2, ISBN 9965-07-036-9. [Google Scholar]
- Plant Identifier of Central Asia. Critical Flora Synopsis; Kovalevskaya, S.S., Ed.; Fan: Tashkent, Uzbekistan, 1971; Volume 2. [Google Scholar]
- Cherepanov, S.K. Vascular Plants of Russia and Neighbouring Countries; Mir i Semya: Saint Petersburg, Russia, 1995. [Google Scholar]
- Gemedzhieva, N.G.; Musaev, K.L.; Karzhaubekova, Z.Z.; Lesova, Z.T.; Ramazanova, M.S.; Kirienko, V.A. Distribution and Reserves of Rheum tataricum L. in the Ili River Valley. News Natl. Acad. Sci. Repub. Kazakhstan. Ser. Biol. Med. Sci. 2016, 2, 72–79. [Google Scholar]
- Seregin, A.P. Digital Herbarium of Moscow State University: Electronic Resource. Available online: https://plant.depo.msu.ru/ (accessed on 15 April 2025).
- Ranges of Medicinal and Related Plants of the USSR (Atlas), 2nd ed.; Schmidt, V.M., Ed.; Leningrad University Press: Leningrad, Russia, 1990. [Google Scholar]
- Brezhnev, D.D.; Korovina, O.N. Wild Relatives of Cultivated Plants in the Flora of the USSR; Kolos: Leningrad, Russia, 1981. [Google Scholar]
- Ivanova, K.V. Genus rheum L.-Rhubarb. In Cultivated Flora of the USSR: Leafy Vegetable Plants; Agropromizdat: Leningrad, Russia, 1988; Volume XII, pp. 30–70. [Google Scholar]
- Flora of Siberia. Salicaceae-Amaranthaceae; Krasnoborov, I.V., Malysheva, L.I., Eds.; Nauka: Novosibirsk, Russia, 1992; Volume 5. [Google Scholar]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A Modified Protocol for Rapid DNA Isolation from Plant Tissues Using Cetyltrimethylammonium Bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef]
- Azmat, M.A.; Khan, I.A.; Cheema, H.M.N.; Rajwana, I.A.; Khan, A.S.; Khan, A.A. Extraction of DNA Suitable for PCR Applications from Mature Leaves of Mangifera indica L. J. Zhejiang Univ. Sci. B 2012, 13, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Turaki, A.A.; Ahmad, B.; Magaji, U.F.; Abdulrazak, U.K.; Yusuf, B.A.; Hamza, A.B. Optimised Cetyltrimethylammonium Bromide (CTAB) DNA Extraction Method of Plant Leaf with High Polysaccharide and Polyphenolic Compounds for Downstream Reliable Molecular Analyses. AJB 2017, 16, 1354–1365. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Kalendar, R.; Flavell, A.J.; Ellis, T.H.N.; Sjakste, T.; Moisy, C.; Schulman, A.H. Analysis of Plant Diversity with Retrotransposon-Based Molecular Markers. Heredity 2011, 106, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Işık, M.İ.; Türkoğlu, A.; Yaman, M.; Haliloğlu, K.; Öztürk, H.İ.; Erkol, Ş.; Tan, M.; Bocianowski, J. Understanding Genetic Diversity and Population Structure in Forage Pea (Pisum sativum Var. Arvense L.) Using Inter-Primer Binding Site (iPBS) Retrotransposon Marker. Genet. Resour. Crop Evol. 2025. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; ISBN 978-0-19-535051-7. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Chen, S.; Dick, J.; Owen, A.B. Consistency of Markov Chain Quasi-Monte Carlo on Continuous State Spaces. Ann. Stat. 2011, 39, 673–701. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A Program for Identifying Clustering Modes and Packaging Population Structure Inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]

| Species | Na | Ne | I | He | uHe | R | NPB |
|---|---|---|---|---|---|---|---|
| Rh. tataricum (ZH) | 1.063 | 1.293 | 0.267 | 0.176 | 0.185 | 0 | 51.25 |
| Rh. tataricum (TUR) | 1.350 | 1.376 | 0.335 | 0.222 | 0.234 | 2 | 65.00 |
| Rh. turkestanicum | 1.025 | 1.245 | 0.231 | 0.150 | 0.158 | 1 | 47.50 |
| Rh. cordatum | 1.050 | 1.253 | 0.230 | 0.151 | 0.158 | 2 | 46.25 |
| Rh. compactum | 0.888 | 1.117 | 0.183 | 0.115 | 0.121 | 0 | 41.25 |
| Rh. altaicum | 1.263 | 1.298 | 0.284 | 0.183 | 0.193 | 2 | 60.00 |
| Rh. nanum | 1.075 | 1.282 | 0.250 | 0.165 | 0.174 | 2 | 50.00 |
| Average | 1.102 | 1.275 | 0.254 | 0.166 | 0.175 | 1.3 | 51.61 |
| Variation | df | SS | MS | Est. Var. | % | PhiPT | p (Rand ≥ Data) |
|---|---|---|---|---|---|---|---|
| Interspecific | 6 | 408.514 | 68.086 | 5.931 | 40% | 0.403 | 0.001 |
| Intraspecific | 69 | 553.000 | 8.778 | 8.778 | 60% | ||
| Total | 69 | 961.514 | 14.709 | 100% |
| Rh. tataricum (ZH) | Rh. tataricum (TUR) | Rh. cordatum | Rh. turkestanicum | Rh. altaicum | Rh. nanum | Rh. compactum | |
|---|---|---|---|---|---|---|---|
| - | 0.922 | 0.771 | 0.914 | 0.920 | 0.832 | 0.893 | Rh. tataricum (ZH) |
| 0.081 | - | 0.766 | 0.892 | 0.889 | 0.809 | 0.907 | Rh. tataricum (TUR) |
| 0.260 | 0.267 | - | 0.714 | 0.830 | 0.904 | 0.763 | Rh. cordatum |
| 0.090 | 0.114 | 0.337 | - | 0.896 | 0.766 | 0.911 | Rh. turkestanicum |
| 0.083 | 0.118 | 0.187 | 0.110 | - | 0.836 | 0.903 | Rh. altaicum |
| 0.184 | 0.212 | 0.101 | 0.267 | 0.179 | - | 0.816 | Rh. nanum |
| 0.113 | 0.098 | 0.271 | 0.094 | 0.102 | 0.203 | - | Rh. compactum |
| No. | Species | GPS Coordinates | Altitude, m a.s.l. | Collection Sites of Rheum L. Species Specimens |
|---|---|---|---|---|
| 1 | Rh. tataricum L. (ZH) | N 43°25′32″ E 70°39′38″ | 441 | Kazakhstan, Zhambyl Region, Talas District—northern shore of Lake Akkol, 8 km west of v. Akkol, within shrub–wormwood–grassland vegetation. |
| 2 | Rh. tataricum L. (TUR) | N 44°36′48″ E 68°44′04″ | 240 | Kazakhstan, Turkestan Region, Sozak District—54 km north of v. Sozak, within saxaul (Haloxylon spp.) vegetation. |
| 3 | Rh. turkestanicum Janisch. | N 42°33′50″ E 67°56′42″ | 191 | Kazakhstan, Turkestan Region, Otyrar District—19 km southwest of v. Koksarai, on sandy terrain. |
| 4 | Rh. cordatum Losinsk. | N 43°21′58″ E 75°02′15″ | 833 | Kazakhstan, Zhambyl Region, Kordai District—6 km south of v. Kenen, on mountainous slopes. |
| 5 | Rh. compactum L. (putative synonym Rh. altaicum Losinsk.) | N 49°14′08″ E 89°11′23″ | 1186 | Kazakhstan, Abay Region, Ulan District—southeastern part of the Kalbinsky Ridge, Kaindy Massif. |
| 6 | Rh. altaicum Losinsk. (putative synonym Rh. compactum L.) | N 49°29′59″ E 83°02′22″ | 830 | Kazakhstan, Abay Region, Ulan District—Kalbinsky Ridge, Tainty Tract, southeast of v. Asybulak. |
| 7 | Rh. nanum Lingelsh. | N 47°34′40″ E 83°41′00″ | 495 | Kazakhstan, Abay Region, Kurchum District—southwestern slope of the Kurchum Ridge, near v. Kalguty. |
| No. | ID | Sequence | Tm (°C) | LC (%) | CG (%) |
|---|---|---|---|---|---|
| 1 | 2217 | ACTTGGATGTCGATACCA | 50.0 | 88 | 44.4 |
| 2 | 2219 | GAACTTATGCCGATACCA | 55.0 | 88 | 44.4 |
| 3 | 2220 | ACCTGGCTCATGATGCCA | 55.0 | 80 | 55.6 |
| 4 | 2221 | ACCTAGCTCACGATGCCA | 58.0 | 88 | 55.6 |
| 5 | 2222 | ACTTGGATGCCGATACCA | 53.0 | 86 | 50.0 |
| 6 | 2224 | ATCCTGGCAATGGAACCA | 53.0 | 83 | 50.0 |
| 7 | 2226 | CGGTGACCTTTGATACCA | 53.0 | 83 | 50.0 |
| 8 | 2228 | CATTGGCTCTTGATACCA | 55.0 | 86 | 44.4 |
| 9 | 2230 | TCTAGGCGTCTGATACCA | 54.0 | 91 | 50.0 |
| 10 | 2231 | ACTTGGATGCTGATACCA | 50.0 | 83 | 44.4 |
| 11 | 2240 | AACCTGGCTCAGATGCCA | 58.9 | 88 | 55.6 |
| 12 | 2246 | ACTAGGCTCTGTATACCA | 50.0 | 88 | 44.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khapilina, O.N.; Turzhanova, A.S.; Gemejieva, N.G.; Sumbembayev, A.A.; Arysbayeva, R.B.; Magzumova, S.; Kudrina, N.O.; Kulmanov, T.E.; Mamirova, A.; Terletskaya, N.V. Exploring Genetic Diversity and Inter-/Intraspecific Polymorphism in Rheum sp. (Polygonaceae) Using the iPBS Retrotransposon Marker System. Int. J. Mol. Sci. 2025, 26, 8943. https://doi.org/10.3390/ijms26188943
Khapilina ON, Turzhanova AS, Gemejieva NG, Sumbembayev AA, Arysbayeva RB, Magzumova S, Kudrina NO, Kulmanov TE, Mamirova A, Terletskaya NV. Exploring Genetic Diversity and Inter-/Intraspecific Polymorphism in Rheum sp. (Polygonaceae) Using the iPBS Retrotransposon Marker System. International Journal of Molecular Sciences. 2025; 26(18):8943. https://doi.org/10.3390/ijms26188943
Chicago/Turabian StyleKhapilina, Oxana N., Ainur S. Turzhanova, Nadezhda G. Gemejieva, Aidar A. Sumbembayev, Raya B. Arysbayeva, Saule Magzumova, Nataliya O. Kudrina, Timur E. Kulmanov, Aigerim Mamirova, and Nina V. Terletskaya. 2025. "Exploring Genetic Diversity and Inter-/Intraspecific Polymorphism in Rheum sp. (Polygonaceae) Using the iPBS Retrotransposon Marker System" International Journal of Molecular Sciences 26, no. 18: 8943. https://doi.org/10.3390/ijms26188943
APA StyleKhapilina, O. N., Turzhanova, A. S., Gemejieva, N. G., Sumbembayev, A. A., Arysbayeva, R. B., Magzumova, S., Kudrina, N. O., Kulmanov, T. E., Mamirova, A., & Terletskaya, N. V. (2025). Exploring Genetic Diversity and Inter-/Intraspecific Polymorphism in Rheum sp. (Polygonaceae) Using the iPBS Retrotransposon Marker System. International Journal of Molecular Sciences, 26(18), 8943. https://doi.org/10.3390/ijms26188943







