Genotype-Phenotype Correlation of Seven Known and Novel β-Globin Gene Variants
Abstract
1. Introduction
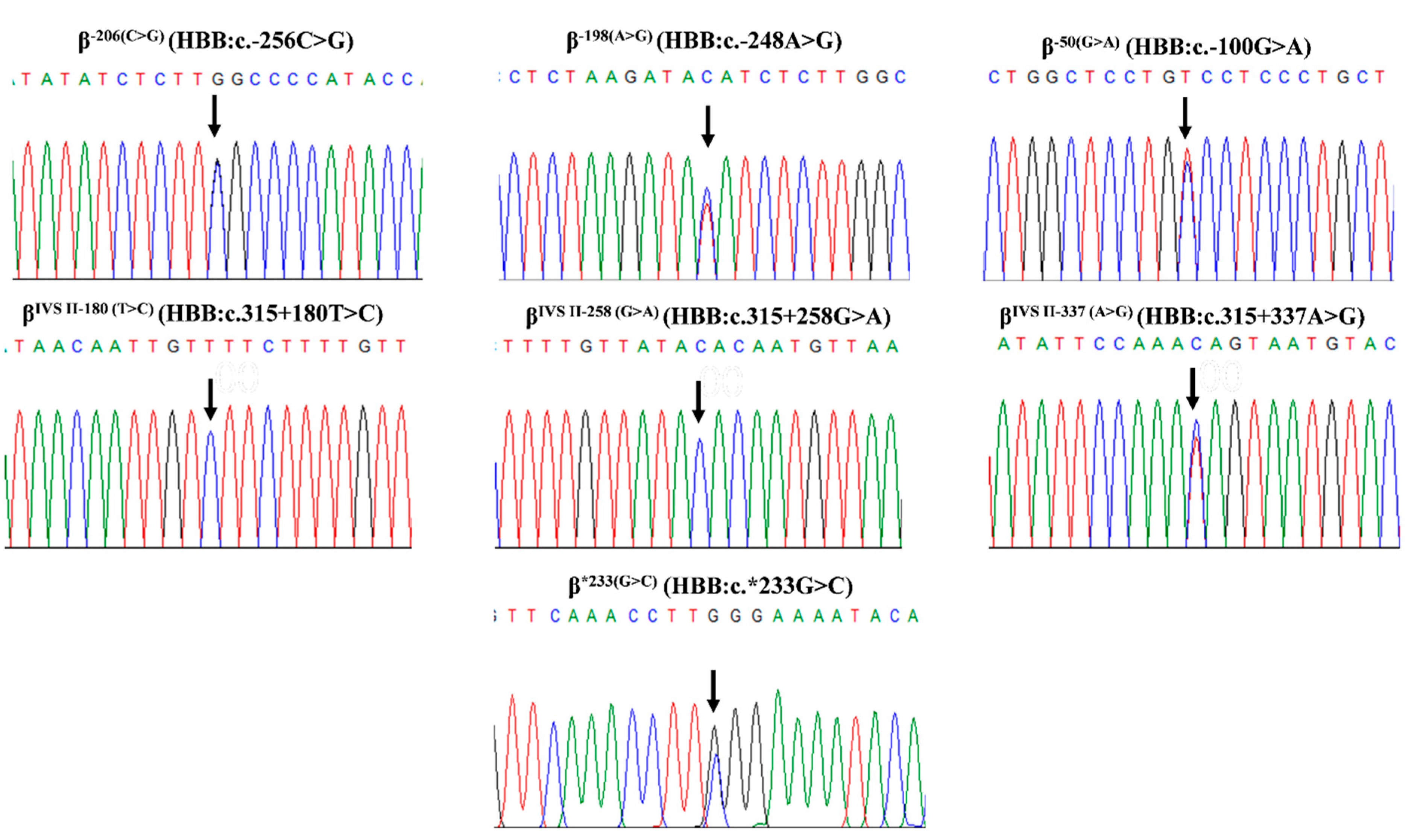
2. Results
3. Discussion
4. Materials and Methods
4.1. Subjects and Hematological Analyses
4.2. Molecular Analysis
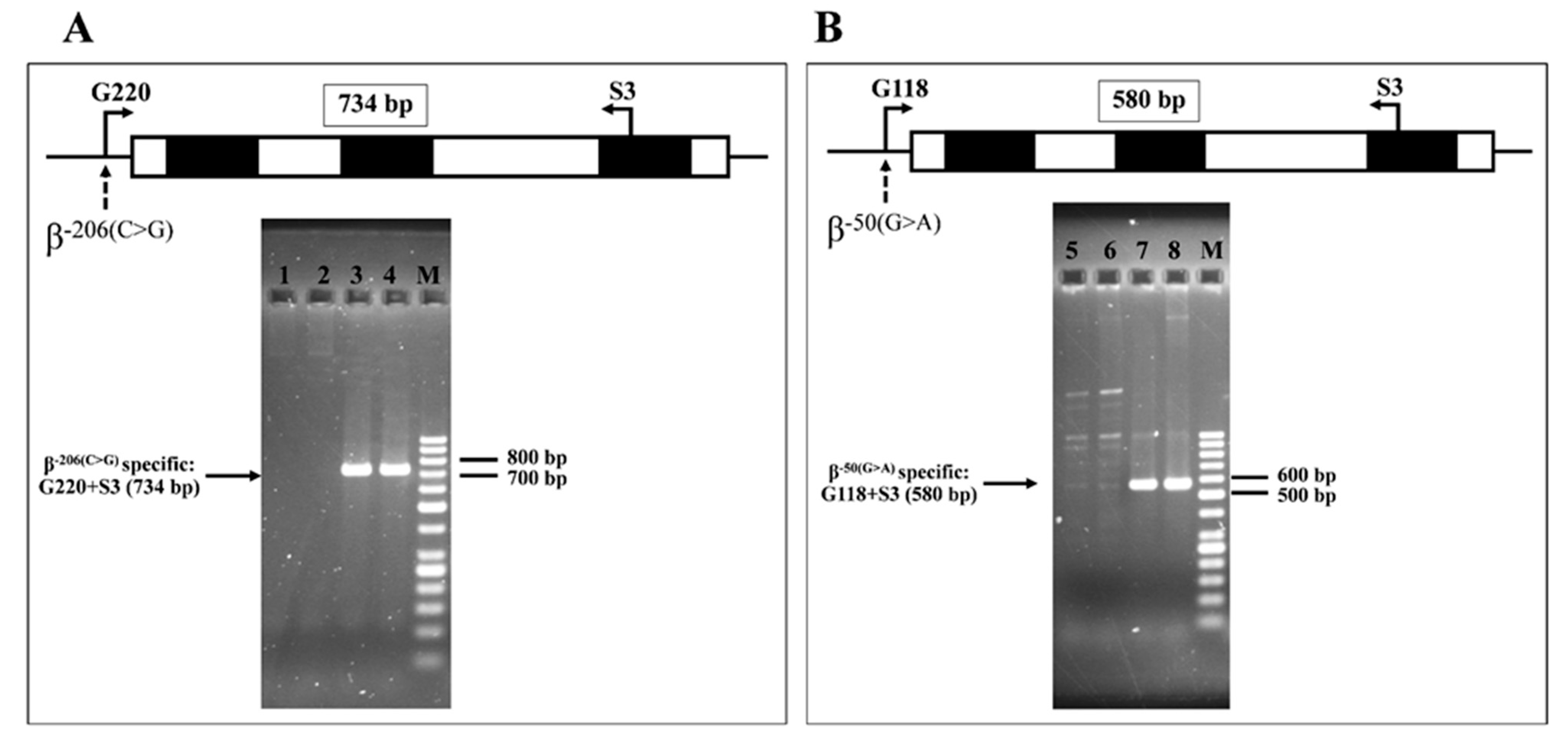
4.3. Molecular Confirmation of the β−206(C>G)) and β−50(G>A)
4.4. Transcription Factor Binding Site Prediction for β−206(C>G), β−198(A>G), and β−50(G>A)
4.5. Splice Site Prediction of βIVSII−180(T>C), βIVSII−258(G>A), and βIVSII−337(A>G)
4.6. In Silico Functional Study Prediction for the Seven β-Globin Variants
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taher, A.T.; Weatherall, D.J.; Cappellini, M.D. Thalassaemia. Lancet 2018, 391, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Kountouris, P.; Lederer, C.W.; Fanis, P.; Feleki, X.; Old, J.; Kleanthous, M. IthaGenes: An interactive database for haemoglobin variations and epidemiology. PLoS ONE 2014, 9, e103020. [Google Scholar] [CrossRef] [PubMed]
- Yamsri, S.; Sanchaisuriya, K.; Fucharoen, G.; Sae-Ung, N.; Ratanasiri, T.; Fucharoen, S. Prevention of severe thalassemia in northeast Thailand: 16 years of experience at a single university center. Prenat. Diagn. 2010, 30, 540–546. [Google Scholar] [CrossRef]
- Kountouris, P.; Stephanou, C.; Lederer, C.W.; Traeger-Synodinos, J.; Bento, C.; Harteveld, C.L.; Fylaktou, E.; Koopmann, T.T.; Halim-Fikri, H.; Michailidou, K.; et al. Adapting the ACMG/AMP variant classification framework: A perspective from the ClinGen Hemoglobinopathy Variant Curation Expert Panel. Hum. Mutat. 2022, 43, 1089–1096. [Google Scholar] [CrossRef]
- Tamana, S.; Xenophontos, M.; Minaidou, A.; Stephanou, C.; Harteveld, C.L.; Bento, C.; Traeger-Synodinos, J.; Fylaktou, I.; Yasin, N.M.; Abdul Hamid, F.S.; et al. Evaluation of in silico predictors on short nucleotide variants in HBA1, HBA2, and HBB associated with haemoglobinopathies. eLife 2022, 11, e79713. [Google Scholar] [CrossRef]
- Bayramov, B.; Aliyeva, G.; Asadov, C.; Mammadova, T.; Karimova, N.; Eynullazadeh, K.; Gafarova, S.; Akbarov, S.; Farhadova, S.; Safarzadeh, Z.; et al. A novel frameshift mutation at codon 2 (-T) (HBB: C.9delT) and first report of three new β-globin mutations from Azerbaijan. Hemoglobin 2019, 43, 280–282. [Google Scholar] [CrossRef]
- Luo, S.; Chen, X.; Zeng, D.; Tang, N.; Yuan, D.; Zhong, Q.; Mao, A.; Xu, R.; Yan, T. The value of single-molecule real-time technology in the diagnosis of rare thalassemia variants and analysis of phenotype-genotype correlation. J. Hum. Genet. 2022, 67, 183–195. [Google Scholar] [CrossRef]
- Li, D.Z.; Liao, C.; Xie, X.M.; Zhou, J.Y. A novel mutation of −50 (G-->A) in the direct repeat element of the beta-globin gene identified in a patient with severe beta-thalassemia. Ann. Hematol. 2009, 88, 1149–1150. [Google Scholar] [CrossRef]
- Sirdah, M.M.; Sievertsen, J.; Al-Yazji, M.S.; Tarazi, I.S.; Al-Haddad, R.M.; Horstmann, R.D.; Timmann, C. The spectrum of β-thalassemia mutations in Gaza Strip, Palestine. Blood Cells Mol. Dis. 2013, 50, 247–251. [Google Scholar] [CrossRef]
- Smith, D.L.; Mitui, M.; Park, J.Y.; Luu, H.S.; Timmons, C.F. Characterization of the HBB: C.*233G > C variant: No evidence of a β-thalassemic phenotype. Hemoglobin 2016, 40, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Mahabhol, C.; Yothindamrongkul, L.; Nuntanajaroenkul, N.; Nawasod, P.; Tepakhan, W.; Karnpean, R.; Sowithayasakul, P.; Makruasi, N.; Trongwongsa, T.; Jomoui, W. The validation of whole β-globin gene sequencing for detecting β-thalassemia mutations found in Thailand using Next-Generation Sequencing (NGS). Ann. Hematol. 2025, 104, 2653–2661. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Chen, Q.; Zhang, Z.; Peng, S.; Liu, J.; Pang, J.; Jia, Z.; Xi, H.; Li, J.; Chen, L.; et al. Identification of rare thalassemia variants using third-generation sequencing. Front. Genet. 2023, 13, 1076035. [Google Scholar] [CrossRef]
- Srivorakun, H.; Thawinan, W.; Fucharoen, G.; Sanchaisuriya, K.; Fucharoen, S. Thalassemia and erythroid transcription factor KLF1 mutations associated with borderline hemoglobin A2 in the Thai population. Arch. Med. Sci. 2020, 18, 112–120. [Google Scholar] [CrossRef]
- Singha, K.; Teawtrakul, N.; Fucharoen, G.; Fucharoen, S. Molecular and haematological characterisation of haemolytic anaemia associated with biallelic KLF1 mutations: A case series. J. Clin. Pathol. 2024, 77, 783–789. [Google Scholar] [CrossRef]
- Songdej, D.; Teawtrakul, N.; Laoaroon, N.; Komvilaisak, P.; Sripornsawan, P.; Surapolchai, P.; Hantaweepant, C.; Tantiworawit, A.; Hantrakool, S.; Lauhasurayotin, S.; et al. Impact of HbE mutation on the clinical severity of HbH disease: A multicentre study from Thailand. Br. J. Haematol. 2025, 206, 703–712. [Google Scholar] [CrossRef]
- Jomoui, W.; Tepakhan, W.; Satthakarn, S.; Panyasai, S. Molecular spectrum of Hb H disease and characterization of rare deletional α-thalassemia found in Thailand. Scand. J. Clin. Lab. Investig. 2020, 80, 528–535. [Google Scholar] [CrossRef]
- Zhao, Y.; Jiang, F.; Li, D.Z. Hematological characteristics of β-globin gene mutation −50 (G>A) (HBB: C.-100G>A) carriers in mainland China. Hemoglobin 2020, 44, 240–243. [Google Scholar] [CrossRef]
- Long, J.; Liu, E. Identification of the β thalassemia allele β−50 and analysis of the hematology data of carriers in a southern Chinese population. Ann. Hum. Genet. 2022, 86, 63–70. [Google Scholar] [CrossRef]
- Mohd Yasin, N.; Abdul Hamid, F.S.; Hassan, S.; Mat Yusoff, Y.; Mohd Sahid, E.N.; Esa, E. An insight of −50 (G>A) mutation in the direct repeat element of the β-globin gene: From Malaysian perspective. Malays. J. Pathol. 2022, 44, 301–302. [Google Scholar] [PubMed]
- Charoenwijitkul, T.; Singha, K.; Fucharoen, G.; Sanchaisuriya, K.; Thepphitak, P.; Wintachai, P.; Karnpean, R.; Fucharoen, S. Molecular characteristics of α+-thalassemia (3.7 kb deletion) in Southeast Asia: Molecular subtypes; haplotypic heterogeneity; multiple founder effects and laboratory diagnostics. Clin. Biochem. 2019, 71, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Chaibunruang, A.; Prommetta, S.; Yamsri, S.; Fucharoen, G.; Sae-Ung, N.; Sanchaisuriya, K.; Fucharoen, S. Molecular and hematological studies in a large cohort of α(0)-thalassemia in northeast Thailand: Data from a single referral center. Blood Cells Mol. Dis. 2013, 51, 89–93. [Google Scholar] [CrossRef]
- Tsunoda, T.; Takagi, T. Estimating transcription factor bindability on DNA. Bioinformatics 1999, 15, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome. Biol. 2006, 7 (Suppl. 1), S10. [Google Scholar] [CrossRef]
- de Sainte Agathe, J.M.; Filser, M.; Isidor, B.; Besnard, T.; Gueguen, P.; Perrin, A.; Van Goethem, C.; Verebi, C.; Masingue, M.; Rendu, J.; et al. SpliceAI-visual: A free online tool to improve SpliceAI splicing variant interpretation. Hum. Genom. 2023, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Byrne, A.B.; Feng, B.J.; Pagel, K.A.; Mooney, S.D.; Karchin, R.; O’Donnell-Luria, A.; Harrison, S.M.; Tavtigian, S.V.; Greenblatt, M.S.; et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am. J. Hum. Genet. 2022, 109, 2163–2177. [Google Scholar] [CrossRef]
- Bergquist, T.; Stenton, S.L.; Nadeau, E.A.W.; Byrne, A.B.; Greenblatt, M.S.; Harrison, S.M.; Tavtigian, S.V.; O’Donnell-Luria, A.; Biesecker, L.G.; Radivojac, P.; et al. Calibration of additional computational tools expands ClinGen recommendation options for variant classification with PP3/BP4 criteria. Genet. Med. 2025, 27, 101402. [Google Scholar] [CrossRef]
| No | Sex a | Hb Type | Hb A2 (%) | Hb E + A2 (%) | Hb F (%) | RBC (1012/L) | Hb (g/dL) | Hct (%) | MCV (fL) | MCH (pg) | MCHC (g/dL) | RDW (%) | β-Globin Genotype | α-Globin Genotype | KLF1 Mutation Identified b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | A2A | 4.4 | - | 0.6 | 6.0 | 16.2 | 46.2 | 77.4 | 27.1 | 35.1 | 14.8 | β−206(C>G)/β−31(A>G) | αCSα/αα | - |
| 2 | M | A2A | 4.5 | - | 0 | 6.1 | 13.3 | 39.1 | 64.5 | 21.9 | 34.0 | 18.3 | β−206(C>G)/β−31(A>G) | αα/αα | - |
| 3 | F | A2A | 5.1 | - | 0.7 | 5.3 | 10.9 | 34.7 | 65.2 | 20.4 | 31.3 | 18.6 | β−206(C>G)/β−50(G>A) | αα/αα | - |
| 4 | F | A2A | 2.9 | - | 0 | 4.7 | 12.9 | 38.7 | 83.0 | 27.5 | 33.2 | 12.1 | β−198(A>G)/βA | −α3.7/αα | - |
| 5 | M | A2A | 2.9 | - | 0 | 4.9 | 14.2 | 42.1 | 86.8 | 29.2 | 33.6 | 13.2 | β−198(A>G)/βA | αα/αα | - |
| 6 | M | A2A | 2.8 | - | 0 | 4.8 | 15.1 | 43.8 | 90.8 | 31.4 | 34.6 | 12.4 | β−198(A>G)/βA | αα/αα | - |
| 7 | M | A2A | 3.6 | - | 0 | Na | Na | Na | 78.4 | 25.5 | 32.5 | Na | β−198(A>G)/βA | −α3.7/αα | A298P |
| 8 | M | A2A | 3.6 | - | 2.2 | Na | Na | Na | 86.6 | 28.8 | 33.3 | Na | β−198(A>G)/βA | αα/αα | G176AfsX179 |
| 9 | F | A2A | 3.9 | - | 2.4 | 5.8 | 13.6 | 42.2 | 72.8 | 23.4 | 32.2 | Na | β−198(A>G)/βA | −α3.7/αα | H299D |
| 10 | F | A2(F)A | 2.0 | - | 8.6 | 4.7 | 11.2 | 35.5 | 75.1 | 23.7 | 31.6 | 17.3 | β−50(G>A)/βA | −α3.7/αα | - |
| 11 | M | A2A | 3.6 | - | 0.3 | Na | Na | Na | Na | Na | Na | Na | β−50(G>A)/βA | Na | - |
| 12 | F | A2A | 3.6 | - | 1.5 | Na | Na | Na | 70.3 | 22.7 | 32.3 | Na | β−50(G>A)/βA | αα/αα | - |
| 13 | F | A2A | 3.7 | - | 0.6 | Na | Na | Na | 80.7 | 26.9 | 33.3 | Na | β−50(G>A)/βA | −α3.7/αα | - |
| 14 | M | A2A | 3.8 | - | 0.3 | 5.2 | 14.0 | 41.0 | 79.0 | 27.0 | 34.2 | 14.4 | β−50(G>A)/βA | αα/αα | - |
| 15 | F | A2A | 3.9 | - | 1.1 | 4.8 | 12.1 | 36.0 | 75.0 | 25.2 | 33.6 | Na | β−50(G>A)/βA | −α3.7/αα | G176AfsX179 |
| 16 | F | A2A | 3.9 | - | 5.2 | 5.4 | 12.5 | 40.0 | 73.7 | 23.1 | 31.3 | 15.1 | β−50(G>A)/βA | −α3.7/αα | R328H |
| 17 | F | A2A | 4.1 | - | 0 | Na | Na | Na | 74.0 | 23.0 | 31.1 | Na | β−50(G>A)/βA | Na | - |
| 18 | M | A2A | 4.1 | - | 2.3 | 5.5 | 14.3 | 42.6 | 77.5 | 26.0 | 33.6 | Na | β−50(G>A)/βA | Na | - |
| 19 | F | A2A | 5.0 | - | 1.2 | 5.5 | 12.0 | 37.0 | 66.2 | 21.7 | 32.8 | 15.4 | β−50(G>A)/βMalay | αα/αα | - |
| 20 | F | A2A | 5.5 | - | 0 | 5.2 | 11.2 | 34.0 | 65.1 | 21.6 | 33.2 | 14.1 | β−50(G>A)/β41/42(-TTCT) | αα/αα | - |
| 21 | F | A2A | 6.1 | - | 1.7 | 4.4 | 8.3 | 26.9 | 61.0 | 18.9 | 31.0 | 17.6 | β−50(G>A)/βIVSI−1(G>T) | αα/αα | - |
| 22 | M | A2A | 6.2 | - | 0.6 | 5.9 | 12.9 | 40.0 | 67.4 | 20.6 | 32.4 | Na | β−50(G>A)/βIVSI−1(G>T) | −α3.7/αα | - |
| 23 | M | CSEABart’s | 2.3 | 16.3 | 1.7 | 5.1 | 8.0 | 29.2 | 57.7 | 15.8 | 27.4 | 23.5 | β−50(G>A)/βE | --SEA/αCSα | - |
| 24 | M | E(F)A | - | 27.7 | 6.8 | 6.2 | 14.2 | 44.7 | 72.0 | 22.7 | 31.5 | 18.2 | β−50(G>A)/βE | −α4.2/αα | - |
| 25 | F | EA | - | 28.2 | 3.3 | 5.0 | 13.3 | 40.0 | 79.5 | 26.5 | 33.3 | 15.0 | β−50(G>A)/βE | αα/αα | - |
| 26 | F | E(F)A | - | 29.7 | 6.2 | Na | Na | Na | Na | Na | Na | Na | β−50(G>A)/βE | αα/αα | - |
| 27 | M | EA | 3.4 | 31.5 | 1.1 | Na | Na | Na | 75.0 | Na | Na | Na | β−50(G>A)/βE | Na | - |
| 28 | F | A2A | 3.6 | - | 0 | Na | Na | Na | 74.9 | 24.5 | 32.7 | Na | βIVSII−180(T>C)/βA | αα/αα | A298P |
| 29 | F | A2A | 3.8 | - | 0 | Na | Na | Na | Na | Na | Na | Na | βIVSII−258(G>A)/βA | αα/αα | A298P |
| 30 | F | A2A | 3.6 | - | 5.0 | Na | Na | Na | 78.5 | Na | Na | Na | βIVSII−337(A>G)/βA | −α3.7/αα | R328H |
| 31 | F | A2A | 2.8 | - | 0 | 4.0 | 12.3 | 38.1 | 94.3 | 30.6 | 32.4 | 13.7 | β*233(G>C)/βA | −α3.7/αα | - |
| 32 | M | A2A | 2.6 | - | 0 | 5.4 | 15.6 | 45.0 | 83.3 | 28.9 | 34.7 | 12.8 | β*233(G>C)/βA | αα/αα | - |
| 33 | F | A2A | 2.7 | - | 0 | 4.2 | 12.4 | 36.3 | 86.3 | 29.4 | 34.1 | 13.3 | β*233(G>C)/βA | αα/αα | - |
| β-Globin Genotype | n | Hb A2 (%) | Hb E + A2 (%) | Hb F (%) | RBC (1012/L) | Hb (g/dL) | MCV (fL) | MCH (pg) | RDW (%) |
|---|---|---|---|---|---|---|---|---|---|
| β−206(C>G)/β−31(A>G) | 2 | 4.4, 4.5 | - | 0.6, 0 | 6.0, 6.1 a | 16.2, 13.3 | 77.4, 64.5 | 27.1, 21.9 | 14.8, 18.3 |
| βA/β−31(A>G) | 41 | 4.7 ± 0.4 | - | 1.2 ± 0.1 | 5.0 ± 0.7 a | 12.9 ± 1.5 | 77.4 ± 4.5 | 25.4 ± 1.3 | 14.7 ± 1.2 |
| β−50(G>A)/βA | 9 | 3.6 ± 0.3 b | - | 2.2 ± 2.9 b | 5.1 ± 0.4 | 12.8 ± 1.3 b | 75.7 ± 3.3 b | 24.7 ± 1.8 b | 15.6 ± 1.5 b |
| βA/βA | 89 | 2.8 ± 0.2 b | - | 0.1 ± 0.2 b | 5.0 ± 0.5 | 14.8 ± 1.6 b | 88.8 ± 3.9 b | 29.8 ± 1.4 b | 12.9 ± 0.9 b |
| β−50(G>A)/βMalay | 1 | 5.0 | - | 1.2 | 5.53 | 12.0 | 66.2 | 21.7 | 15.4 |
| βA/βMalay | 29 | 4.4 ± 0.4 | - | 1.3 ± 1.4 | 5.2 ± 0.8 | 12.0 ± 1.6 | 70.2 ± 5.1 | 23.4 ± 2.5 | 15.6 ± 1.8 |
| β−50(G>A)/β0 | 3 | 5.9 ± 0.4 | - | 0.8 ± 0.9 | 5.2 ± 0.8 | 10.8 ± 2.3 | 64.5 ± 3.2 | 20.4 ± 1.4 | 15.9 ± 2.5 |
| βA/β0 | 309 | 5.7 ± 0.7 | - | 1.4 ± 1.1 | 5.5 ± 0.9 | 11.4 ± 1.9 | 63.6 ± 4.0 | 20.6 ± 1.8 | 17.2 ± 2.0 |
| β−50(G>A)/βE | 4 | - | 29.3 ± 1.7 d | 4.4 ± 2.7 c,d | 5.6 ± 0.8 | 13.8 ± 0.6 d | 75.5 ± 3.8 d | 24.6 ± 2.7 d | 16.6 ± 2.3 d |
| βA/βE | 112 | - | 29.1 ± 3.0 | 1.4 ± 1.3 c | 5.1 ± 0.9 | 13.0 ± 1.7 | 77.3 ± 5.2 | 25.8 ± 2.2 | 14.7 ± 1.2 |
| β−28(G>A)/βE | 143 | - | 56.2 ± 6.7 d | 15.0 ± 8.2 d | 5.1 ± 0.9 | 9.7 ± 1.4 d | 60.8 ± 5.8 d | 19.2 ± 1.7 d | 21.9 ± 3.3 d |
| Variants | HGVS Name (HBB) | RS Number | Normal Thai Subjects | Global (gnomAD v4-Genomes) | ||||
|---|---|---|---|---|---|---|---|---|
| N | Reference Allele | Alternative Allele | N | Reference Allele | Alternative Allele | |||
| −206(C>G) | c.−256C>G | rs376005360 | 178 | 1.00 (n = 178) | 0.00 (n = 0) | 149,210 | 0.999906 | 0.000094 |
| −198(A>G) | c.−248A>G | rs76306358 | 178 | 0.9831 (n = 175) | 0.0169 (n = 3) | 149,276 | 0.999571 | 0.000429 |
| −50(G>A) | c.−100G>A | rs281864524 | 178 | 1.00 (n = 178) | 0.00 (n = 0) | 149,276 | 0.999967 | 0.000033 |
| CD 2 (CAT>CAC) | c.9T>C | rs713040 | 178 | 0.4213 (n = 75) | 0.5787 (n = 103) | 149,134 | 0.184794 | 0.815206 |
| IVS II-16 (G>C) | c.315 + 16G>C | rs10768683 | 178 | 0.4213 (n = 75) | 0.5787 (n = 103) | 149,086 | 0.182519 | 0.817481 |
| IVS II-74 (T>G) | c.315 + 74T>G | rs7480526 | 178 | 0.7921 (n = 141) | 0.2079 (n = 37) | 149,020 | 0.560589 | 0.439411 |
| IVS II-81 (C>T) | c.315 + 81C>T | rs7946748 | 178 | 0.9831 (n = 175) | 0.0169 (n = 3) | 149,168 | 0.893891 | 0.106109 |
| IVS II-180 (T>C) | c.315 + 180T>C | rs529931134 | 178 | 1.00 (n = 178) | 0.00 (n = 0) | 149,304 | 0.999906 | 0.000094 |
| IVS II-258 (G>A) | c.315 + 258G>A | rs1029410290 | 178 | 1.00 (n = 178) | 0.00 (n = 0) | 145,536 | 0.999993 | 0.000007 |
| IVS II-337 (A>G) | c.315 + 337A>G | rs561258571 | 178 | 1.00 (n = 178) | 0.00 (n = 0) | 149,146 | 0.999980 | 0.000020 |
| IVS II-666 (C>T) | c.316 − 185C>T | rs1609812 | 178 | 0.4157 (n = 74) | 0.5843 (n = 104) | 149,128 | 0.185056 | 0.814944 |
| *233(G>C) | c.*233G>C | rs12788013 | 178 | 0.9831 (n = 175) | 0.0169 (n = 3) | 149,150 | 0.893979 | 0.106021 |
| Variants | SpliceAI a | CADD b | PhyloP b | PromoterAI c |
|---|---|---|---|---|
| −206(C>G) | 0.02 | 2.08 (Moderate Benign) | −0.029 (Moderate Benign) | −0.03 |
| −198(A>G) | 0.01 | 2.75 (Moderate Benign) | 0.427 (Supporting Benign) | −0.03 |
| −50(G>A) | 0.01 | 11.9 (Moderate Benign) | 1.74 (Supporting Benign) | −0.13 |
| IVS II-180 (T>C) | 0.01 | 5.22 (Moderate Benign) | 0.7 (Supporting Benign) | - |
| IVS II-258 (G>A) | 0 | 0.234 (Moderate Benign) | −0.341 (Moderate Benign) | - |
| IVS II-337 (A>G) | 0 | 0.607 (Moderate Benign) | 0.002 (Moderate Benign) | - |
| TTS +99 (G>C) | 0.11 | 7.73 (Moderate Benign) | −0.836 (Moderate Benign) | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singha, K.; Pansuwan, A.; Fucharoen, G.; Fucharoen, S. Genotype-Phenotype Correlation of Seven Known and Novel β-Globin Gene Variants. Int. J. Mol. Sci. 2025, 26, 8872. https://doi.org/10.3390/ijms26188872
Singha K, Pansuwan A, Fucharoen G, Fucharoen S. Genotype-Phenotype Correlation of Seven Known and Novel β-Globin Gene Variants. International Journal of Molecular Sciences. 2025; 26(18):8872. https://doi.org/10.3390/ijms26188872
Chicago/Turabian StyleSingha, Kritsada, Anupong Pansuwan, Goonnapa Fucharoen, and Supan Fucharoen. 2025. "Genotype-Phenotype Correlation of Seven Known and Novel β-Globin Gene Variants" International Journal of Molecular Sciences 26, no. 18: 8872. https://doi.org/10.3390/ijms26188872
APA StyleSingha, K., Pansuwan, A., Fucharoen, G., & Fucharoen, S. (2025). Genotype-Phenotype Correlation of Seven Known and Novel β-Globin Gene Variants. International Journal of Molecular Sciences, 26(18), 8872. https://doi.org/10.3390/ijms26188872






