Rules of Engagement for Components of Membrane Protein Biogenesis at the Human Endoplasmic Reticulum
Abstract
1. Introduction

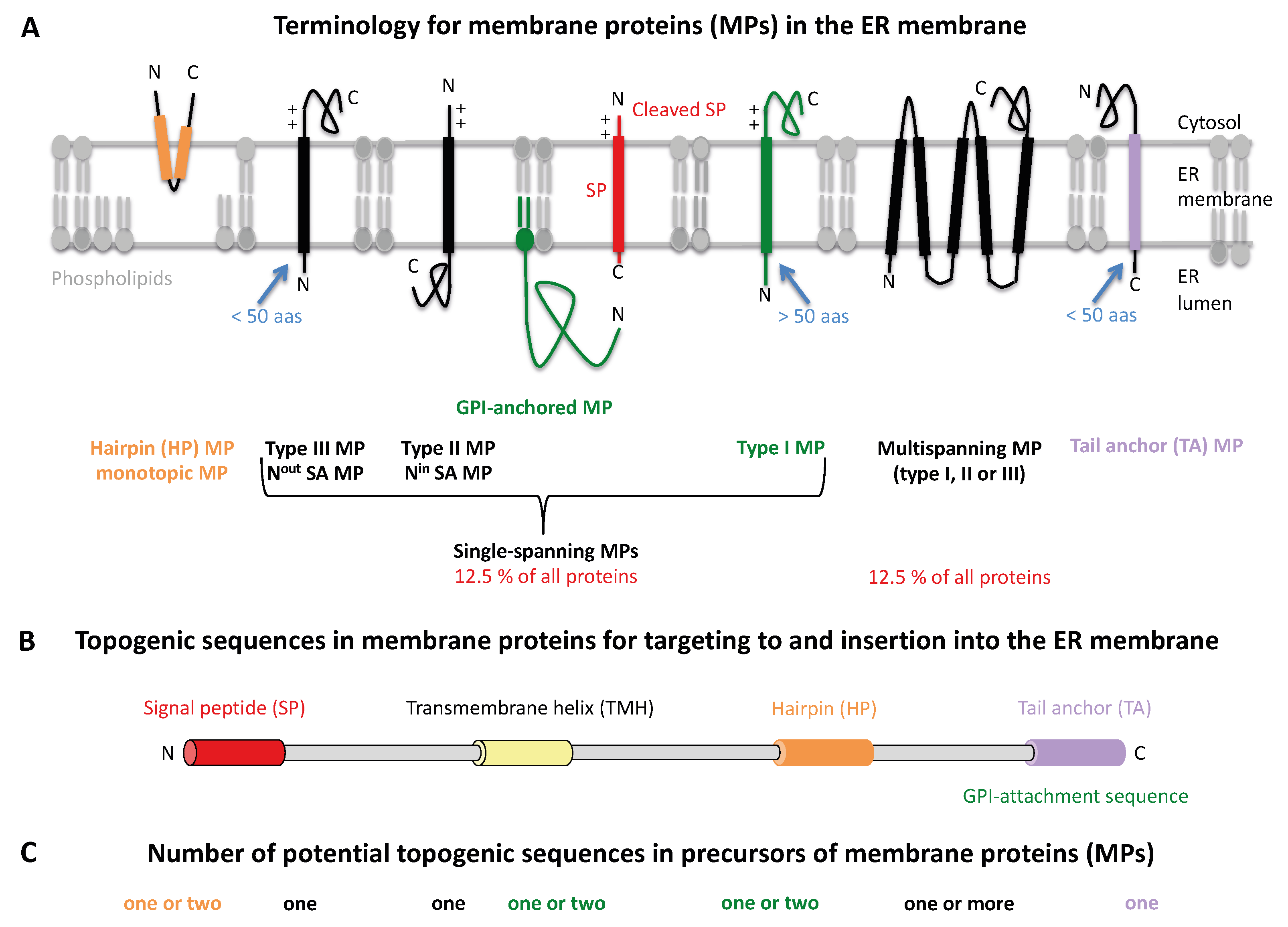
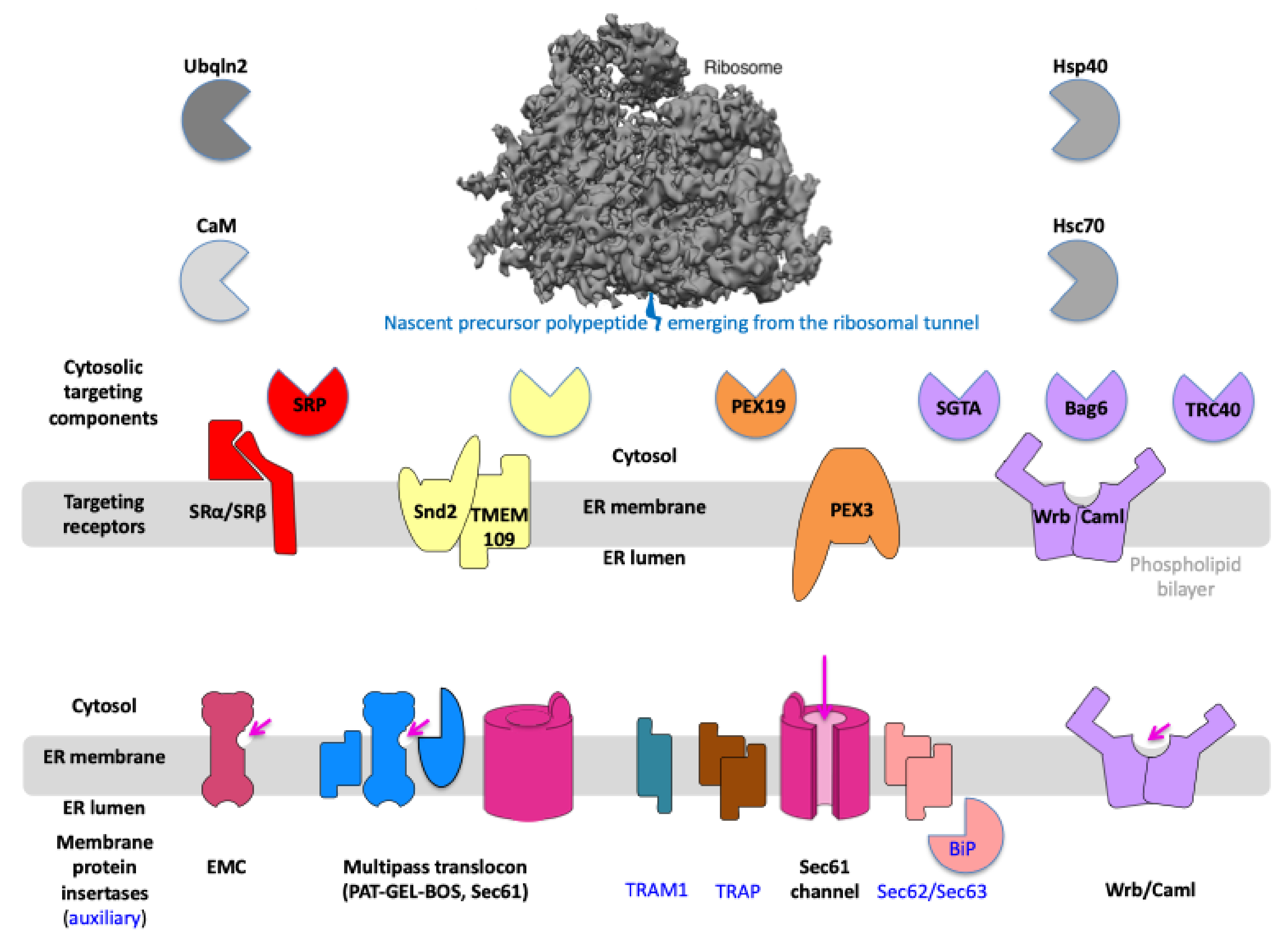

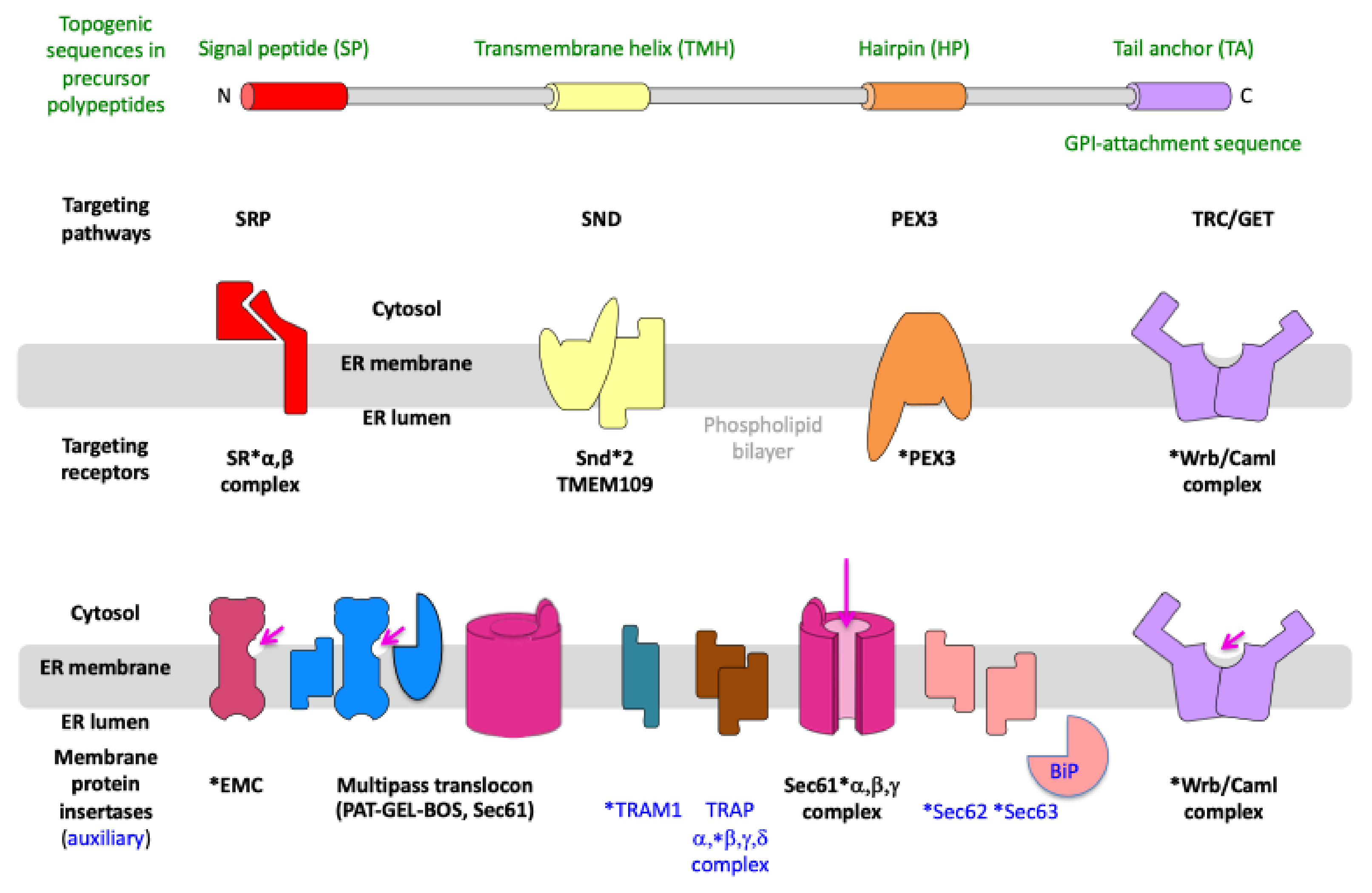
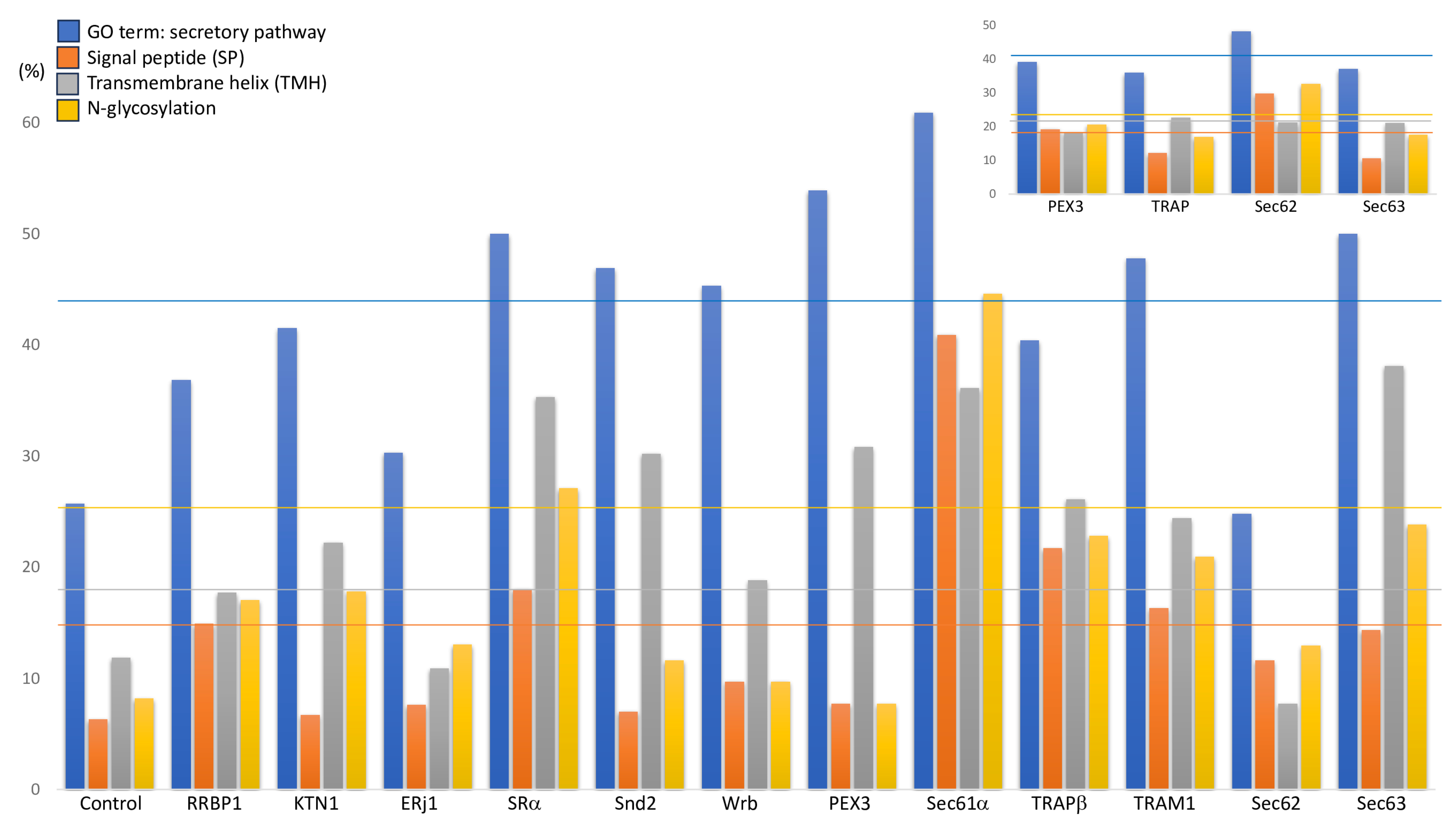
2. Results
2.1. Identification and Classification of Putative Clients
2.2. Weaknesses of the Experimental Approach
2.3. Clients in Precursor Polypeptide Targeting to the Human Endoplasmic Reticulum
2.3.1. The SRP/SR Targeting Pathway
2.3.2. The SRP-Independent Targeting Pathways
2.4. Clients in Insertion of Precursor Polypeptides into the Membrane of the Human Endoplasmic Reticulum
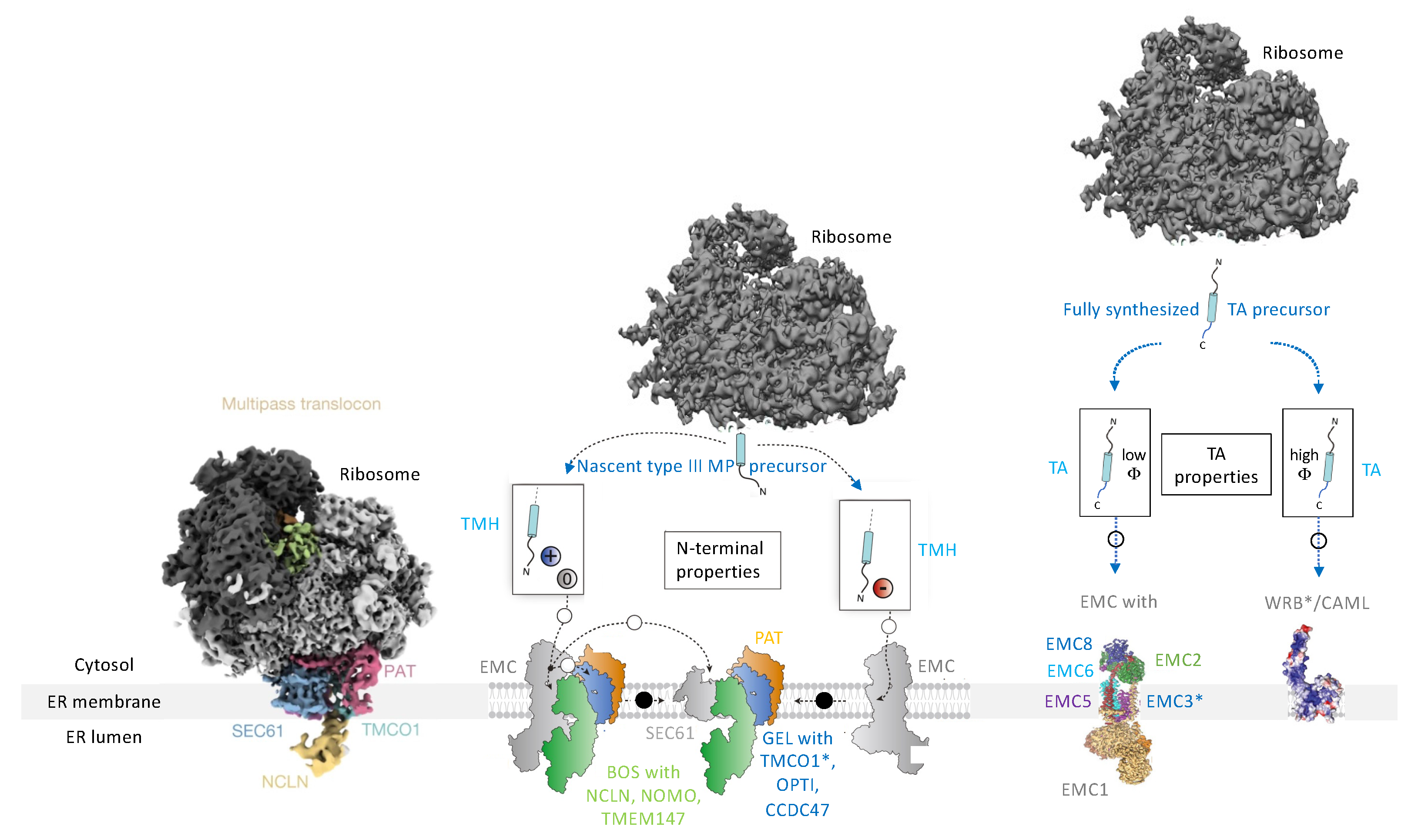
2.4.1. The Sec61 Complex as the Central Entry Gateway into the ER Membrane
2.4.2. The Auxiliary Sec61 Components: TRAP, TRAM1, Sec62, and Sec63
2.4.3. The ER Membrane Complex or EMC
2.5. N-Terminal Methionine Excision and Acetylation of Membrane Protein Precursors
3. Discussion
3.1. Unexpected Insights into General Protein Biogenesis at the ER, Revisited
3.1.1. mRNA- and/or Ribosome-Receptors and the TIGER Domain
3.1.2. PEX3 and ER Exit Sites (ERES)
3.2. Unexpected Results Regarding Membrane Protein Biogenesis at the ER Membrane
3.2.1. Membrane Protein Insertases in the ER Membrane: How Many Are There?
3.2.2. GPI-Anchored MPs
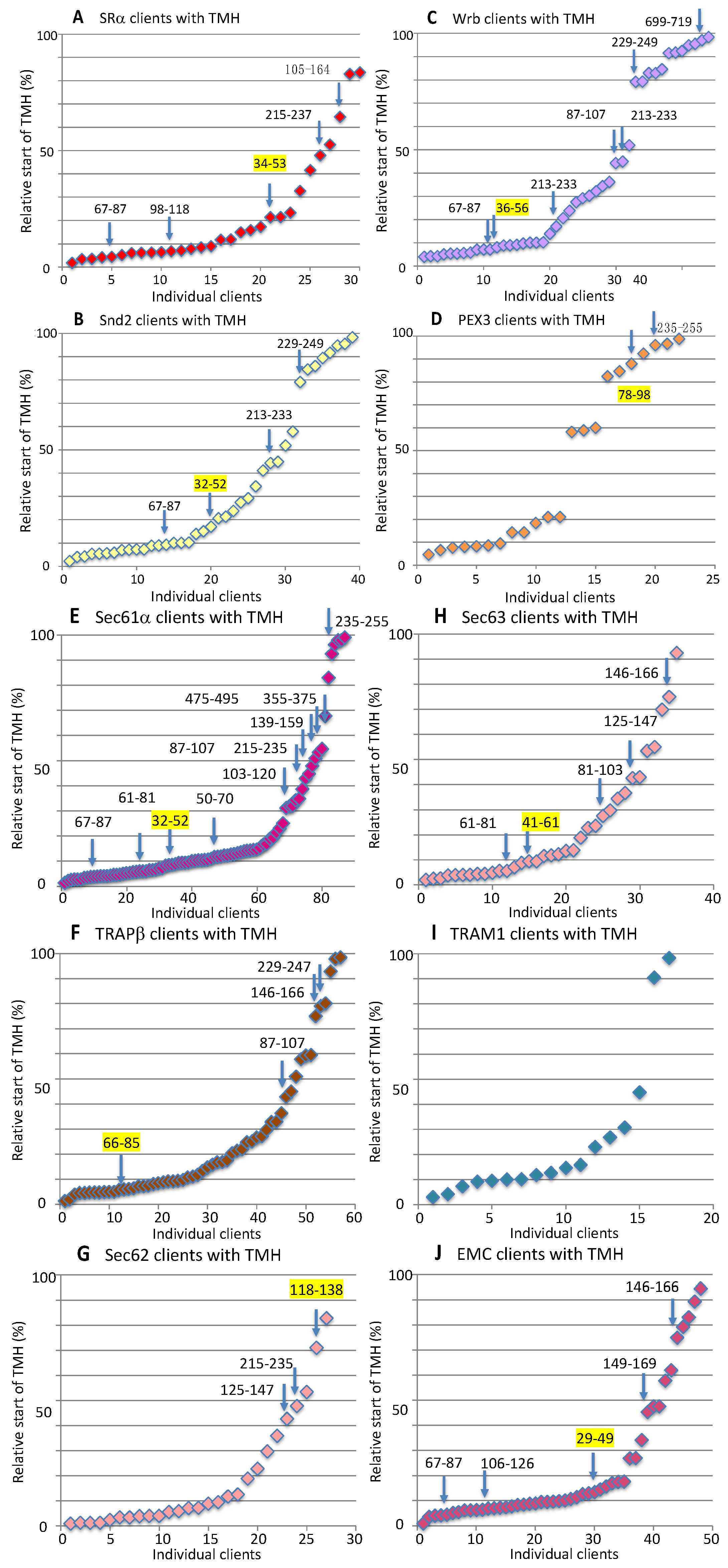
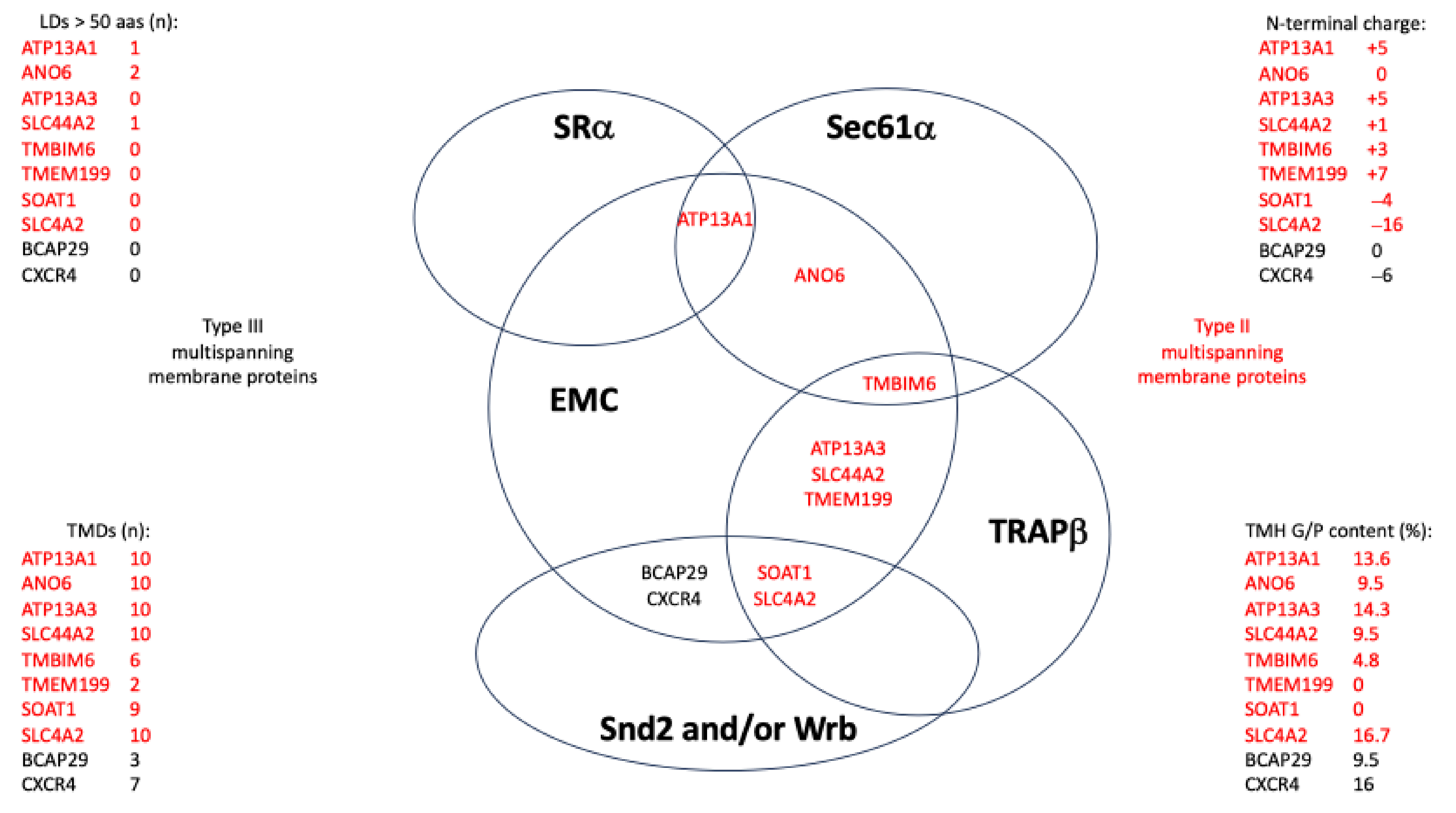
3.2.3. Examples for Client Pathways or System Redundancy
3.2.4. Putative Regulatory Mechanisms
3.2.5. EMC Clients
3.3. Medical Aspects
3.4. Novel Aspects: N-Terminal Methionine Excision and N-Acetylation of Membrane Protein Precursors
3.5. Limitations and Critical Review of the Experimental Approach
3.5.1. Limitations of the Experimental Approach
3.5.2. Critical Review of the Experimental Approach
4. Materials and Methods
5. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BiP | Immunoglobulin heavy chain binding protein, an HSP70 protein family member |
| BOS | Back of Sec61 complex |
| CAML | Calcium modulating ligand |
| EMC | ER membrane complex, comprising an Oxa1 superfamily membrane protein insertase |
| ER | Endoplasmic reticulum |
| ERj | ER J-domain protein, an HSP40 protein family member |
| GEL | GET- and EMC-like, comprising an Oxa1 superfamily membrane protein insertase |
| GET | Guided entry of TA proteins |
| GO | Gene ontology |
| GPI | Glycosylphosphatidylinositol |
| GPI-T | GPI-transamidase |
| HP | Hairpin |
| HSP | Heat-shock protein |
| KTN | Kinectin |
| METAP | Methionine aminopeptidase |
| MP | Membrane protein |
| NAC | Nascent polypeptide-associated complex |
| MPT | Multipass translocon |
| NAT | N-acetyltransferase |
| OST | Oligosaccharyl transferase |
| PAT | PAT 10 comprising complex |
| PDI | Protein disulfide isomerase |
| PEX | Peroxisomal protein |
| PPI | Peptidyl-prolyl cis/trans isomerase |
| RNC | Ribosome-nascent chain |
| SEC | Protein involved in secretion |
| SND | SRP-independent |
| SP | Signal peptide |
| SPC | Signal peptidase complex |
| SR | SRP-receptor |
| SRP | Signal recognition particle |
| TA | Tail anchor |
| TMD | Transmembrane domain |
| TMEM | Transmembrane protein |
| TMH | Most N-terminal transmembrane helix |
| TRAM | Translocating chain-associated membrane protein |
| TRAP | Translocon-associated protein |
| TRC | Transmembrane recognition complex |
| WRB | Tryptophan-rich basic protein, an Oxa1 superfamily membrane protein insertase |
References
- Palade, G. Intracellular aspects of protein synthesis. Science 1975, 189, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Palade, G.; Porter, K.R. Studies on the endoplasmic reticulum. J. Exp. Med. 1954, 100, 641–656. [Google Scholar] [CrossRef] [PubMed]
- Blobel, G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA 1980, 77, 1496–1500. [Google Scholar] [CrossRef]
- Schekman, R.; Novick, P. 23 genes, 23 years later. Cell 2004, 116, 13–15. [Google Scholar] [CrossRef] [PubMed]
- Egea, P.F.; Stroud, R.M.; Walter, P. Targeting proteins to membranes: Structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005, 15, 213–220. [Google Scholar] [CrossRef]
- Aviram, N.; Schuldiner, M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017, 130, 4079–4085. [Google Scholar] [CrossRef]
- Jansen, R.L.M.; Van der Klei, I.J. The peroxisome biogenesis factors Pex3 and Pex19: Multitasking proteins with disputed functions. FEBS Lett. 2019, 593, 457–474. [Google Scholar] [CrossRef]
- Dhiman, R.; Caesar, S.; Thiam, A.R.; Schrul, B. Mechanisms of protein targeting to lipid droplets: A unified cell biological and biophysical perspective. Semin. Cell Dev. Biol. 2020, 108, 4–13. [Google Scholar] [CrossRef]
- Hansen, K.G.; Aviram, N.; Laborenz, J.; Bibi, C.; Meyer, M.; Spang, A.; Schuldiner, M.; Herrmann, J.M. An ER surface retrieval pathway safeguaerds the import of mitochondrial membrane proteins in yeast. Science 2018, 361, 1118–1122. [Google Scholar] [CrossRef]
- Koch, C.; Schuldiner, M.; Herrmann, J.M. ER-SURF: Riding the endoplasmic reticulum SURFace to mitochondria. Int. J. Mol. Sci. 2021, 22, 9655. [Google Scholar] [CrossRef]
- Blobel, G.; Dobberstein, B. Transfer of proteins across membranes: I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 1975, 67, 835–851. [Google Scholar] [CrossRef]
- Blobel, G.; Dobberstein, B. Transfer of proteins across membranes: II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 1975, 67, 852–862. [Google Scholar] [CrossRef]
- von Heijne, G. Signal sequences. J. Mol. Biol. 1985, 184, 99–105. [Google Scholar] [CrossRef]
- Ng, D.T.; Brown, J.D.; Walter, P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 1996, 134, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Hegde, R.S.; Bernstein, H. The surprising complexity of signal sequences. Trends Biochem. Sci. 2006, 31, 563–571. [Google Scholar]
- Goder, V.; Spiess, M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 2003, 22, 3645–3653. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gislason, M.K.; Pihl, S.I.; Tsiigos, K.D.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. Signal 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Nguyen, D.; Bhadra, P.; Jung, M.; Helms, V.; Zimmermann, R. Signal peptide features determining the substrate specificities of targeting and translocation components in human ER protein import. Front. Physiol. 2022, 13, 833540. [Google Scholar] [CrossRef]
- Chen, X.; Van Valkenburgh, C.; Liang, H.; Fang, H.; Green, N. Signal peptidase and oligosaccharyltransferase interact in a sequential and dependent manner within the endoplasmic reticulum. J. Biol. Chem. 2001, 276, 2411–2416. [Google Scholar] [CrossRef]
- Liaci, A.M.; Steigenberger, B.; Tamara, S.; de Souza, P.T.; Gröllers-Mulderij, M.; Ogrissek, P.; Marrink, S.-J.; Scheltema, R.A.; Förster, F. Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. Mol. Cell 2021, 81, 3934–3948. [Google Scholar] [CrossRef]
- Braunger, K.; Pfeffer, S.; Shrimal, S.; Gilmore, R.; Berninhausen, O.; Mandon, E.C.; Becker, T.; Förster, F.; Beckmann, R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 2018, 360, 215–219. [Google Scholar] [CrossRef]
- Wild, R.; Kowal, J.; Eyring, J.; Ngwa, E.M.; Aebi, M.; Locher, K.P. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science 2018, 359, 545–550. [Google Scholar] [CrossRef]
- Neupert, W. A mitochondrial odyssey. Annu. Rev. Biochem. 2012, 81, 1–33. [Google Scholar] [CrossRef]
- Schatz, G. The fires of life. Annu. Rev. Biochem. 2012, 81, 34–59. [Google Scholar] [CrossRef] [PubMed]
- Busch, J.D.; Fielden, L.F.; Pfanner, N.; Wiedemann, N. Mitochondrial protein transport: Versatility of translocases and mechanisms. Mol. Cell 2023, 83, 1931–1952. [Google Scholar] [CrossRef]
- von Heijne, G.; Gavel, Y. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 1988, 174, 671–678. [Google Scholar] [CrossRef]
- Baker, J.A.; Wong, W.-C.; Eisenhaber, B.; Warwicker, J.; Eisenhaber, F. Charged residues next to transmembrane regions revisited: “Positive-inside rule” is complemented by the “negative inside depletion/outside enrichment rule”. BMC Biol. 2017, 15, 66. [Google Scholar] [CrossRef]
- Hegde, R.S.; Keenan, R.J. The mechanism of integral membrane protein biogenesis. Nat. Rev. Mol. Cell Biol. 2022, 23, 107–124. [Google Scholar] [CrossRef]
- O’Keefe, S.; Pool, M.R.; High, S. Membrane protein biogenesis at the ER: The highways and byways. FEBS J. 2022, 289, 6835–6862. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, T.A. A life of translocations. Annu. Rev. Biochem. 2023, 93, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kizmaz, B.; Herrmann, J.M. Membrane insertases at a glance. J. Cell Sci. 2023, 136, jcs261219. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, X.; Yang, Y.; Li, L. Protein translocation through α-helical channels and insertases. Structure 2025, 33, 15–28. [Google Scholar] [CrossRef]
- Kutay, U.; Hartmann, E.; Rapoport, T.A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993, 3, 72–75. [Google Scholar] [CrossRef]
- Schuldiner, M.; Metz, J.; Schmid, V.; Denic, V.; Rakwalska, M.; Schmitt, H.D.; Schwappach, B.; Weissman, J.S. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 2008, 134, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Rabu, C.; Schmid, V.; Schwappach, B.; High, S. Biogenesis of tail-anchored proteins: The beginning for the end? J. Cell Sci. 2009, 122, 3605–3612. [Google Scholar] [CrossRef]
- Ast, T.; Cohen, G.; Schuldiner, M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 2013, 152, 1134–1145. [Google Scholar] [CrossRef] [PubMed]
- Schrul, B.; Kopito, R.R. Peroxin-dependent targeting of a lipid-droplet-destined membrane protein to ER subdomains. Nat. Cell Biol. 2016, 18, 740–751. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Sakisaka, T. The peroxisome biogenesis factors posttranslationally target reticulon homology-domain containing proteins to the endoplasmic reticulum membrane. Sci. Rep. 2018, 8, 2322. [Google Scholar] [CrossRef] [PubMed]
- Borgese, N.; Coy-Vergara, J.; Colombo, S.F.; Schwappach, B. The ways of tails: The GET pathway and more. Proteins 2019, 38, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Leznicki, P.; Schneider, H.O.; Harvey, J.V.; Shi, W.Q.; High, S. Co-translational biogenesis of lipid droplet integral membrane proteins. J. Cell Sci. 2021, 132, jcs.259220. [Google Scholar] [CrossRef]
- Kinoshita, T. Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 2020, 10, 190290. [Google Scholar] [CrossRef]
- Sinning, I.; McDowell, M.A. Cryo-EM insights into tail-anchored membrane protein biogenesis. Curr. Opin. Struct. Biol. 2022, 75, 102428. [Google Scholar] [CrossRef]
- Jung, M.; Zimmermann, R. Quantitative mass spectrometry characterizes client spectra of components for targeting of membrane proteins to and their insertion into the membrane of the human ER. Int. J. Mol. Sci. 2023, 24, 14166. [Google Scholar] [CrossRef]
- Gemmer, M.; Chaillet, M.; Förster, F. Exploring the molecular composition of the multipass translocon in its native membrane environment. Life Sci. Alliance 2024, 7, e202302496. [Google Scholar] [CrossRef]
- Vismpas, D.; Förster, F. RAMPing up the knowledge of the translocon. eLife 2024, 13, e98548. [Google Scholar] [CrossRef]
- Lang, S.; Pfeffer, S.; Lee, P.-H.; Cavalié, A.; Helms, V.; Förster, F.; Zimmermann, R. An update on Sec61 channel function, mechanisms, and related diseases. Front. Physiol. 2017, 8, 887. [Google Scholar] [CrossRef] [PubMed]
- Sicking, M.; Lang, S.; Bochen, F.; Drenth, J.P.H.; Zacharia, M.; Zimmermann, R.; Roos, A.; Linxweiler, M. Complexity and specificity of Sec61 channelopathies: Human diseases affecting gating of the Sec61 complex. Cells 2021, 10, 1036. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Ibrahimi, I.; Blobel, G. Translocation of proteins across the endoplasmic reticulum, I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981, 91, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Blobel, G. Translocation of proteins across the endoplasmic reticulum, II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981, 91, 551–556. [Google Scholar] [CrossRef]
- Siegel, V.; Walter, P. Functional dissection of the signal recognition particle. Trends Biochem. Sci. 1988, 13, 314–316. [Google Scholar] [CrossRef]
- Hann, B.C.; Walter, P. The signal recognition particle in Saccharomyces cerevisiae. Cell 1991, 67, 131–144. [Google Scholar] [CrossRef]
- Hann, B.C.; Stirling, C.J.; Walter, P. SEC65 gene product is a subunit of the yeast signal recognition particle required for its integrity. Nature 1992, 356, 532–533. [Google Scholar] [CrossRef]
- Halic, M.; Beckmann, R. The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Biol. 2005, 15, 116–125. [Google Scholar] [CrossRef]
- Halic, M.; Blau, M.; Becker, T.; Mielke, T.; Pool, M.R.; Wild, K.; Sinning, I.; Beckmann, R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 2006, 444, 507–511. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Hegde, R.S. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife 2015, 4, e07975. [Google Scholar] [CrossRef]
- Gamerdinger, M.; Hanebuth, M.A.; Frickey, T.; Deuerling, E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 2015, 348, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.-H.; Lee, J.H.; Chandrasekar, S.; Shan, S.O. A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun. 2020, 11, 5840. [Google Scholar] [CrossRef]
- Jomaa, A.; Eitzinger, S.; Zhu, Z.; Chandrasekar, S.; Kobajashi, K.; Shan, S.-O.; Ban, N. Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle. Cell Rep. 2021, 36, 109350. [Google Scholar] [CrossRef] [PubMed]
- Jomaa, A.; Gamerdinger, M.; Hsieh, H.-H.; Wallisch, A.; Chandrasekaran, V.; Ulusoy, Z.; Scaiola, A.; Hegde, R.S.; Shan, S.-O.; Ban, N.; et al. Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting. Science 2022, 375, 839–844. [Google Scholar] [CrossRef]
- Meyer, D.I.; Dobberstein, B. A membrane component essential for vectorial translocation of nascent proteins across the endoplasmic reticulum: Requirements for its extraction and reassociation with the membrane. J. Cell Biol. 1980, 87, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, R.; Blobel, G.; Walter, P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982, 95, 463–469. [Google Scholar] [CrossRef]
- Tajima, S.; Lauffer, L.; Rath, V.L.; Walter, P. The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J. Cell Biol. 1986, 103, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Jan, C.H.; Williams, C.C.; Weissman, J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 2014, 346, 1257521. [Google Scholar] [CrossRef]
- Chartron, J.W.; Hunt, K.C.L.; Frydman, J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 2016, 536, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hegde, R.S. Identification of a factor that accelerates substrate release from the signal recognition particle. Science 2024, 386, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Koch, B.D.; Werner-Washburne, M.; Craig, E.A.; Schekman, R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 1988, 332, 800–805. [Google Scholar] [CrossRef]
- Rothblatt, J.A.; Deshaies, R.J.; Sanders, S.L.; Daum, G.; Schekman, R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J. Cell Biol. 1989, 109, 2641–2652. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Sanders, S.L.; Feldheim, D.A.; Schekman, R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 1991, 349, 806–808. [Google Scholar] [CrossRef]
- Sanders, S.L.; Whitfield, K.M.; Vogel, J.P.; Rose, M.D.; Schekman, R. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 1992, 69, 353–365. [Google Scholar] [CrossRef]
- Brodsky, J.L.; Goeckeler, J.; Schekman, R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 1995, 92, 9643–9646. [Google Scholar] [CrossRef]
- Lyman, S.K.; Schekman, R. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J. Cell Biol. 1995, 131, 1163–1171. [Google Scholar] [CrossRef]
- Craven, R.A.; Egerton, M.; Stirling, C.J. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996, 15, 2640–2650. [Google Scholar] [CrossRef]
- Lyman, S.K.; Schekman, R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell 1997, 88, 85–96. [Google Scholar] [CrossRef]
- Young, B.P.; Craven, R.A.; Reid, P.J.; Willer, M.; Stirling, C.J. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001, 20, 262–271. [Google Scholar] [CrossRef]
- Jung, S.-j.; Kim, J.E.H.; Reithinger, J.H.; Kim, H. The Sec62–Sec63 translocon facilitates translocation of the C-terminus of membrane proteins. J. Cell Sci. 2014, 127, 4270–4278. [Google Scholar]
- Cohen, N.; Aviram, N.; Schuldiner, M. A systematic proximity ligation approach to studying protein-substrate specificity identifies the substrate spectrum of the Ssh1 translocon. EMBO J. 2023, 43, e113385. [Google Scholar] [CrossRef]
- Schlenstedt, G.; Zimmermann, R. Import of frog prepropeptide GLa into microsomes requires ATP but does not involve docking protein or ribosomes. EMBO J. 1987, 6, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Müller, G.; Zimmermann, R. Import of honeybee prepromelittin into the endoplasmic reticulum: Structural basis for independence of SRP and docking protein. EMBO J. 1987, 6, 2099–2107. [Google Scholar] [CrossRef]
- Müller, G.; Zimmermann, R. Import of honeybee prepromelittin into the endoplasmic reticulum: Energy requirements for membrane insertion. EMBO J. 1988, 7, 639–648. [Google Scholar] [CrossRef]
- Wiech, H.; Sagstetter, M.; Müller, G.; Zimmermann, R. The ATP requiring step in the assembly of M 13 procoat protein into microsomes is related to preservation of transport competence of the precursor protein. EMBO J. 1987, 6, 1011–1016. [Google Scholar] [CrossRef]
- Zimmermann, R.; Sagstetter, M.; Lewis, M.J.; Pelham, H.R.B. Seventy-kilodalton heat shock proteins and an additional component from reticulocyte lysate stimulate import of M 13 procoat protein into microsomes. EMBO J. 1988, 7, 2875–2880. [Google Scholar] [CrossRef]
- Schlenstedt, G.; Gudmundsson, G.H.; Boman, H.G.; Zimmermann, R. A large presecretory protein translocates both cotranslationally, using signal recognition particle and ribosome, and posttranslationally, without these ribonucleoparticles, when synthesized in the presence of mammalian microsomes. J. Biol. Chem. 1990, 265, 13960–13968. [Google Scholar] [CrossRef]
- Klappa, P.; Mayinger, P.; Pipkorn, R.; Zimmermann, M.; Zimmermann, R. A microsomal protein is involved in ATP-dependent transport of presecretory proteins into mammalian microsomes. EMBO J. 1991, 10, 2795–2803. [Google Scholar] [CrossRef]
- Klappa, P.; Freedman, R.; Zimmermann, R. Protein disulfide isomerase and a lumenal cyclophilin-type peptidyl prolyl cis-trans isomerase are in transient contact with secretory proteins during late stages of translocation. Eur. J. Biochem. 1995, 232, 755–764. [Google Scholar]
- Dierks, T.; Volkmer, J.; Schlenstedt, G.; Jung, C.; Sandholzer, U.; Zachmann, K.; Schlotterhose, P.; Neifer, K.; Schmidt, B.; Zimmermann, R. A microsomal ATP-binding protein involved in efficient protein transport into the mammalian endoplasmic reticulum. EMBO J. 1996, 15, 6931–6942. [Google Scholar] [CrossRef]
- Tyedmers, J.; Lerner, M.; Bies, C.; Dudek, J.; Skowronek, M.H.; Haas, I.G.; Heim, N.; Nastainczyk, W.; Volkmer, J.; Zimmermann, R. Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc. Natl. Acad. Sci. USA 2000, 97, 7214–7219. [Google Scholar] [CrossRef]
- Mayer, H.-A.; Grau, H.; Kraft, R.; Prehn, S.; Kalies, K.-U.; Hartmann, E. Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 2000, 275, 14550–14557. [Google Scholar] [CrossRef]
- Tyedmers, J.; Lerner, M.; Wiedmann, M.; Volkmer, J.; Zimmermann, R. Polypeptide chain binding proteins mediate completion of cotranslational protein translocation into the mammalian endoplasmic reticulum. EMBO Rep. 2005, 4, 505–510. [Google Scholar] [CrossRef]
- Müller, L.; Diaz de Escauriaza, M.; Lajoie, P.; Theis, M.; Jung, M.; Müller, A.; Burgard, C.; Greiner, M.; Snapp, E.L.; Dudek, J.; et al. Evolutionary gain of function of the ER membrane protein Sec62 from yeast to humans. Mol. Biol. Cell 2010, 21, 691–703. [Google Scholar] [CrossRef]
- Shao, S.; Hegde, R.S. A calmodulin-dependent translocation pathway for small secretory proteins. Cell 2011, 147, 1576–1588. [Google Scholar] [CrossRef]
- Schäuble, N.; Lang, S.; Jung, M.; Cappel, S.; Schorr, S.; Ulucan, Ö.; Linxweiler, J.; Dudek, J.; Blum, R.; Helms, V.; et al. BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. EMBO J. 2012, 31, 3282–3296. [Google Scholar] [CrossRef]
- Lakkaraju, A.K.K.; Thankappan, R.; Mary, C.; Garrison, J.L.; Taunton, J.; Strub, K. Efficient secretion of small proteins in mammalian cells relies on Sec62-dependent posttranslational translocation. Mol. Biol. Cell 2012, 23, 2712–2722. [Google Scholar] [CrossRef]
- Lang, S.; Benedix, J.; Fedeles, S.V.; Schorr, S.; Schirra, C.; Schäuble, N.; Jalal, C.; Greiner, M.; Haßdenteufel, S.; Tatzelt, J.; et al. Different effects of Sec61α-, Sec62 and Sec63-depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J. Cell Sci. 2012, 125, 1958–1969. [Google Scholar]
- Johnson, N.; Vilardi, F.; Lang, S.; Leznicki, P.; Zimmermann, R.; High, S. TRC-40 can deliver short secretory proteins to the Sec61 translocon. J. Cell Sci. 2012, 125, 3612–3620. [Google Scholar] [CrossRef]
- Haßdenteufel, S.; Johnson, N.; Paton, A.W.; Paton, J.C.; High, S.; Zimmermann, R. Chaperone-mediated Sec61 channel gating during ER import of small precursor proteins overcomes Sec61 inhibitor-reinforced energy barrier. Cell Rep. 2018, 23, 1373–1386. [Google Scholar] [CrossRef]
- Haßdenteufel, S.; Nguyen, D.; Helms, V.; Lang, S.; Zimmermann, R. Components and mechanisms for ER import of small human presecretory proteins. FEBS Lett. 2019, 593, 2506–2524. [Google Scholar] [CrossRef]
- Ziska, A.; Tatzelt, J.; Dudek, J.; Paton, A.W.; Paton, J.C.; Zimmermann, R.; Haßdenteufel, S. The signal peptide plus a cluster of positive charges in prion protein dictate chaperone-mediated Sec61-channel gating. Biol. Open 2019, 8, bio040691. [Google Scholar] [CrossRef]
- Schorr, S.; Nguyen, D.; Haßdenteufel, S.; Nagaraj, N.; Cavalié, A.; Greiner, M.; Weissgerber, P.; Loi, M.; Paton, A.W.; Paton, J.C.; et al. Proteomics identifies signal peptide features determining the substrate specificity in human Sec62/Sec63-dependent ER protein import. FEBS J. 2020, 287, 4612–4640. [Google Scholar] [CrossRef]
- Sun, S.; Li, X.; Mariappan, M. Signal sequences encode information for protein folding in the endoplasmic reticulum. J. Cell Biol. 2023, 222, e202203070. [Google Scholar] [CrossRef]
- Simon, S.M.; Blobel, G. A protein-conducting channel in the endoplasmic reticulum. Cell 1991, 65, 371–380. [Google Scholar] [CrossRef]
- Görlich, D.; Hartmann, E.; Prehn, S.; Rapoport, T.A. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature 1992, 357, 47–52. [Google Scholar] [CrossRef]
- Görlich, D.; Prehn, S.; Hartmann, E.; Kalies, K.-U.; Rapoport, T.A. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 1992, 71, 489–503. [Google Scholar] [CrossRef]
- Görlich, D.; Rapoport, T.A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 1993, 75, 615–630. [Google Scholar] [CrossRef]
- Kalies, K.-U.; Rapoport, T.A.; Hartmann, E. The beta-subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J. Cell Biol. 1998, 141, 887–894. [Google Scholar] [CrossRef]
- Beckmann, R.; Spahn, C.M.; Eswar, N.; Helmers, J.; Penczek, P.A.; Sali, A.; Frank, J.; Blobel, G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 2001, 107, 361–372. [Google Scholar] [CrossRef]
- Wirth, A.; Jung, M.; Bies, C.; Frien, M.; Tyedmers, J.; Zimmermann, R.; Wagner, R. The Sec61p complex is a dynamic precursor activated channel. Mol. Cell 2003, 12, 261–268. [Google Scholar] [CrossRef]
- Van den Berg, B.; Clemons, W.M.; Collinson, I.; Modis, Y.; Hartmann, E.; Harrison, S.C.; Rapoport, T.A. X-ray structure of a protein-conducting channel. Nature 2004, 427, 36–44. [Google Scholar] [CrossRef]
- Devaraneni, P.K.; Conti, B.; Matsumara, Y.; Yang, Z.; Johnson, A.E.; Skach, W.R. Stepwise insertion and inversion of a type II signal anchor sequence in the ribosome-Sec61 translocon complex. Cell 2011, 146, 134–147. [Google Scholar] [CrossRef]
- Pfeffer, S.; Brandt, F.; Hrabe, T.; Lang, S.; Eibauer, M.; Zimmermann, R.; Förster, F. Structure and 3D arrangement of ER-membrane associated ribosomes. Structure 2012, 20, 1508–1518. [Google Scholar] [CrossRef]
- Pfeffer, S.; Dudek, J.; Gogala, M.; Schorr, S.; Linxweiler, J.; Lang, S.; Becker, T.; Beckmann, R.; Zimmermann, R.; Förster, F. Structure of the mammalian oligosaccharyltransferase in the native ER protein translocon. Nat. Commun. 2014, 5, 3072. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Fernández, I.S.; Scheres, S.H.W.; Hegde, R.S. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 2014, 157, 1632–1643. [Google Scholar] [CrossRef]
- Jadhav, B.; McKenna, M.; Johnson, N.; High, S.; Sinning, I.; Pool, M.R. Mammalian SRP receptor switches the Sec61 translocase from Sec62 to SRP-dependent translocation. Nat. Commun. 2015, 6, 10133. [Google Scholar] [CrossRef] [PubMed]
- Conti, B.J.; Devaraneni, P.K.; Yang, Z.; David, L.L.; Skach, W.R. Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol. Cell 2015, 58, 269–283. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Hegde, R.S. Structure of the Sec61 channel opened by a signal peptide. Science 2016, 351, 88–91. [Google Scholar] [CrossRef]
- Pfeffer, S.; Burbaum, L.; Unverdorben, P.; Pech, M.; Chen, Y.; Zimmermann, R.; Beckmann, R.; Förster, F. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 2015, 6, 8403. [Google Scholar] [CrossRef]
- Mahamid, J.; Pfeffer, S.; Schaffer, M.; Villa, E.; Danev, R.; Kuhn Cuellar, L.; Förster, F.; Hyman, A.A.; Plitzko, J.M.; Baumeister, W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 2016, 351, 969–972. [Google Scholar] [CrossRef] [PubMed]
- Gemmer, M.; Förster, F. A clearer picture of the ER translocon complex. J. Cell Sci. 2020, 133, jcs231340. [Google Scholar] [CrossRef]
- Gemmer, M.; Chaillet, M.; van Loenhout, J.; Arenas, R.C.; Vismpas, D.; Gröllers-Mulderji, M.; Kohl, F.A.; Albanese, P.; Scheltema, R.A.; Howes, S.C.; et al. Visualization of translation and protein biogenesis at the ER membrane. Nature 2023, 614, 160–167. [Google Scholar] [CrossRef]
- Mariappan, M.; Li, X.; Stefanovic, S.; Sharma, A.; Mateja, A.; Keenan, R.J.; Hegde, R.S. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 2010, 466, 1120–1124. [Google Scholar] [CrossRef]
- Leznicki, P.; Clancy, A.; Schwappach, B.; High, S. Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 2010, 123, 2170–2178. [Google Scholar] [CrossRef] [PubMed]
- Borgese, N.; Fasana, E. Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta 2011, 1808, 937–946. [Google Scholar] [CrossRef]
- Haßdenteufel, S.; Schäuble, N.; Cassella, P.; Leznicki, P.; Müller, A.; High, S.; Jung, M.; Zimmermann, R. Calcium-calmodulin inhibits tail-anchored protein insertion into the mammalian endoplasmic reticulum membrane. FEBS Lett. 2011, 585, 3485–3490. [Google Scholar] [CrossRef] [PubMed]
- Vilardi, F.; Lorenz, H.; Dobberstein, B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J. Cell Sci. 2011, 124, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Leznicki, P.; Warwicker, J.; High, S. A biochemical analysis of the constraints of tail-anchored protein biogenesis. Biochem. J. 2011, 436, 719–727. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Sakisaka, T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol. Cell 2012, 48, 387–397. [Google Scholar] [CrossRef]
- Leznicki, P.; High, S. SGTA antagonizes BAG6-mediated protein triage. Proc. Natl. Acad. Sci. USA 2012, 109, 19214–19219. [Google Scholar] [CrossRef]
- Hegde, R.S.; Keenan, R.J. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2013, 12, 787–798. [Google Scholar] [CrossRef]
- Vilardi, F.; Stephan, M.; Clancy, A.; Janshoff, A.; Schwappach, B. WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS ONE 2014, 9, e85033. [Google Scholar] [CrossRef]
- Wang, F.; Chan, C.; Weir, N.R.; Denic, V. The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature 2014, 512, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Casson, J.; McKenna, M.; Haßdenteufel, S.; Aviram, N.; Zimmerman, R.; High, S. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 2017, 130, 3851–3861. [Google Scholar] [CrossRef]
- Zhang, Y.; De Laurentiis, E.; Bohnsack, K.E.; Wahlig, M.; Ranjan, N.; Gruseck, S.; Hackert, P.; Wölfle, T.; Rodnina, M.; Schwappach, B.; et al. Ribosome-bound Get4/5 facilitate the capture of tail-anchored proteins by Sgt2 in yeast. Nat. Commun. 2021, 12, 782. [Google Scholar] [CrossRef]
- Carvalho, H.J.F.; Del Bondio, A.; Maltecca, F.; Colombo, S.F.; Borgese, N. The WRB subunit of the Get3 receptor is required for the correct integration of its partner CAML into the ER. Sci. Rep. 2019, 9, 11887. [Google Scholar] [CrossRef]
- McDowell, M.A.; Heimes, M.; Fiorentino, F.; Mehmood, S.; Farka, A.; Coy-Vergara, J.; Wu, D.; Bolla, J.R.; Schmid, V.; Heinze, R.; et al. Structural basis of tail-anchored membrane protein biogenesis by the GET insertase complex. Mol. Cell 2020, 80, 72–86. [Google Scholar] [CrossRef]
- Leznicki, P.; High, S. SGTA associates with nascent membrane protein precursors. EMBO Rep. 2020, 21, e48835. [Google Scholar] [CrossRef]
- Farkas, A.; Urlaub, H.; Bohnsack, K.E.; Schwappach, B. Regulated targeting of the monotopic hairpin membrane protein Erg1 requires the GET pathway. J. Cell Biol. 2022, 221, e202201036. [Google Scholar] [CrossRef]
- Wiedmann, M.; Kurzchalia, T.V.; Hartmann, E.; Rapoport, T.A. A signal sequence receptor in the endoplasmic reticulum membrane. Nature 1987, 328, 830–833. [Google Scholar] [CrossRef] [PubMed]
- Fons, R.D.; Bogert, B.A.; Hegde, R.S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003, 160, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Sommer, N.; Junne, T.; Kalies, K.-U.; Spiess, M.; Hartmann, E. TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim. Biophys. Acta 2013, 1833, 3104–3111. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Dudek, J.; Ng, B.; Schaffa, M.; Albert, S.; Plitzko, J.; Baumeister, W.; Zimmermann, R.; Freeze, H.; Engel, B.D.; et al. Dissecting the molecular organization of the translocon-associatecd protein complex. Nat. Commun. 2017, 8, 14516. [Google Scholar] [CrossRef] [PubMed]
- Jaskolowski, M.; Jomaa, A.; Gamerdinger, M.; Shresta, S.; Leibundgut, M.; Deuerling, E.; Ban, N. Molecular basis of the TRAP complex function in ER protein biogenesis. Nat. Struct. Mol. Biol. 2023, 30, 770–777. [Google Scholar] [CrossRef]
- Pauwels, E.; Shewakramani, N.R.; De Wijngaert, B.; Camps, A.; Provinciael, B.; Stroobants, J.; Kalies, K.-U.; Hartmann, E.; Maes, P.; Vermeire, K.; et al. Structural insights into TRAP association with ribosome-Sec61 complex and translocon inhibition by a CADA derivative. Sci. Adv. 2023, 9, eadf0797. [Google Scholar] [CrossRef]
- Wilson, R.; Allen, A.J.; Oliver, J.; Brookman, J.L.; High, S.; Bulleid, N. The translocation, folding, assembly, and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem. J. 1995, 387, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Haßdenteufel, S.; Klein, M.-C.; Melnyk, A.; Zimmermann, R. Protein transport into the human ER and related diseases: Sec61-channelopathies. Biochem. Cell Biol. 2014, 92, 499–509. [Google Scholar] [CrossRef]
- Schubert, D.; Klein, M.-C.; Haßdenteufel, S.; Caballero-Oteyza, A.; Yang, L.; Proietti, M.; Bulashevska, A.; Kemming, J.; Kühn, J.; Winzer, S.; et al. Plasma cell deficiency in human subjects with heterozygous mutations in Sec61 translocon alpha 1 (SEC61A1). J. Allergy Clin. Immunol. 2018, 141, 1427–1438. [Google Scholar] [CrossRef]
- Van Nieuwenhove, E.; Barber, J.; Smeets, E.; Neumann, J.; Willemsen, M.; Pasciuto, E.; Prezzemolo, T.; Lagou, V.; Seldeslachts, L.; Malengier-Devlies, B.; et al. Defective Sec61α1 underlies a novel cause of autosomal dominant severe congenital neutropenia. J. Allergy Clin. Immunol. 2020, 146, 1180–1192. [Google Scholar] [CrossRef]
- Bolar, N.A.; Golzio, C.; Živná, M.; Hayot, G.; Van Hemelrijk, C.; Schepers, D.; Vandeweyer, G.; Hoischen, A.; Huyghe, J.R.; Raes, A.; et al. Heterozygous Loss-of-Function SEC61A1 Mutations Cause Autosomal-Dominant Tubulo-Interstitial and Glomerulocystic Kidney Disease with Anemia. Am. J. Hum. Genet. 2016, 99, 174–187. [Google Scholar] [CrossRef]
- Fedeles, S.V.; Tian, X.; Gallagher, A.-R.; Mitobe, M.; Nishio, S.; Lee, S.H.; Cai, Y.; Geng, L.; Crews, C.M.; Somlo, S. A genetic interaction network of five genes for human polycystic kidney and liver disease defines polycystin-1 as the central determinant of cyst formation. Nat. Genet. 2011, 43, 639–647. [Google Scholar] [CrossRef]
- Besse, W.; Dong, K.; Choi, J.; Punia, S.; Fedeles, S.V.; Choi, M.; Gallagher, A.-R.; Huang, E.B.; Gulati, A.; Knight, J.; et al. Isolated polycystic liver disease genes define effector s of polycystin-1 function. J. Clin. Investig. 2017, 127, 1772–1785. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.P.; Durin, Z.; Unal, Ö.; Ng, B.G.; Marrecau, T.; Keldermans, L.; Souche, E.; Rymen, D.; Gündüz, M.; Köse, G.; et al. CAMLG-CDG: A novel Congenital Disorder of Glycosylation linked to defective membrane trafficking. Hum. Mol. Genet. 2022, 31, 2571–2581. [Google Scholar] [CrossRef] [PubMed]
- Weiand, M.; Sandfort, V.; Nadzemova, O.; Schierwagen, R.; Trebicka, J.; Schlevogt, B.; Kabar, I.; Schmidt, H.; Zibert, A. Comparative analysis of SEC61A1 mutant R236C in two patient-derived cellular platforms. Sci. Rep. 2024, 15, 9506. [Google Scholar] [CrossRef]
- Ng, B.G.; Freeze, H.F.; Himmelreich, N.; Blau, N.; Ferreira, C.R. Clinical and biochemical footprints of congenital disorders of glycosylation: Proposed nosology. Mol. Genet. Metab. 2024, 142, 108476. [Google Scholar] [CrossRef]
- Sicking, M.; Živná, M.; Bhadra, P.; Barešová, V.; Tirincsi, A.; Hadzibeganovic, D.; Hodaňová, K.; Vyleťal, P.; Sovová, J.; Jedličkova, I.; et al. Phenylbutyrate rescues the transport defect of the Sec61α mutations V67G and T185A for renin. Life Sci. Alliance 2022, 5, e202101150. [Google Scholar] [CrossRef]
- Erdmann, F.; Schäuble, N.; Lang, S.; Jung, M.; Honigmann, A.; Ahmad, M.; Dudek, J.; Benedix, J.; Harsman, A.; Kopp, A.; et al. Interaction of calmodulin with Sec61a limits Ca2+ leakage from the endoplasmic reticulum. EMBO J. 2011, 30, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Aviram, N.; Ast, T.; Costa, E.A.; Arakel, E.; Chuartzman, S.G.; Jan, C.H.; Haßdenteufel, S.; Dudek, J.; Jung, M.; Schorr, S.; et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 2016, 540, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Haßdenteufel, S.; Sicking, M.; Schorr, S.; Aviram, N.; Fecher-Trost, C.; Schuldiner, M.; Jung, M.; Zimmermann, R.; Lang, S. hSnd2 protein represents an alternative targeting factor to the endoplasmic reticulum in human cells. FEBS Lett. 2017, 591, 3211–3224. [Google Scholar] [CrossRef]
- Tirincsi, A.; O’Keefe, S.; Nguyen, D.; Sicking, M.; Dudek, J.; Förster, F.; Jung, M.; Hadzibeganovic, D.; Helms, V.; High, S.; et al. Proteomics identifies substrates and a novel component in hSnd2-dependent ER protein targeting. Cells 2022, 11, 2925. [Google Scholar] [CrossRef]
- Zimmermann, R.; Lang, S.; Lerner, M.; Förster, F.; Nguyen, D.; Helms, V.; Schrul, B. Quantitative proteomics and differential protein abundance ananalysis after depletion of PEX3 from human cells identifies additional aspects of protein targeting to the ER. Int. J. Mol. Sci. 2021, 22, 13028. [Google Scholar] [CrossRef]
- Lang, S.; Erdmann, F.; Jung, M.; Wagner, R.; Cavalié, A.; Zimmermann, R. Sec61 complexes form ubiquitous ER Ca2+ leak channels. Channels 2011, 5, 228–235. [Google Scholar] [CrossRef]
- Schorr, S.; Klein, M.-C.; Gamayun, I.; Melnyk, A.; Jung, M.; Schäuble, N.; Wang, Q.; Hemmis, B.; Bochen, F.; Greiner, M.; et al. Co-chaperone specificity in gating of the polypeptide conducting channel in the membrane of the human endoplasmic reticulum. J. Biol. Chem. 2015, 290, 18621–18635. [Google Scholar] [CrossRef] [PubMed]
- Linxweiler, M.; Schorr, S.; Jung, M.; Schäuble, N.; Linxweiler, J.; Langer, F.; Schäfers, H.-J.; Cavalié, A.; Zimmermann, R.; Greiner, M. Targeting cell migration and the ER stress response with calmodulin antagonists: A clinically tested small molecule phenocopy of SEC62 gene silencing in human tumor cells. BMC Cancer 2013, 13, 574. [Google Scholar] [CrossRef]
- Gumbart, J.; Schulten, K. Structural determinants of lateral gate opening in the protein translocon. Biochemistry 2007, 46, 11147–11157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Miller, T.F., III. Long-timescale dynamics and regulation of Sec-facilitated protein translocation. Cell Rep. 2012, 2, 927–937. [Google Scholar] [CrossRef]
- Savitz, A.J.; Meyer, D.I. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature 1990, 346, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Seiser, R.M.; Nicchitta, C.V. The fate of membrane-bound ribosomes following the termination of protein synthesis. J. Biol. Chem. 2000, 275, 33820–33827. [Google Scholar] [CrossRef]
- Potter, M.D.; Seiser, R.M.; Nicchitta, C.V. Ribosome exchange revisited: A mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 2001, 11, 112–115. [Google Scholar] [CrossRef]
- Berkovits, B.D.; Mayr, C. Alternative 3′UTRs act as scaffolds to regulate membrane protein localization. Nature 2015, 522, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Mayr, C. A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein-protein interactions. Cell 2018, 175, 1492–1506. [Google Scholar] [CrossRef]
- Hsu, J.C.-C.; Reid, D.W.; Hoffman, A.M.; Sarkar, D.; Nicchitta, C.V. Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA 2018, 24, 688–703. [Google Scholar]
- Hannigan, M.M.; Hoffman, A.M.; Thompson, J.W.; Zheng, T.; Nicchitta, C.V. Quantitative proteomics links the LRRC59 interactome to mRNA translation on the ER membrane. Mol. Cell. Proteom. 2020, 19, 1826–1849. [Google Scholar] [CrossRef]
- Bhadra, P.; Schorr, S.; Lerner, M.; Nguyen, D.; Dudek, J.; Förster, F.; Helms, V.; Lang, S.; Zimmermann, R. Quantitative proteomics and differential protein abundance analysis after depletion of putative mRNA receptors in the ER membrane of human cells identifies novel aspects of mRNA targeting to the ER. Molecules 2021, 26, 3591. [Google Scholar] [CrossRef]
- Horste, E.L.; Fansler, M.M.; Cai, T.; Chen, X.; Mitschka, S.; Zhen, G.; Lee, F.C.Y.; Ule, J.; Mayr, C. Subcytoplasmic location of translation controls protein output. Mol. Cell 2023, 83, 4509–4523. [Google Scholar] [CrossRef] [PubMed]
- Child, J.R.; Hofler, A.C.; Chen, Q.; Yang, B.H.; Kristofich, J.; Zheng, T.; Hannigan, M.M.; Elles, A.L.; Reid, D.W.; Nicchitta, C.V. Examining SRP pathway function in mRNA localizytion to the endoplasmic reticulum. RNA 2023, 29, 1703–17124. [Google Scholar] [CrossRef]
- Hirata, T.; Yang, J.; Tomida, S.; Tokoro, Y.; Kinoshita, T.; Fujita, M.; Kizuka, Y. ER entry pathway and glycosylation of GPI-anchored proteins are determined by N-terminal signal sequence and C-terminal GPI-attachment sequence. J. Biol. Chem. 2022, 298, 102444. [Google Scholar] [CrossRef]
- Talbot, B.E.; Vandorpe, D.H.; Stotter, B.R.; Alper, S.L.; Schlondorff, J. Transmembrane insertases and N-glycosylation crtically determine synthesis, trafficking, and activity of the nonselective cation channel TRPC6. J. Biol. Chem. 2019, 294, 12655–12669. [Google Scholar] [CrossRef]
- Yang, J.; Hirata, T.; Liu, Y.S.; Guo, X.Y.; Gao, X.-D.; Kinoshita, T.; Fujita, M. Human SND2 mediates ER targeting of GPI-anchored proteins with low hydrophobic GPI attachment signals. FEBS Lett. 2021, 595, 1542–1558. [Google Scholar] [CrossRef]
- Anghel, S.A.; McGilvray, P.T.; Hegde, R.S.; Keenan, R.J. Identification of Oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Rep. 2017, 21, 3708–3716. [Google Scholar] [CrossRef]
- Chitwood, P.J.; Juszkiewicz, S.; Guna, A.; Shao, S.; Hegde, R.S. EMC is required to initiate accurate membrane protein topogenesis. Cell 2018, 175, 1507–1519. [Google Scholar] [CrossRef]
- Shurtleff, M.J.; Itzhak, D.N.; Hussmann, J.A.; Schirle Oakdale, N.T.; Costa, E.A.; Jonikas, M.; Weibezahn, J.; Popova, K.D.; Jan, C.H.; Sinitcyn, P.; et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. eLife 2018, 7, e37018. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wu, Q.; Zhou, B.; Choi, M.Y.; Ding, B.; Yang, W.; Dong, M. Proteomic analysis indentifies membrane proteins dependent on the ER membrane protein complex. Cell Rep. 2019, 28, 2517–2526. [Google Scholar] [CrossRef]
- McGilvray, P.T.; Anghel, S.A.; Sundaram, A.; Zhong, F.; Trnka, M.J.; Fuller, J.R.; Hu, H.; Burlingame, A.L.; Keenan, R.J. An ER translocon for multi-pass mambrane protein biogenesis. eLife 2020, 9, e56889. [Google Scholar] [CrossRef] [PubMed]
- O’Donnel, J.P.; Philips, B.P.; Yagita, Y.; Juszkiewicz, S.; Wagner, A.; Malinverni, D.; Keenan, R.J.; Mille, E.A.; Hegde, R.S. The architecture of EMC reveals a path for membrane protein insertion. eLife 2020, 9, e57887. [Google Scholar] [CrossRef] [PubMed]
- Pleiner, T.; Tomaleri, G.P.; Januszyk, K.; Inglis, A.J.; Hazu, M.; Voorhees, R.M. Structural basis for membrane insertion by the human ER membrane protein complex. Science 2020, 369, 433–436. [Google Scholar] [CrossRef]
- Bai, L.; You, Q.; Feng, X.; Kovach, A.; Li, H. Structure of the ER membrane complex, a transmembrane insertase. Nature 2020, 584, 475–478. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, S.; Zong, G.; Duah, K.B.; Andrews, L.E.; Shi, W.Q.; High, S. An alternative pathway for membrane protein biogenesis at the endoplasmic reticulum. Commun. Biol. 2021, 4, 828. [Google Scholar] [CrossRef]
- Kizmaz, B.; Flohr, T.; Garg, S.G.; Herrmann, J.M. The ER membrane complex (EMC) can functionally replace the Oxa1 insertase in mitochondria. PLoS Biol. 2022, 20, e3001380. [Google Scholar]
- Meacock, S.L.; Lecomte, F.J.L.; Crawshaw, S.G.; High, S. Different transmembrane domains associate with distinct endoplasmic reticulum components during membrane integration of a polytopic protein. Mol. Biol. Cell 2002, 13, 4114–4129. [Google Scholar] [CrossRef]
- Sundaram, A.; Yamsek, M.; Zhong, F.; Hooda, Y.; Hegde, R.S.; Keenan, R.J. Substrate-driven assembly of a translocon for multipass membrane proteins. Nature 2022, 611, 167–172. [Google Scholar] [CrossRef]
- Samlinskaite, L.; Kim, M.K.; Lewis, A.J.O.; Keenan, R.J.; Hegde, R.S. Mechanism of an intramembrane chaperone for multipass membrane proteins. Nature 2022, 611, 161–166. [Google Scholar] [CrossRef]
- Wu, H.; Hegde, R.S. Mechanism of signal-anchor triage during early steps of membrane protein insertion. Mol. Cell 2023, 83, 961–973. [Google Scholar] [CrossRef]
- Page, K.R.; Nguyen, V.N.; Pleiner, T.; Tomaleri, G.P.; Wang, M.L.; Guna, A.; Hazu, M.; Wang, T.-Y.; Chou, T.-F.; Voorhees, R.M. Role of the holo-insertase complex in the biogensis of biophysically diverse ER membrane proteins. Mol. Cell 2024, 84, 3302–3319. [Google Scholar] [CrossRef]
- Wiedmann, B.; Saki, H.; Davis, T.A.; Wiedmann, M. A protein complex required for signal-sequence-specific sorting and translocation. Nature 1994, 370, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Moeller, I.; Jung, M.; Beatrix, B.; Levy, R.; Kreibich, G.; Zimmermann, R.; Wiedmann, M.; Lauring, B. A general mechanism for regulation of access to the translocon: Competition for a membrane attachment site on ribosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 13425–13430. [Google Scholar] [CrossRef]
- Gamerdinger, M.; Kobayashi, K.; Wallisch, A.; Kreft, S.G.; Sailer, C.; Schlömer, R.; Sachs, N.; Jomaa, A.; Stengel, F.; Ban, N.; et al. Early scanning of nascent polypeptides inside the ribosomal tunnel by NAC. Mol. Cell 2019, 75, 996–1006. [Google Scholar] [CrossRef]
- Gamerdinger, M.; Deuerling, E. Cotranslational sorting and processing of newly synthesized proteins in eukaryotes. Trends Biochem. Sci. 2024, 49, 105–118. [Google Scholar] [CrossRef]
- Forte, G.M.; Pool, M.R.; Stirling, C.J. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 2011, 9, e1001073. [Google Scholar] [CrossRef]
- Aksnes, H.; Van Damme, P.; Goris, M.; Starheim, K.K.; Marie, M.; Stove, S.I.; Hoel, C.; Kalvik, T.V.; Hole, K.; Glomes, N.; et al. An organellar N-acetyltransferase Naa60, acetylates cytosolic N termini of transmembrane proteins and maintains Golgi integrity. Cell Rep. 2015, 10, 1362–1374. [Google Scholar] [CrossRef]
- Aksnes, H.; Drazic, A.; Marie, M.; Arnesen, T. First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem. Sci. 2016, 41, 746–760. [Google Scholar] [CrossRef]
- Knorr, A.G.; Schmidt, C.; Tesina, P.; Berninghausen, O.; Becker, T.; Beatrix, B.; Beckmann, R. Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Nat. Struct. Mol. Biol. 2019, 26, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Tanco, S.; Jonckheere, V.; Tharkeshwar, A.K.; Bogaert, A.; Gevaert, K.; Van Damme, P. Proximal partners of organellar N-terminal acetyltransferase NAA60: Insights into Golgi structure and transmembrane topology. Open Biol. 2025, 15, 240225. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Stutz, R.; Schorr, S.; Lang, S.; Pfeffer, S.; Freeze, H.F.; Förster, F.; Helms, V.; Dudek, J.; Zimmermann, R. Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat. Commun. 2018, 9, 37639. [Google Scholar] [CrossRef]
- Klein, M.-C.; Lerner, M.; Nguyen, D.; Pfeffer, S.; Dudek, J.; Förster, F.; Helms, V.; Lang, S.; Zimmermann, R. TRAM1 protein may support ER protein import by modulating the phospholipid bilayer near the lateral gate of the Sec61 channel. Channels 2020, 14, 28–44. [Google Scholar] [CrossRef] [PubMed]
- High, S.; Martoglio, B.; Görlich, D.; Andersen, S.S.L.; Ashford, A.A.; Giner, A.; Hartmann, E.; Prehn, S.; Rapoport, T.A.; Dobberstein, B.; et al. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J. Biol. Chem. 1993, 268, 26745–26751. [Google Scholar] [CrossRef] [PubMed]
- Hegde, R.S.; Voigt, S.; Rapoport, T.A.; Lingappa, V.R. TRAM regulates the exposure of nascent secretory proteins to the cytosol during translocation into the endoplasmic reticulum. Cell 1998, 92, 621–631. [Google Scholar] [CrossRef]
- Voigt, S.; Jungnickel, B.; Hartmann, E.; Rapoport, T.A. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 1996, 134, 25–35. [Google Scholar] [CrossRef]
- Sauri, A.; McCormick, P.J.; Johnson, A.E.; Mingarro, I. Sec61alpha and TRAM are sequentially adjacent to a nascent viral membrane protein during its ER integration. J. Mol. Biol. 2007, 366, 366–374. [Google Scholar] [CrossRef]
- Itskanov, S.; Park, E. Structure of the posttranslational Sec protein-translocation channel complex from yeast. Science 2019, 363, 84–87. [Google Scholar] [CrossRef]
- Wu, X.; Cabanos, C.; Rapoport, T.A. Structure of the post-translational protein translocation machinery of the ER membrane. Nature 2019, 566, 136–139. [Google Scholar] [CrossRef]
- Itskanov, S.; Kuo, K.M.; Gumbart, J.C.; Park, E. Stepwise gating of the Sec61 protein-conducting channel by Sec62 and Sec63. Nat. Struct. Mol. Biol. 2021, 28, 162–172. [Google Scholar] [CrossRef]
- Weng, T.-H.; Steinchen, W.; Beatrix, B.; Berninghausen, O.; Becker, T.; Bange, G.; Cheng, J.; Beckmann, R. Architecture of the active post-translational SEC translocon. EMBO J. 2021, 40, e105643. [Google Scholar] [CrossRef] [PubMed]
- Dudek, J.; Volkmer, J.; Bies, C.; Guth, S.; Müller, A.; Lerner, M.; Feick, P.; Schäfer, K.H.; Morgenstern, E.; Hennessy, F.; et al. A novel type of cochaperone mediates transmembrane recruitment of DnaK-like chaperones to ribosomes. EMBO J. 2002, 21, 2958–2967. [Google Scholar] [CrossRef]
- Dudek, J.; Greiner, M.; Müller, A.; Hendershot, L.M.; Kopsch, K.; Nastainczyk, W.; Zimmermann, R. ERj1p plays a basic role in protein biogenesis at the endoplasmic reticulum. Nat. Struct. Mol. Biol. 2005, 12, 1008–1014. [Google Scholar] [CrossRef]
- Blau, M.; Mullapudi, S.; Becker, T.; Dudek, J.; Zimmermann, R.; Penczek, P.A.; Beckmann, R. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat. Struct. Mol. Biol. 2005, 12, 1015–1016. [Google Scholar] [CrossRef] [PubMed]
- Benedix, J.; Lajoie, P.; Jaiswal, H.; Burgard, C.; Greiner, M.; Zimmermann, R.; Rospert, S.; Snapp, E.L.; Dudek, J. BiP modulates the affinity of its co-chaperone ERj1 to ribosomes. J. Biol. Chem. 2010, 285, 36427–36433. [Google Scholar] [CrossRef]
- Song, J.; Mizrak, A.; Lee, C.W.; Cicconet, M.; Lai, Z.W.; Tang, W.-C.; Mohr, S.E.; Farese, R.V., Jr.; Walther, T.C. Identification of two pathways mediating protein targeting from ER to lipid droplets. Nat. Cell Biol. 2022, 24, 1364–1377. [Google Scholar] [CrossRef]
- Voeltz, G.K.; Prinz, W.A.; Shibata, Y.; Rist, J.M.; Rapoport, T.A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 2006, 124, 573–586. [Google Scholar] [CrossRef]
- Allen, K.N.; Entova, S.; Ray, L.C.; Imperiali, B. Monotopic membrane proteins join the fold. Trends Biochem. Sci. 2019, 44, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.-J.; Mukherjee, S.; Langer, J.D.; Hummer, G.; McDowell, M.A. SND3 is the membrane insertase within a fungal multipass translocon. bioRxiv 2025. [Google Scholar] [CrossRef]
- Javaneinen, M.; Simek, J.; Tranter, D.; O’Keefe, S.; Karki, S.; Biriukov, D.; Sachl, R.; Paavvilainen, V.O. Lipid scrambling pathways in the Sec61 translocon complex. J. Am. Chem. Soc. 2025, 147, 15970–15984. [Google Scholar] [CrossRef] [PubMed]
- Pool, M. Targeting of proteins for translocation at the endoplasmic reticulum. Int. J. Mol. Sci. 2022, 23, 3773. [Google Scholar] [CrossRef]
- Hsieh, H.-H.; Shan, S.-O. Fidelity of cotranslational protein targeting to the endoplasmic reticulum. Int. J. Mol. Sci. 2022, 23, 281. [Google Scholar] [CrossRef]
- Bhadra, P.; Helms, V. Molecular modeling of signal peptide recognition by eukaryotic Sec complexes. Int. J. Mol. Sci. 2021, 22, 10705. [Google Scholar] [CrossRef]
- Liaci, A.M.; Förster, F. Take me home, protein roads: Structural insigths into signal peptide interactions during ER translocation. Int. J. Mol. Sci. 2021, 22, 11871. [Google Scholar] [CrossRef]
- Jung, S.-j.; Kim, H. Emerging view on the molecular functions of Sec62 and Sec63 in protein translocation. Int. J. Mol. Sci. 2021, 22, 12757. [Google Scholar] [CrossRef]
- Tirincsi, A.; Sicking, M.; Hadzibeganovic, D.; Haßdenteufel, S.; Lang, S. The molecular biodiversity of protein targeting and protein transport related to the endoplasmic reticulum. Int. J. Mol. Sci. 2021, 23, 143. [Google Scholar] [CrossRef]
- Whitley, P.; Grau, B.; Gumbart, J.C.; Martinez-Gil, L.; Mingarro, I. Folding and insertion of transmembrane helices at the ER. Int. J. Mol. Sci. 2021, 22, 12778. [Google Scholar] [CrossRef]
- Sanchez, W.N.; Driessen, A.J.M.; Wilson, C.A.M. Protein targeting to the ER membrane: Multiple pathways and shared machinery. Crit. Rev. Biochem. Mol. Biol. 2025, 60, 33–79. [Google Scholar] [CrossRef]
- Gruss, O.J.; Feick, P.; Frank, R.; Dobberstein, B. Phsophorylation of components of the ER translocation site. Eur. J. Biochem. 1999, 260, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Faust, M.; Jung, M.; Günther, J.; Zimmermann, R.; Montenarh, M. Localization of individual subunits of protein kinase CK2 to the endoplasmic reticulum and to the Golgi apparatus. Mol. Cell. Biochem. 2001, 227, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Götz, C.; Müller, A.; Montenarh, M.; Zimmermann, R.; Dudek, J. ERj1 is a substrate of phosphorylation by CK2. Biochem. Biophys. Res. Commun. 2009, 388, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Ampofo, E.; Welker, S.; Jung, M.; Müller, L.; Greiner, M.; Zimmermann, R.; Montenarh, M. CK2 phosphorylation of human Sec63 regulates its interaction with Sec62. Biochim. Biophys. Acta 2013, 1830, 2938–2945. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, A.; Bai, Y.; Chen, Y.; Cates, K.; Kerr, C.; Bermudez, A.; Susanto, T.T.; Wysong, K.; Marquez, F.J.G.; et al. A subcellular map of translational machinery composition and regulation at the single-molecule level. Science 2015, 387, eadn2623. [Google Scholar] [CrossRef]
- Garrison, J.L.; Kunkel, E.J.; Hegde, R.S.; Taunton, J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature 2005, 436, 285–289. [Google Scholar] [CrossRef]
- Besemer, J.; Harent, H.; Wang, S.; Oberhauser, B.; Marquardt, K.; Foster, C.A.; Schreiner, E.P.; de Vries, J.E.; Dascher-Nadel, C.; Lindley, I.J.D. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature 2005, 436, 290–293. [Google Scholar] [CrossRef]
- Cross, B.C.S.; McKibbin, C.; Callan, A.C.; Roboti, P.; Piacenti, M.; Rabu, C.; Wilson, C.M.; Whitehead, R.; Flitsch, S.L.; Pool, M.R.; et al. Eeyarestatin I inhibits Sec61-mediated protein translocation at the endoplasmic reticulum. J. Cell Sci. 2009, 122, 4393–4400. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.S.; Hill, K.; McKenna, M.; Ogbechi, J.; High, S.; Willis, A.E.; Simmonds, R.E. The pathogenic mechanism of the Mycobacterium ulcerans virulence factor, Mycolactone, depends on blockade of protein translocation into the ER. PloS Pathog. 2014, 10, e1004061. [Google Scholar] [CrossRef] [PubMed]
- MacKinnon, A.L.; Paavilainen, V.O.; Sharma, A.; Hegde, R.S.; Taunton, J. An allosteric Sec61 inhibitor traps nascent transmembrane helices at the lateral gate. eLife 2014, 3, e01483. [Google Scholar] [CrossRef] [PubMed]
- Paatero, A.O.; Kellosalo, J.; Dunyak, B.M.; Almaliti, J.; Gestwicki, J.E.; Gerwick, W.H.; Taunton, J.; Paavilainen, V.O. Apratoxin kills cells by direct blockade of the Sec61 protein translocation channel. Cell Chem. Biol. 2016, 23, 561–566. [Google Scholar] [CrossRef]
- Baron, L.; Paatero, A.O.; Morel, J.-D.; Impens, F.; Guenin-Macé, L.; Saint-Auret, S.; Blanchard, N.; Dillmann, R.; Niang, F.; Pellegrini, S.; et al. Maycolactone subervts immunity by selectively blocking the Sec61 translocon. J. Exp. Med. 2016, 213, 2885–2896. [Google Scholar] [CrossRef]
- Pauwels, E.; Schülein, R.; Vermeire, K. Inhibitors of the Sec61 complex and novel high throughput screening strategies to target the protein translocation pathway. Int. J. Mol. Sci. 2021, 22, 12007. [Google Scholar] [CrossRef]
- Pauwels, E.; Provinciael, B.; Camps, A.; Hartmann, E.; Vermeire, K. Reduced DNAJC3 expression affects protein translocation across the ER membrane and attenuates the down-modulating effect of the translocation inhibitor cyclotriazadisulfonamide. Int. J. Mol. Sci. 2022, 23, 584. [Google Scholar] [CrossRef]
- Zimmermann, J.S.M.; Linxweiler, J.; Radosa, J.; Linxweiler, M.; Zimmermann, R. The ER membrane protein Sec62 as potential therapeutic target in SEC62 overexpressing tumors. Front. Physiol. 2022, 13, 1014271. [Google Scholar] [CrossRef]
- Aksnes, H.; Ree, R.; Arnesen, T. Co-translational, post-translational, and non-catalytic roles of N-acetyltransferases. Mol. Cell 2019, 73, 1098–1114. [Google Scholar] [CrossRef]
- Lentzsch, A.M.; Yudin, D.; Gamerdinger, M.; Chandrasekar, S.; Rabl, L.; Scaiola, A.; Deuerling, E.; Ban, N.; Shan, S.-O. NAC guides a ribosomal multienzyme complex for nascent protein processing. Nature 2024, 633, 718–724. [Google Scholar] [CrossRef]
- Pfeiffer, N.V.; Dirndorfer, D.; Lang, S.; Resenberger, U.K.; Restelli, L.M.; Hemion, C.; Miesbauer, M.; Frank, S.; Neutzner, A.; Zimmermann, R.; et al. Structural features within the nascent chain regulate alternative targeting of secretory proteins to mitochondria. EMBO J. 2013, 32, 1036–1051. [Google Scholar] [CrossRef]
- Mick, D.U.; Rodriguez, R.B.; Leib, R.D.; Adams, C.M.; Chien, A.S.; Gygi, S.P.; Nachuri, M.V. Proteomics of primary cilia by proximity labeling. Dev. Cell 2015, 35, 497–512. [Google Scholar] [CrossRef]
- May, E.A.; Kalocsay, M.; D’Auriac, I.G.; Schuster, P.S.; Gygi, S.P.; Nachury, M.V.; Mick, D.U. Time-resolved proteomics profiling of the ciliary hedgehog response. J. Cell Biol. 2021, 220, e202007207. [Google Scholar] [CrossRef] [PubMed]
- Go, C.D.; Knight, J.D.R.; Rajasekharan, A.; Rathod, B.; Hesketh, G.G.; Abe, K.T.; Youn, J.-Y.; Samavarchi-Tehrani, P.; Zhang, H.; Zhu, L.Y.; et al. A proximity-dependent biotinylation map of a human cell. Nature 2021, 595, 120–124, Correction in: Nature 2022, 602, E16. [Google Scholar] [CrossRef] [PubMed]
- Sicking, M.; Jung, M.; Lang, S. Lights, camera, interaction: Studying protein-protein interactions of the ER protein translocase in living cells. Int. J. Mol. Sci. 2021, 22, 10358. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zimmermann, R. Rules of Engagement for Components of Membrane Protein Biogenesis at the Human Endoplasmic Reticulum. Int. J. Mol. Sci. 2025, 26, 8823. https://doi.org/10.3390/ijms26188823
Zimmermann R. Rules of Engagement for Components of Membrane Protein Biogenesis at the Human Endoplasmic Reticulum. International Journal of Molecular Sciences. 2025; 26(18):8823. https://doi.org/10.3390/ijms26188823
Chicago/Turabian StyleZimmermann, Richard. 2025. "Rules of Engagement for Components of Membrane Protein Biogenesis at the Human Endoplasmic Reticulum" International Journal of Molecular Sciences 26, no. 18: 8823. https://doi.org/10.3390/ijms26188823
APA StyleZimmermann, R. (2025). Rules of Engagement for Components of Membrane Protein Biogenesis at the Human Endoplasmic Reticulum. International Journal of Molecular Sciences, 26(18), 8823. https://doi.org/10.3390/ijms26188823





