Orphan Three-Finger Toxins from Snake Venoms: Unexplored Library of Novel Biological Ligands with Potential New Structures and Functions
Abstract
1. Introduction
2. De-Orphaned 3FTxs
2.1. Orphan Group I: Righalexin-Related Toxins (Extrinsic Tenase Inhibitors—Exins)
2.2. Orphan Group II (Antagonists of nAChRs and mAChRs)
2.3. Orphan Group III: Bucain-Related Toxins
2.4. Orphan Group IV: Candoxin-Related Toxins (Antagonists of nAChRs)
2.5. Orphan Group V: γ-Bungaratoxin-Related Toxins
2.6. Orphan Group VIII: Haditoxin-Related Toxins (Dimeric Antagonists of nAChRs)
2.7. Orphan Group XV: Cytotoxin-A5- and μ-EPTX-Na1a-Related Toxins (Integrin/Sodium Channel Inhibitors)
2.8. Orphan Group XVII: BM8/BM14/Bulongin-Related Toxins (Antagonists of mAChRs and nAChRs)
2.9. Orphan Group XIX: Bucandin/Actiflagelin-Related Toxins
2.10. Orphan Group XX
2.11. Other Orphan Groups
3. Novel Classes of 3FTxs
3.1. Colubrid 3FTxs
3.1.1. Denmotoxin
3.1.2. Irditoxin
3.2. Adrenoceptor Inhibitors
3.2.1. β-Cardiotoxin
3.2.2. AdTx1 (ρ-Da1a)
3.2.3. ρ-Da1b
3.2.4. α-Adrenergic/Muscarinic/Dopaminergic/Aminergic Toxins
3.3. Viperid 3FTxs
3.4. Acid-Sensing Ion Channels (ASICs) and Pain-Modifying Toxins
3.5. GABAA Receptor Modulators
3.6. Ω-Neurotoxins, a New Class of nAChR Antagonists
3.7. δ-Elapitoxins (Calliotoxin and Related Sodium Channel Activators)
3.8. Potassium Channel Activator
3.9. αδ-Bungarotoxins
3.10. Σ-Neurotoxins
4. Omics Technologies in the Discovery of 3FTxs
5. Identification of New Orphan 3FTxs
5.1. N-Terminal Variants
5.2. Disulfide Bridge Variants
5.3. Additional Spare Cys Residues
6. Fascinating Three-Ring Circus in 3FTxs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kini, R.M. Molecular moulds with multiple missions: Functional sites in three-finger toxins. Clin. Exp. Pharmacol. Physiol. 2002, 29, 815–822. [Google Scholar] [CrossRef]
- Fry, B.G.; Wuster, W.; Kini, R.M.; Brusic, V.; Khan, A.; Venkataraman, D.; Rooney, A.P. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 2003, 57, 110–129. [Google Scholar] [CrossRef]
- Kini, R.M.; Doley, R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Nirthanan, S.; Gwee, M.C. Three-finger alpha-neurotoxins and the nicotinic acetylcholine receptor, forty years on. J. Pharmacol. Sci. 2004, 94, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kessler, P.; Marchot, P.; Silva, M.; Servent, D. The three-finger toxin fold: A multifunctional structural scaffold able to modulate cholinergic functions. J. Neurochem. 2017, 142 (Suppl. S2), 7–18. [Google Scholar] [CrossRef]
- Servent, D.; Fruchart-Gaillard, C. Muscarinic toxins: Tools for the study of the pharmacological and functional properties of muscarinic receptors. J. Neurochem. 2009, 109, 1193–1202. [Google Scholar] [CrossRef]
- Blanchet, G.; Collet, G.; Mourier, G.; Gilles, N.; Fruchart-Gaillard, C.; Marcon, E.; Servent, D. Polypharmacology profiles and phylogenetic analysis of three-finger toxins from mamba venom: Case of aminergic toxins. Biochimie 2014, 103, 109–117. [Google Scholar] [CrossRef]
- Shi, M.; Zhang, S.; Zhu, A.; Toyoda, Y.; Kong, F.; Sun, X.; Xu, X.; Yan, C.; Liu, X. Molecular mechanism of human alpha(1A)-adrenoceptor inhibition by Mamba snake toxin AdTx1. Commun. Biol. 2025, 8, 1055. [Google Scholar] [CrossRef]
- Zhong, Y.; Tao, H.; Zhang, Y.; He, B.; Jiao, H.; Wang, D.; Teng, M.; Hu, H.; Guo, Q.; Tao, Y. Structure-guided engineering of snake toxins for selective modulation of adrenergic and muscarinic receptors. Nat. Commun. 2025, 16, 6478. [Google Scholar] [CrossRef]
- Maeda, S.; Xu, J.; FM, N.K.; Clark, M.J.; Zhao, J.; Tsutsumi, N.; Aoki, J.; Sunahara, R.K.; Inoue, A.; Garcia, K.C.; et al. Structure and selectivity engineering of the M(1) muscarinic receptor toxin complex. Science 2020, 369, 161–167. [Google Scholar] [CrossRef]
- De Weille, J.R.; Schweitz, H.; Maes, P.; Tartar, A.; Lazdunski, M. Calciseptine, a peptide isolated from black mamba venom, is a specific blocker of the L-type calcium channel. Proc. Natl. Acad. Sci. USA 1991, 88, 2437–2440. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Yao, X.; Chen, J.; Huang, G.; Fan, X.; Xue, L.; Li, Z.; Wu, T.; Zheng, Y.; Huang, J.; et al. Structural basis for human Ca(v)1.2 inhibition by multiple drugs and the neurotoxin calciseptine. Cell 2023, 186, 5363–5374.e16. [Google Scholar] [CrossRef] [PubMed]
- Diochot, S.; Baron, A.; Salinas, M.; Douguet, D.; Scarzello, S.; Dabert-Gay, A.S.; Debayle, D.; Friend, V.; Alloui, A.; Lazdunski, M.; et al. Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature 2012, 490, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Liu, S.; Li, S.; Zhang, M.; Yang, F.; Wen, M.; Shi, P.; Wang, T.; Pan, M.; Chang, S.; et al. Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1. eLife 2020, 9, e57096. [Google Scholar] [CrossRef]
- Rosso, J.P.; Schwarz, J.R.; Diaz-Bustamante, M.; Ceard, B.; Gutierrez, J.M.; Kneussel, M.; Pongs, O.; Bosmans, F.; Bougis, P.E. MmTX1 and MmTX2 from coral snake venom potently modulate GABAA receptor activity. Proc. Natl. Acad. Sci. USA 2015, 112, E891–E900. [Google Scholar]
- McDowell, R.S.; Dennis, M.S.; Louie, A.; Shuster, M.; Mulkerrin, M.G.; Lazarus, R.A. Mambin, a potent glycoprotein IIb-IIIa antagonist and platelet aggregation inhibitor structurally related to the short neurotoxins. Biochemistry 1992, 31, 4766–4772. [Google Scholar] [CrossRef]
- Shiu, J.H.; Chen, C.Y.; Chang, L.S.; Chen, Y.C.; Chen, Y.C.; Lo, Y.H.; Liu, Y.C.; Chuang, W.J. Solution structure of gamma-bungarotoxin: The functional significance of amino acid residues flanking the RGD motif in integrin binding. Proteins 2004, 57, 839–849. [Google Scholar]
- Sutcliffe, M.J.; Jaseja, M.; Hyde, E.I.; Lu, X.; Williams, J.A. Three-dimensional structure of the RGD-containing neurotoxin homologue dendroaspin. Nat. Struct. Biol. 1994, 1, 802–807. [Google Scholar] [CrossRef]
- Banerjee, Y.; Mizuguchi, J.; Iwanaga, S.; Kini, R.M. Hemextin AB complex--a snake venom anticoagulant protein complex that inhibits factor VIIa activity. Pathophysiol. Haemost. Thromb. 2005, 34, 184–187. [Google Scholar]
- Choudhury, M.; McCleary, R.J.R.; Kini, R.M.; Velmurugan, D. Orphan Three-Finger Toxins Bind at Tissue Factor-Factor VIIa Interface to Inhibit Factor X Activation: Identification of Functional Site by Docking. TH Open 2018, 2, e303–e314. [Google Scholar] [CrossRef][Green Version]
- Anderson, A.J.; Harvey, A.L.; Mbugua, P.M. Effects of fasciculin 2, an anticholinesterase polypeptide from green mamba venom, on neuromuscular transmission in mouse diaphragm preparations. Neurosci. Lett. 1985, 54, 123–128. [Google Scholar] [CrossRef]
- Cheung, J.; Rudolph, M.J.; Burshteyn, F.; Cassidy, M.S.; Gary, E.N.; Love, J.; Franklin, M.C.; Height, J.J. Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J. Med. Chem. 2012, 55, 10282–10286. [Google Scholar] [CrossRef]
- Gasanov, S.E.; Dagda, R.K.; Rael, E.D. Snake Venom Cytotoxins, Phospholipase A(2)s, and Zn(2+)-dependent Metalloproteinases: Mechanisms of Action and Pharmacological Relevance. J. Clin. Toxicol. 2014, 4, 1000181. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Folch, B.; Letourneau, M.; Vaudry, D.; Truong, N.H.; Doucet, N.; Chatenet, D.; Fournier, A. Cardiotoxin-I: An unexpectedly potent insulinotropic agent. Chembiochem 2012, 13, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Aziz, T.M.; Al Khoury, S.; Jaquillard, L.; Triquigneaux, M.; Martinez, G.; Bourgoin-Voillard, S.; Seve, M.; Arnoult, C.; Beroud, R.; De Waard, M. Actiflagelin, a new sperm activator isolated from Walterinnesia aegyptia venom using phenotypic screening. J. Venom. Anim. Toxins Incl. Trop. Dis. 2018, 24, 2. [Google Scholar] [CrossRef]
- Lewis, R.J.; Garcia, M.L. Therapeutic potential of venom peptides. Nat. Rev. Drug Discov. 2003, 2, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.Y.; Kini, R.M. Exogenous Factors from Venomous and Hematophagous Animals in Drugs and Diagnostic Developments for Cardiovascular and Neurovascular Diseases. Cardiovasc. Hematol. Disord. Drug Targets 2019, 19, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M.; Koh, C.Y. Snake venom three-finger toxins and their potential in drug development targeting cardiovascular diseases. Biochem. Pharmacol. 2020, 181, 114105. [Google Scholar] [CrossRef]
- Osipov, A.V.; Kasheverov, I.E.; Makarova, Y.V.; Starkov, V.G.; Vorontsova, O.V.; Ziganshin, R.; Andreeva, T.V.; Serebryakova, M.V.; Benoit, A.; Hogg, R.C.; et al. Naturally occurring disulfide-bound dimers of three-fingered toxins: A paradigm for biological activity diversification. J. Biol. Chem. 2008, 283, 14571–14580. [Google Scholar] [CrossRef]
- Osipov, A.V.; Rucktooa, P.; Kasheverov, I.E.; Filkin, S.Y.; Starkov, V.G.; Andreeva, T.V.; Sixma, T.K.; Bertrand, D.; Utkin, Y.N.; Tsetlin, V.I. Dimeric alpha-cobratoxin X-ray structure: Localization of intermolecular disulfides and possible mode of binding to nicotinic acetylcholine receptors. J. Biol. Chem. 2012, 287, 6725–6734. [Google Scholar] [CrossRef]
- Pawlak, J.; Mackessy, S.P.; Sixberry, N.M.; Stura, E.A.; Le Du, M.H.; Menez, R.; Foo, C.S.; Menez, A.; Nirthanan, S.; Kini, R.M. Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J. 2009, 23, 534–545. [Google Scholar] [CrossRef]
- Roy, A.; Zhou, X.; Chong, M.Z.; D’Hoedt, D.; Foo, C.S.; Rajagopalan, N.; Nirthanan, S.; Bertrand, D.; Sivaraman, J.; Kini, R.M. Structural and functional characterization of a novel homodimeric three-finger neurotoxin from the venom of Ophiophagus hannah (king cobra). J. Biol. Chem. 2010, 285, 8302–8315. [Google Scholar] [CrossRef]
- Foo, C.S.; Jobichen, C.; Hassan-Puttaswamy, V.; Dekan, Z.; Tae, H.S.; Bertrand, D.; Adams, D.J.; Alewood, P.F.; Sivaraman, J.; Nirthanan, S.; et al. Fulditoxin, representing a new class of dimeric snake toxins, defines novel pharmacology at nicotinic ACh receptors. Br. J. Pharmacol. 2020, 177, 1822–1840. [Google Scholar] [CrossRef]
- Nirthanan, S.; Charpantier, E.; Gopalakrishnakone, P.; Gwee, M.C.; Khoo, H.E.; Cheah, L.S.; Kini, R.M.; Bertrand, D. Neuromuscular effects of candoxin, a novel toxin from the venom of the Malayan krait (Bungarus candidus). Br. J. Pharmacol. 2003, 139, 832–844. [Google Scholar] [CrossRef]
- Hassan-Puttaswamy, V.; Adams, D.J.; Kini, R.M. A Distinct Functional Site in Omega-Neurotoxins: Novel Antagonists of Nicotinic Acetylcholine Receptors from Snake Venom. ACS Chem. Biol. 2015, 10, 2805–2815. [Google Scholar] [CrossRef]
- Kini, R.M.; Koh, C.Y. Variations in “Functional Site” Residues and Classification of Three-Finger Neurotoxins in Snake Venoms. Toxins 2025, 17, 364. [Google Scholar] [CrossRef] [PubMed]
- Barnwal, B.; Jobichen, C.; Girish, V.M.; Foo, C.S.; Sivaraman, J.; Kini, R.M. Ringhalexin from Hemachatus haemachatus: A novel inhibitor of extrinsic tenase complex. Sci. Rep. 2016, 6, 25935. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, M.; McCleary, R.J.R.; Kesherwani, M.; Kini, R.M.; Velmurugan, D. Comparison of proteomic profiles of the venoms of two of the ‘Big Four’ snakes of India, the Indian cobra (Naja naja) and the common krait (Bungarus caeruleus), and analyses of their toxins. Toxicon 2017, 135, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.; Kerkkamp, H.M.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef]
- Utkin, Y.N.; Kukhtina, V.V.; Kryukova, E.V.; Chiodini, F.; Bertrand, D.; Methfessel, C.; Tsetlin, V.I. “Weak toxin” from Naja kaouthia is a nontoxic antagonist of alpha 7 and muscle-type nicotinic acetylcholine receptors. J. Biol. Chem. 2001, 276, 15810–15815. [Google Scholar] [CrossRef]
- Utkin, Y.N.; Kukhtina, V.V.; Maslennikov, I.V.; Eletsky, A.V.; Starkov, V.G.; Weise, C.; Franke, P.; Hucho, F.; Tsetlin, V.I. First tryptophan-containing weak neurotoxin from cobra venom. Toxicon 2001, 39, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, F.H. Snake venom toxins. The primary structure of protein S4C11. A neurotoxin homologue from the venom of forest cobra (Naja melanoleuca). Biochim. Biophys. Acta 1975, 400, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Joubert, F.J. The purification and amino acid sequence of toxin CM-13b from Naja haje annulifera (Egyptian cobra) venom. Hoppe Seylers Z. Physiol. Chem. 1975, 356, 1901–1908. [Google Scholar] [PubMed]
- Joubert, F.J.; Taljaard, N. Snake venoms. The amino acid sequences of two Melanoleuca-type toxins. Hoppe Seylers Z. Physiol. Chem. 1980, 361, 425–436. [Google Scholar] [CrossRef]
- Shafqat, J.; Siddiqi, A.R.; Zaidi, Z.H.; Jornvall, H. Extensive multiplicity of the miscellaneous type of neurotoxins from the venom of the cobra Naja naja naja and structural characterization of major components. FEBS Lett. 1991, 284, 70–72. [Google Scholar] [CrossRef]
- Mordvintsev, D.Y.; Polyak, Y.L.; Rodionov, D.I.; Jakubik, J.; Dolezal, V.; Karlsson, E.; Tsetlin, V.I.; Utkin, Y.N. Weak toxin WTX from Naja kaouthia cobra venom interacts with both nicotinic and muscarinic acetylcholine receptors. FEBS J. 2009, 276, 5065–5075. [Google Scholar] [CrossRef]
- Lyukmanova, E.N.; Shenkarev, Z.O.; Shulepko, M.A.; Paramonov, A.S.; Chugunov, A.O.; Janickova, H.; Dolejsi, E.; Dolezal, V.; Utkin, Y.N.; Tsetlin, V.I.; et al. Structural Insight into Specificity of Interactions between Nonconventional Three-finger Weak Toxin from Naja kaouthia (WTX) and Muscarinic Acetylcholine Receptors. J. Biol. Chem. 2015, 290, 23616–23630. [Google Scholar] [CrossRef]
- Lyukmanova, E.N.; Shulepko, M.A.; Shenkarev, Z.O.; Kasheverov, I.E.; Chugunov, A.O.; Kulbatskii, D.S.; Myshkin, M.Y.; Utkin, Y.N.; Efremov, R.G.; Tsetlin, V.I.; et al. Central loop of non-conventional toxin WTX from Naja kaouthia is important for interaction with nicotinic acetylcholine receptors. Toxicon 2016, 119, 274–279. [Google Scholar] [CrossRef]
- Murakami, M.T.; Kini, R.M.; Arni, R.K. Crystal structure of bucain, a three-fingered toxin from the venom of the Malayan krait (Bungarus candidus). Protein Pept. Lett. 2009, 16, 1473–1477. [Google Scholar] [CrossRef]
- Nirthanan, S.; Charpantier, E.; Gopalakrishnakone, P.; Gwee, M.C.; Khoo, H.E.; Cheah, L.S.; Bertrand, D.; Kini, R.M. Candoxin, a novel toxin from Bungarus candidus, is a reversible antagonist of muscle (alphabetagammadelta) but a poorly reversible antagonist of neuronal alpha 7 nicotinic acetylcholine receptors. J. Biol. Chem. 2002, 277, 17811–17820. [Google Scholar] [CrossRef]
- V, R.P.; Chary, K.V.R.; Kini, R.M.; Govil, G. Solution structure of candoxin, a novel three-finger toxin from the venom of Bungarus candidus. Arkivoc 2006, 2006, 1–16. [Google Scholar] [CrossRef]
- Pachiappan, A.; Thwin, M.M.; Manikandan, J.; Gopalakrishnakone, P. Glial inflammation and neurodegeneration induced by candoxin, a novel neurotoxin from Bungarus candidus venom: Global gene expression analysis using microarray. Toxicon 2005, 46, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Zhan, C.; Li, B.; Hu, L.; Wei, X.; Feng, L.; Fu, W.; Lu, W. Micelle-based brain-targeted drug delivery enabled by a nicotine acetylcholine receptor ligand. Angew. Chem. Int. Ed. Engl. 2011, 50, 5482–5485. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Gao, J.; Zhan, C.; Xie, C.; Chai, Z.; Ran, D.; Ying, M.; Zheng, P.; Lu, W. Liposome-based glioma targeted drug delivery enabled by stable peptide ligands. J. Control. Release 2015, 218, 13–21. [Google Scholar] [CrossRef]
- Wei, X.; Zhan, C.; Shen, Q.; Fu, W.; Xie, C.; Gao, J.; Peng, C.; Zheng, P.; Lu, W. A D-peptide ligand of nicotine acetylcholine receptors for brain-targeted drug delivery. Angew. Chem. Int. Ed. Engl. 2015, 54, 3023–3027. [Google Scholar]
- Chai, Z.; Hu, X.; Wei, X.; Zhan, C.; Lu, L.; Jiang, K.; Su, B.; Ruan, H.; Ran, D.; Fang, R.H.; et al. A facile approach to functionalizing cell membrane-coated nanoparticles with neurotoxin-derived peptide for brain-targeted drug delivery. J. Control. Release 2017, 264, 102–111. [Google Scholar]
- Nirthanan, S.; Gopalakrishnakone, P.; Gwee, M.C.; Khoo, H.E.; Kini, R.M. Non-conventional toxins from Elapid venoms. Toxicon 2003, 41, 397–407. [Google Scholar] [CrossRef]
- Aird, S.D.; Womble, G.C.; Yates, J.R., 3rd; Griffin, P.R. Primary structure of gamma-bungarotoxin, a new postsynaptic neurotoxin from venom of Bungarus multicinctus. Toxicon 1999, 37, 609–625. [Google Scholar]
- Chen, W.; Yu, H.; Sun, C.; Dong, M.; Zhao, N.; Wang, Y.; Yu, K.; Zhang, J.; Xu, N.; Liu, W. gamma-Bungarotoxin impairs the vascular endothelial barrier function by inhibiting integrin alpha5. Toxicol. Lett. 2023, 383, 177–191. [Google Scholar]
- Pillet, L.; Tremeau, O.; Ducancel, F.; Drevet, P.; Zinn-Justin, S.; Pinkasfeld, S.; Boulain, J.C.; Menez, A. Genetic engineering of snake toxins. Role of invariant residues in the structural and functional properties of a curaremimetic toxin, as probed by site-directed mutagenesis. J. Biol. Chem. 1993, 268, 909–916. [Google Scholar] [CrossRef]
- Antil, S.; Servent, D.; Menez, A. Variability among the sites by which curaremimetic toxins bind to torpedo acetylcholine receptor, as revealed by identification of the functional residues of alpha-cobratoxin. J. Biol. Chem. 1999, 274, 34851–34858. [Google Scholar] [CrossRef]
- Dewan, J.C.; Grant, G.A.; Sacchettini, J.C. Crystal structure of kappa-bungarotoxin at 2.3-A resolution. Biochemistry 1994, 33, 13147–13154. [Google Scholar] [CrossRef]
- Wu, P.L.; Lee, S.C.; Chuang, C.C.; Mori, S.; Akakura, N.; Wu, W.G.; Takada, Y. Non-cytotoxic cobra cardiotoxin A5 binds to alpha(v)beta3 integrin and inhibits bone resorption. Identification of cardiotoxins as non-RGD integrin-binding proteins of the Ly-6 family. J. Biol. Chem. 2006, 281, 7937–7945. [Google Scholar] [CrossRef]
- Teitelbaum, S.L. Bone resorption by osteoclasts. Science 2000, 289, 1504–1508. [Google Scholar] [CrossRef] [PubMed]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Engleman, V.W.; Nickols, G.A.; Ross, F.P.; Horton, M.A.; Griggs, D.W.; Settle, S.L.; Ruminski, P.G.; Teitelbaum, S.L. A peptidomimetic antagonist of the alpha(v)beta3 integrin inhibits bone resorption in vitro and prevents osteoporosis in vivo. J. Clin. Investig. 1997, 99, 2284–2292. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Fisher, J.E.; Gentile, M.; Seedor, J.G.; Leu, C.T.; Rodan, S.B.; Rodan, G.A. The integrin ligand echistatin prevents bone loss in ovariectomized mice and rats. Endocrinology 1998, 139, 1411–1419. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, C.; Xu, X.; Zhang, Y.; Gong, X.; Yang, Z.; Zhang, H.; Tang, D.; Liang, S.; Liu, Z. Naja atra venom peptide reduces pain by selectively blocking the voltage-gated sodium channel Nav1.8. J. Biol. Chem. 2019, 294, 7324–7334. [Google Scholar] [CrossRef]
- Bierhaus, A.; Fleming, T.; Stoyanov, S.; Leffler, A.; Babes, A.; Neacsu, C.; Sauer, S.K.; Eberhardt, M.; Schnolzer, M.; Lasitschka, F.; et al. Methylglyoxal modification of Nav1.8 facilitates nociceptive neuron firing and causes hyperalgesia in diabetic neuropathy. Nat. Med. 2012, 18, 926–933, Erratum in Nat. Med. 2012, 18, 1445. [Google Scholar] [CrossRef]
- Blasius, A.L.; Dubin, A.E.; Petrus, M.J.; Lim, B.K.; Narezkina, A.; Criado, J.R.; Wills, D.N.; Xia, Y.; Moresco, E.M.; Ehlers, C.; et al. Hypermorphic mutation of the voltage-gated sodium channel encoding gene Scn10a causes a dramatic stimulus-dependent neurobehavioral phenotype. Proc. Natl. Acad. Sci. USA 2011, 108, 19413–19418. [Google Scholar]
- Rowe, A.H.; Xiao, Y.; Rowe, M.P.; Cummins, T.R.; Zakon, H.H. Voltage-gated sodium channel in grasshopper mice defends against bark scorpion toxin. Science 2013, 342, 441–446. [Google Scholar] [CrossRef]
- Maeshima, Y.; Manfredi, M.; Reimer, C.; Holthaus, K.A.; Hopfer, H.; Chandamuri, B.R.; Kharbanda, S.; Kalluri, R. Identification of the anti-angiogenic site within vascular basement membrane-derived tumstatin. J. Biol. Chem. 2001, 276, 15240–15248. [Google Scholar] [CrossRef]
- Wang, W.J. Agglucetin, a tetrameric C-type lectin-like venom protein, regulates endothelial cell survival and promotes angiogenesis by activating integrin alphavbeta3 signaling. Biochem. Biophys. Res. Commun. 2008, 369, 753–760. [Google Scholar] [CrossRef]
- Wang, W.J. Acurhagin-C, an ECD disintegrin, inhibits integrin alphavbeta3-mediated human endothelial cell functions by inducing apoptosis via caspase-3 activation. Br. J. Pharmacol. 2010, 160, 1338–1351. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.C.; Fan, C.Y.; Gong, Y.; Yang, S.L. cDNA sequence analysis of a novel member of the three loop protein family from the Chinese continental banded krait. Biosci. Biotechnol. Biochem. 1999, 63, 940–942. [Google Scholar] [CrossRef]
- Chung, C.; Wu, B.N.; Yang, C.C.; Chang, L.S. Muscarinic toxin-like proteins from Taiwan banded krait (Bungarus multicinctus) venom: Purification, characterization and gene organization. Biol. Chem. 2002, 383, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Utkin, Y.N.; Kasheverov, I.E.; Kudryavtsev, D.S.; Andreeva, T.V.; Starkov, V.G.; Ziganshin, R.H.; Kuznetsov, D.V.; Anh, H.N.; Thao, N.T.; Khoa, N.C.; et al. Nonconventional three-finger toxin BMLCL from krait Bungarus multicinctus venom with high affinity interacts with nicotinic acetylcholine receptors. Dokl. Biochem. Biophys. 2015, 464, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, P.; Deacon, A.M.; Comoso, S.; Rajaseger, G.; Kini, R.M.; Uson, I.; Kolatkar, P.R. The atomic resolution structure of bucandin, a novel toxin isolated from the Malayan krait, determined by direct methods. Acta Crystallogr. D Biol. Crystallogr. 2000, 56 Pt 11, 1401–1407. [Google Scholar]
- Torres, A.M.; Kini, R.M.; Selvanayagam, N.; Kuchel, P.W. NMR structure of bucandin, a neurotoxin from the venom of the Malayan krait (Bungarus candidus). Biochem. J. 2001, 360 Pt 3, 539–548. [Google Scholar] [CrossRef]
- Girish, V.M.; Kini, R.M. Exactin: A specific inhibitor of Factor X activation by extrinsic tenase complex from the venom of Hemachatus haemachatus. Sci. Rep. 2016, 6, 32036. [Google Scholar] [CrossRef]
- Osipov, A.V.; Meshcheryakova, A.V.; Starkov, V.G.; Ziganshin, R.K.; Oustitch, T.L.; Peters, L.E.; Tsetlin, V.I.; Utkin, Y.N. New paradoxical three-finger toxin from the cobra Naja kaouthia venom: Isolation and characterization. Dokl. Biochem. Biophys. 2017, 475, 264–266. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Lumsden, N.G.; Wuster, W.; Wickramaratna, J.C.; Hodgson, W.C.; Kini, R.M. Isolation of a neurotoxin (alpha-colubritoxin) from a nonvenomous colubrid: Evidence for early origin of venom in snakes. J. Mol. Evol. 2003, 57, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Wuster, W.; Ryan Ramjan, S.F.; Jackson, T.; Martelli, P.; Kini, R.M. Analysis of Colubroidea snake venoms by liquid chromatography with mass spectrometry: Evolutionary and toxinological implications. Rapid Commun. Mass. Spectrom. 2003, 17, 2047–2062. [Google Scholar] [CrossRef] [PubMed]
- Lumsden, N.G.; Fry, B.G.; Manjunatha Kini, R.; Hodgson, W.C. In vitro neuromuscular activity of ‘colubrid’ venoms: Clinical and evolutionary implications. Toxicon 2004, 43, 819–827. [Google Scholar] [CrossRef]
- Lumsden, N.G.; Fry, B.G.; Ventura, S.; Kini, R.M.; Hodgson, W.C. The in vitro and in vivo pharmacological activity of Boiga dendrophila (mangrove catsnake) venom. Auton. Autacoid Pharmacol. 2004, 24, 107–113. [Google Scholar] [CrossRef]
- Pawlak, J.; Mackessy, S.P.; Fry, B.G.; Bhatia, M.; Mourier, G.; Fruchart-Gaillard, C.; Servent, D.; Menez, R.; Stura, E.; Menez, A.; et al. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity. J. Biol. Chem. 2006, 281, 29030–29041. [Google Scholar] [CrossRef]
- Bourne, Y.; Talley, T.T.; Hansen, S.B.; Taylor, P.; Marchot, P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. EMBO J. 2005, 24, 1512–1522. [Google Scholar]
- Pawlak, J.; Kini, R.M. Unique gene organization of colubrid three-finger toxins: Complete cDNA and gene sequences of denmotoxin, a bird-specific toxin from colubrid snake Boiga dendrophila (Mangrove Catsnake). Biochimie 2008, 90, 868–877. [Google Scholar]
- Pawlak, J.; Manjunatha Kini, R. Snake venom glutaminyl cyclase. Toxicon 2006, 48, 278–286. [Google Scholar] [CrossRef]
- Heyborne, W.H.; Mackessy, S.P. Identification and characterization of a taxon-specific three-finger toxin from the venom of the Green Vinesnake (Oxybelis fulgidus; family Colubridae). Biochimie 2013, 95, 1923–1932. [Google Scholar]
- Modahl, C.M.; Mrinalini; Frietze, S.; Mackessy, S.P. Adaptive evolution of distinct prey-specific toxin genes in rear-fanged snake venom. Proc. Biol. Sci. 2018, 285, 20181003, Erratum in Proc. Biol. Sci. 2018, 285, 20181838. [Google Scholar]
- Dufton, M.J.; Hider, R.C. Structure and pharmacology of elapid cytotoxins. Pharmacol. Ther. 1988, 36, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Endo, T. Structure-function relationships of postsynaptic neurotoxins from snake venoms. Snake Toxins 1991, 165–222. [Google Scholar]
- Alexander, S.P.; Mathie, A.; Peters, J.A. Guide to Receptors and Channels (GRAC), 3rd edition. Br. J. Pharmacol. 2008, 153 (Suppl. S2), S1–S209. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, N.; Pung, Y.F.; Zhu, Y.Z.; Wong, P.T.; Kumar, P.P.; Kini, R.M. Beta-cardiotoxin: A new three-finger toxin from Ophiophagus hannah (king cobra) venom with beta-blocker activity. FASEB J. 2007, 21, 3685–3695. [Google Scholar] [CrossRef]
- Xiang, Y.; Kobilka, B.K. Myocyte adrenoceptor signaling pathways. Science 2003, 300, 1530–1532. [Google Scholar] [CrossRef]
- Hollenberg, N.K. The role of beta-blockers as a cornerstone of cardiovascular therapy. Am. J. Hypertens. 2005, 18 Pt 2, 165S–168S. [Google Scholar] [CrossRef]
- Thom, T.; Haase, N.; Rosamond, W.; Howard, V.J.; Rumsfeld, J.; Manolio, T.; Zheng, Z.J.; Flegal, K.; O’Donnell, C.; Kittner, S.; et al. Heart disease and stroke statistics—2006 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2006, 113, e85–e151. [Google Scholar]
- Quinton, L.; Girard, E.; Maiga, A.; Rekik, M.; Lluel, P.; Masuyer, G.; Larregola, M.; Marquer, C.; Ciolek, J.; Magnin, T.; et al. Isolation and pharmacological characterization of AdTx1, a natural peptide displaying specific insurmountable antagonism of the alpha1A-adrenoceptor. Br. J. Pharmacol. 2010, 159, 316–325. [Google Scholar] [CrossRef]
- Delaflotte, S.; Auguet, M.; Chabrier, P.E. Pharmacological evidence that different alpha 1 adrenoceptor subtypes mediate contraction in rabbit prostate and hypogastric artery. Acta Physiol. Scand. 1996, 158, 241–251. [Google Scholar] [CrossRef]
- Palea, S.; Maiga, A.; Guilloteau, V.; Rekik, M.; Guerard, M.; Rouget, C.; Rischmann, P.; Botto, H.; Camparo, P.; Lluel, P.; et al. Effects of rho-Da1a a peptidic alpha(1) (A)-adrenoceptor antagonist in human isolated prostatic adenoma and anaesthetized rats. Br. J. Pharmacol. 2013, 168, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Noble, A.J.; Chess-Williams, R.; Couldwell, C.; Furukawa, K.; Uchyiuma, T.; Korstanje, C.; Chapple, C.R. The effects of tamsulosin, a high affinity antagonist at functional alpha 1A- and alpha 1D-adrenoceptor subtypes. Br. J. Pharmacol. 1997, 120, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Chapple, C.; Andersson, K.E. Tamsulosin: An overview. World J. Urol. 2002, 19, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Rouget, C.; Quinton, L.; Maiga, A.; Gales, C.; Masuyer, G.; Malosse, C.; Chamot-Rooke, J.; Thai, R.; Mourier, G.; De Pauw, E.; et al. Identification of a novel snake peptide toxin displaying high affinity and antagonist behaviour for the alpha2-adrenoceptors. Br. J. Pharmacol. 2010, 161, 1361–1374. [Google Scholar] [CrossRef]
- Brotchie, J.M. Nondopaminergic mechanisms in levodopa-induced dyskinesia. Mov. Disord. 2005, 20, 919–931. [Google Scholar] [CrossRef]
- Perez, D.M. alpha(1)-Adrenergic Receptors in Neurotransmission, Synaptic Plasticity, and Cognition. Front. Pharmacol. 2020, 11, 581098. [Google Scholar] [CrossRef]
- Rosengren, A.H.; Jokubka, R.; Tojjar, D.; Granhall, C.; Hansson, O.; Li, D.Q.; Nagaraj, V.; Reinbothe, T.M.; Tuncel, J.; Eliasson, L.; et al. Overexpression of alpha2A-adrenergic receptors contributes to type 2 diabetes. Science 2010, 327, 217–220. [Google Scholar] [CrossRef]
- Blandizzi, C. Enteric alpha-2 adrenoceptors: Pathophysiological implications in functional and inflammatory bowel disorders. Neurochem. Int. 2007, 51, 282–288. [Google Scholar] [CrossRef]
- Pang, C.C. Autonomic control of the venous system in health and disease: Effects of drugs. Pharmacol. Ther. 2001, 90, 179–230. [Google Scholar] [CrossRef]
- Woodcock, E.A.; Du, X.J.; Reichelt, M.E.; Graham, R.M. Cardiac alpha 1-adrenergic drive in pathological remodelling. Cardiovasc. Res. 2008, 77, 452–462. [Google Scholar] [CrossRef]
- Bekker, A.; Sturaitis, M.K. Dexmedetomidine for neurological surgery. Neurosurgery 2005, 57 (Suppl. S1), 1–10; discussion 1–10. [Google Scholar]
- Koivula, K.; Rondinelli, S.; Nasman, J. The three-finger toxin MTalpha is a selective alpha(2B)-adrenoceptor antagonist. Toxicon 2010, 56, 440–447. [Google Scholar] [PubMed]
- Nareoja, K.; Kukkonen, J.P.; Rondinelli, S.; Toivola, D.M.; Meriluoto, J.; Nasman, J. Adrenoceptor activity of muscarinic toxins identified from mamba venoms. Br. J. Pharmacol. 2011, 164, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Blanchet, G.; Upert, G.; Mourier, G.; Gilquin, B.; Gilles, N.; Servent, D. New alpha-adrenergic property for synthetic MTbeta and CM-3 three-finger fold toxins from black mamba. Toxicon 2013, 75, 160–167. [Google Scholar] [PubMed]
- Fruchart-Gaillard, C.; Mourier, G.; Blanchet, G.; Vera, L.; Gilles, N.; Menez, R.; Marcon, E.; Stura, E.A.; Servent, D. Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors. PLoS ONE 2012, 7, e39166. [Google Scholar] [CrossRef]
- Pahari, S.; Mackessy, S.P.; Kini, R.M. The venom gland transcriptome of the Desert Massasauga rattlesnake (Sistrurus catenatus edwardsii): Towards an understanding of venom composition among advanced snakes (Superfamily Colubroidea). BMC Mol. Biol. 2007, 8, 115. [Google Scholar] [CrossRef]
- Doley, R.; Pahari, S.; Mackessy, S.P.; Kini, R.M. Accelerated exchange of exon segments in Viperid three-finger toxin genes (Sistrurus catenatus edwardsii; Desert Massasauga). BMC Evol. Biol. 2008, 8, 196. [Google Scholar] [CrossRef]
- Doley, R.; Mackessy, S.P.; Kini, R.M. Role of accelerated segment switch in exons to alter targeting (ASSET) in the molecular evolution of snake venom proteins. BMC Evol. Biol. 2009, 9, 146. [Google Scholar] [CrossRef]
- Junqueira-de-Azevedo, I.L.; Ching, A.T.; Carvalho, E.; Faria, F.; Nishiyama, M.Y., Jr.; Ho, P.L.; Diniz, M.R. Lachesis muta (Viperidae) cDNAs reveal diverging pit viper molecules and scaffolds typical of cobra (Elapidae) venoms: Implications for snake toxin repertoire evolution. Genetics 2006, 173, 877–889. [Google Scholar] [CrossRef]
- Hargreaves, A.D.; Swain, M.T.; Logan, D.W.; Mulley, J.F. Testing the Toxicofera: Comparative transcriptomics casts doubt on the single, early evolution of the reptile venom system. Toxicon 2014, 92, 140–156. [Google Scholar] [CrossRef]
- Amorim, F.G.; Morandi-Filho, R.; Fujimura, P.T.; Ueira-Vieira, C.; Sampaio, S.V. New findings from the first transcriptome of the Bothrops moojeni snake venom gland. Toxicon 2017, 140, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Chijiwa, T.; Oda-Ueda, N.; Nakamura, H.; Yamaguchi, K.; Hattori, S.; Matsubara, K.; Matsuda, Y.; Yamashita, A.; Isomoto, A.; et al. The habu genome reveals accelerated evolution of venom protein genes. Sci. Rep. 2018, 8, 11300. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, E.P.; Rautsaw, R.M.; Strickland, J.L.; Holding, M.L.; Hogan, M.P.; Mason, A.J.; Rokyta, D.R.; Parkinson, C.L. Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Sci. Rep. 2018, 8, 15534. [Google Scholar] [CrossRef] [PubMed]
- Wiezel, G.A.; Shibao, P.Y.T.; Cologna, C.T.; Morandi Filho, R.; Ueira-Vieira, C.; De Pauw, E.; Quinton, L.; Arantes, E.C. In-Depth Venome of the Brazilian Rattlesnake Crotalus durissus terrificus: An Integrative Approach Combining Its Venom Gland Transcriptome and Venom Proteome. J. Proteome Res. 2018, 17, 3941–3958. [Google Scholar] [CrossRef]
- Schield, D.R.; Card, D.C.; Hales, N.R.; Perry, B.W.; Pasquesi, G.M.; Blackmon, H.; Adams, R.H.; Corbin, A.B.; Smith, C.F.; Ramesh, B.; et al. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 2019, 29, 590–601. [Google Scholar] [CrossRef]
- Margres, M.J.; Rautsaw, R.M.; Strickland, J.L.; Mason, A.J.; Schramer, T.D.; Hofmann, E.P.; Stiers, E.; Ellsworth, S.A.; Nystrom, G.S.; Hogan, M.P.; et al. The Tiger Rattlesnake genome reveals a complex genotype underlying a simple venom phenotype. Proc. Natl. Acad. Sci. USA 2021, 118, e2014634118. [Google Scholar] [CrossRef]
- Hogan, M.P.; Holding, M.L.; Nystrom, G.S.; Colston, T.J.; Bartlett, D.A.; Mason, A.J.; Ellsworth, S.A.; Rautsaw, R.M.; Lawrence, K.C.; Strickland, J.L.; et al. The genetic regulatory architecture and epigenomic basis for age-related changes in rattlesnake venom. Proc. Natl. Acad. Sci. USA 2024, 121, e2313440121, Erratum in Proc. Natl. Acad. Sci. USA 2025, 122, e2502598122. [Google Scholar] [CrossRef]
- Deval, E.; Gasull, X.; Noel, J.; Salinas, M.; Baron, A.; Diochot, S.; Lingueglia, E. Acid-sensing ion channels (ASICs): Pharmacology and implication in pain. Pharmacol. Ther. 2010, 128, 549–558. [Google Scholar] [CrossRef]
- Kellenberger, S.; Schild, L. International Union of Basic and Clinical Pharmacology. XCI. structure, function, and pharmacology of acid-sensing ion channels and the epithelial Na+ channel. Pharmacol. Rev. 2015, 67, 1–35. [Google Scholar] [CrossRef]
- Gründer, S. Acid-Sensing Ion Channels. In The Oxford Handbook of Neuronal Ion Channels; Bhattacharjee, A., Ed.; Oxford University Press: Oxford, UK, 2020; pp. 646–695. [Google Scholar]
- Vullo, S.; Kellenberger, S. A molecular view of the function and pharmacology of acid-sensing ion channels. Pharmacol. Res. 2020, 154, 104166. [Google Scholar] [CrossRef]
- Heusser, S.A.; Pless, S.A. Acid-sensing ion channels as potential therapeutic targets. Trends Pharmacol. Sci. 2021, 42, 1035–1050. [Google Scholar] [CrossRef]
- Jones, N.G.; Slater, R.; Cadiou, H.; McNaughton, P.; McMahon, S.B. Acid-induced pain and its modulation in humans. J. Neurosci. 2004, 24, 10974–10979. [Google Scholar] [CrossRef]
- Hung, C.H.; Chin, Y.; Fong, Y.O.; Lee, C.H.; Han, D.S.; Lin, J.H.; Sun, W.H.; Chen, C.C. Acidosis-related pain and its receptors as targets for chronic pain. Pharmacol. Ther. 2023, 247, 108444. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Fu, W.; Vasylyev, D.; Waxman, S.G.; Liu, C.J. Ion channels in osteoarthritis: Emerging roles and potential targets. Nat. Rev. Rheumatol. 2024, 20, 545–564. [Google Scholar] [CrossRef] [PubMed]
- Bohlen, C.J.; Chesler, A.T.; Sharif-Naeini, R.; Medzihradszky, K.F.; Zhou, S.; King, D.; Sanchez, E.E.; Burlingame, A.L.; Basbaum, A.I.; Julius, D. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature 2011, 479, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Baron, A.; Diochot, S.; Salinas, M.; Deval, E.; Noel, J.; Lingueglia, E. Venom toxins in the exploration of molecular, physiological and pathophysiological functions of acid-sensing ion channels. Toxicon 2013, 75, 187–204. [Google Scholar] [CrossRef]
- Verkest, C.; Salinas, M.; Diochot, S.; Deval, E.; Lingueglia, E.; Baron, A. Mechanisms of Action of the Peptide Toxins Targeting Human and Rodent Acid-Sensing Ion Channels and Relevance to Their In Vivo Analgesic Effects. Toxins 2022, 14, 709. [Google Scholar] [CrossRef]
- Diochot, S.; Alloui, A.; Rodrigues, P.; Dauvois, M.; Friend, V.; Aissouni, Y.; Eschalier, A.; Lingueglia, E.; Baron, A. Analgesic effects of mambalgin peptide inhibitors of acid-sensing ion channels in inflammatory and neuropathic pain. Pain 2016, 157, 552–559. [Google Scholar] [CrossRef]
- Mazzuca, M.; Heurteaux, C.; Alloui, A.; Diochot, S.; Baron, A.; Voilley, N.; Blondeau, N.; Escoubas, P.; Gelot, A.; Cupo, A.; et al. A tarantula peptide against pain via ASIC1a channels and opioid mechanisms. Nat. Neurosci. 2007, 10, 943–945. [Google Scholar] [CrossRef]
- Schroeder, C.I.; Rash, L.D.; Vila-Farres, X.; Rosengren, K.J.; Mobli, M.; King, G.F.; Alewood, P.F.; Craik, D.J.; Durek, T. Chemical synthesis, 3D structure, and ASIC binding site of the toxin mambalgin-2. Angew. Chem. Int. Ed. Engl. 2014, 53, 1017–1020. [Google Scholar] [CrossRef]
- Pan, M.; He, Y.; Wen, M.; Wu, F.; Sun, D.; Li, S.; Zhang, L.; Li, Y.; Tian, C. One-pot hydrazide-based native chemical ligation for efficient chemical synthesis and structure determination of toxin Mambalgin-1. Chem. Commun. 2014, 50, 5837–5839. [Google Scholar] [CrossRef] [PubMed]
- Mourier, G.; Salinas, M.; Kessler, P.; Stura, E.A.; Leblanc, M.; Tepshi, L.; Besson, T.; Diochot, S.; Baron, A.; Douguet, D.; et al. Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition. J. Biol. Chem. 2016, 291, 2616–2629. [Google Scholar] [CrossRef]
- Lan, H.; Wu, K.; Zheng, Y.; Pan, M.; Huang, Y.C.; Gao, S.; Zheng, Q.Y.; Zheng, J.S.; Li, Y.M.; Xiao, B.; et al. Total synthesis of mambalgin-1/2/3 by two-segment hydrazide-based native chemical ligation. J. Pept. Sci. 2016, 22, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Yu, Y.; Xue, X.; Pan, M.; Wen, M.; Li, S.; Qu, Q.; Li, X.; Zhang, L.; Li, X.; et al. Cryo-EM structure of the ASIC1a-mambalgin-1 complex reveals that the peptide toxin mambalgin-1 inhibits acid-sensing ion channels through an unusual allosteric effect. Cell Discov. 2018, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Cestele, S.; Qu, Y.; Rogers, J.C.; Rochat, H.; Scheuer, T.; Catterall, W.A. Voltage sensor-trapping: Enhanced activation of sodium channels by beta-scorpion toxin bound to the S3-S4 loop in domain II. Neuron 1998, 21, 919–931. [Google Scholar]
- Catterall, W.A.; Cestele, S.; Yarov-Yarovoy, V.; Yu, F.H.; Konoki, K.; Scheuer, T. Voltage-gated ion channels and gating modifier toxins. Toxicon 2007, 49, 124–141. [Google Scholar] [CrossRef]
- Salinas, M.; Kessler, P.; Douguet, D.; Sarraf, D.; Tonali, N.; Thai, R.; Servent, D.; Lingueglia, E. Mambalgin-1 pain-relieving peptide locks the hinge between alpha4 and alpha5 helices to inhibit rat acid-sensing ion channel 1a. Neuropharmacology 2021, 185, 108453. [Google Scholar] [CrossRef]
- Cristofori-Armstrong, B.; Budusan, E.; Rash, L.D. Mambalgin-3 potentiates human acid-sensing ion channel 1b under mild to moderate acidosis: Implications as an analgesic lead. Proc. Natl. Acad. Sci. USA 2021, 118, e2021581118. [Google Scholar]
- Cristofori-Armstrong, B.; Budusan, E.; Smith, J.J.; Reynaud, S.; Voll, K.; Chassagnon, I.R.; Durek, T.; Rash, L.D. Revealing molecular determinants governing mambalgin-3 pharmacology at acid-sensing ion channel 1 variants. Cell Mol. Life Sci. 2024, 81, 266. [Google Scholar] [CrossRef]
- Jin, C.; Ye, Q.H.; Yuan, F.L.; Gu, Y.L.; Li, J.P.; Shi, Y.H.; Shen, X.M.; Bo, L.; Lin, Z.H. Involvement of acid-sensing ion channel 1alpha in hepatic carcinoma cell migration and invasion. Tumour Biol. 2015, 36, 4309–4317. [Google Scholar] [CrossRef]
- Zhu, S.; Zhou, H.Y.; Deng, S.C.; Deng, S.J.; He, C.; Li, X.; Chen, J.Y.; Jin, Y.; Hu, Z.L.; Wang, F.; et al. ASIC1 and ASIC3 contribute to acidity-induced EMT of pancreatic cancer through activating Ca(2+)/RhoA pathway. Cell Death Dis. 2017, 8, e2806. [Google Scholar] [CrossRef]
- Sheng, Y.; Wu, B.; Leng, T.; Zhu, L.; Xiong, Z. Acid-sensing ion channel 1 (ASIC1) mediates weak acid-induced migration of human malignant glioma cells. Am. J. Cancer Res. 2021, 11, 997–1008. [Google Scholar]
- Bychkov, M.; Shulepko, M.; Osmakov, D.; Andreev, Y.; Sudarikova, A.; Vasileva, V.; Pavlyukov, M.S.; Latyshev, Y.A.; Potapov, A.A.; Kirpichnikov, M.; et al. Mambalgin-2 Induces Cell Cycle Arrest and Apoptosis in Glioma Cells via Interaction with ASIC1a. Cancers 2020, 12, 1837. [Google Scholar] [CrossRef] [PubMed]
- Bychkov, M.L.; Kirichenko, A.V.; Shulepko, M.A.; Mikhaylova, I.N.; Kirpichnikov, M.P.; Lyukmanova, E.N. Mambalgin-2 Inhibits Growth, Migration, and Invasion of Metastatic Melanoma Cells by Targeting the Channels Containing an ASIC1a Subunit Whose Up-Regulation Correlates with Poor Survival Prognosis. Biomedicines 2021, 9, 1324. [Google Scholar] [CrossRef] [PubMed]
- Sudarikova, A.V.; Bychkov, M.L.; Kulbatskii, D.S.; Chubinskiy-Nadezhdin, V.I.; Shlepova, O.V.; Shulepko, M.A.; Koshelev, S.G.; Kirpichnikov, M.P.; Lyukmanova, E.N. Mambalgin-2 Inhibits Lung Adenocarcinoma Growth and Migration by Selective Interaction With ASIC1/alpha-ENaC/gamma-ENaC Heterotrimer. Front. Oncol. 2022, 12, 904742. [Google Scholar] [CrossRef] [PubMed]
- Lyukmanova, E.N.; Zaigraev, M.M.; Kulbatskii, D.S.; Isaev, A.B.; Kukushkin, I.D.; Bychkov, M.L.; Shulepko, M.A.; Chugunov, A.O.; Kirpichnikov, M.P. Molecular Basis for Mambalgin-2 Interaction with Heterotrimeric alpha-ENaC/ASIC1a/gamma-ENaC Channels in Cancer Cells. Toxins 2023, 15, 612. [Google Scholar] [CrossRef]
- Sine, S.M.; Engel, A.G. Recent advances in Cys-loop receptor structure and function. Nature 2006, 440, 448–455. [Google Scholar] [CrossRef]
- D’Hulst, C.; Atack, J.R.; Kooy, R.F. The complexity of the GABAA receptor shapes unique pharmacological profiles. Drug Discov. Today 2009, 14, 866–875. [Google Scholar] [CrossRef]
- Olsen, R.W.; Sieghart, W. GABA A receptors: Subtypes provide diversity of function and pharmacology. Neuropharmacology 2009, 56, 141–148. [Google Scholar] [CrossRef]
- Sigel, E.; Steinmann, M.E. Structure, function, and modulation of GABA(A) receptors. J. Biol. Chem. 2012, 287, 40224–40231. [Google Scholar] [CrossRef]
- Miller, P.S.; Aricescu, A.R. Crystal structure of a human GABAA receptor. Nature 2014, 512, 270–275. [Google Scholar] [CrossRef]
- Ben-Ari, Y.; Khalilov, I.; Kahle, K.T.; Cherubini, E. The GABA excitatory/inhibitory shift in brain maturation and neurological disorders. Neuroscientist 2012, 18, 467–486. [Google Scholar] [CrossRef] [PubMed]
- Hines, R.M.; Davies, P.A.; Moss, S.J.; Maguire, J. Functional regulation of GABAA receptors in nervous system pathologies. Curr. Opin. Neurobiol. 2012, 22, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Schuske, K.; Beg, A.A.; Jorgensen, E.M. The GABA nervous system in C. elegans. Trends Neurosci. 2004, 27, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Mohler, H. GABA(A) receptor diversity and pharmacology. Cell Tissue Res. 2006, 326, 505–516. [Google Scholar] [CrossRef]
- Farrant, M.; Nusser, Z. Variations on an inhibitory theme: Phasic and tonic activation of GABA(A) receptors. Nat. Rev. Neurosci. 2005, 6, 215–229. [Google Scholar] [CrossRef]
- Kudryavtsev, D.S.; Shelukhina, I.V.; Son, L.V.; Ojomoko, L.O.; Kryukova, E.V.; Lyukmanova, E.N.; Zhmak, M.N.; Dolgikh, D.A.; Ivanov, I.A.; Kasheverov, I.E.; et al. Neurotoxins from snake venoms and alpha-conotoxin ImI inhibit functionally active ionotropic gamma-aminobutyric acid (GABA) receptors. J. Biol. Chem. 2015, 290, 22747–22758. [Google Scholar] [CrossRef]
- Kasaragod, V.B.; Mortensen, M.; Hardwick, S.W.; Wahid, A.A.; Dorovykh, V.; Chirgadze, D.Y.; Smart, T.G.; Miller, P.S. Mechanisms of inhibition and activation of extrasynaptic alphabeta GABA(A) receptors. Nature 2022, 602, 529–533. [Google Scholar] [CrossRef]
- Rahman, M.M.; Teng, J.; Worrell, B.T.; Noviello, C.M.; Lee, M.; Karlin, A.; Stowell, M.H.B.; Hibbs, R.E. Structure of the Native Muscle-type Nicotinic Receptor and Inhibition by Snake Venom Toxins. Neuron 2020, 106, 952–962.e955. [Google Scholar] [CrossRef]
- Chang, L.S.; Liou, J.C.; Lin, S.R.; Huang, H.B. Purification and characterization of a neurotoxin from the venom of Ophiophagus hannah (king cobra). Biochem. Biophys. Res. Commun. 2002, 294, 574–578. [Google Scholar] [CrossRef]
- Yang, D.C.; Deuis, J.R.; Dashevsky, D.; Dobson, J.; Jackson, T.N.; Brust, A.; Xie, B.; Koludarov, I.; Debono, J.; Hendrikx, I.; et al. The Snake with the Scorpion’s Sting: Novel Three-Finger Toxin Sodium Channel Activators from the Venom of the Long-Glanded Blue Coral Snake (Calliophis bivirgatus). Toxins 2016, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, C.; Yoshida, H.; Shimazu, T.; Teruuchi, T.; Toriba, M.; Tamiya, N. Studies on the venom components of the long-glanded coral snake, Maticora bivirgata. Toxicon 1991, 29, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Fung, S.Y.; Yap, M.K.; Leong, P.K.; Liew, J.L.; Tan, N.H. Unveiling the elusive and exotic: Venomics of the Malayan blue coral snake (Calliophis bivirgata flaviceps). J. Proteom. 2016, 132, 1–12. [Google Scholar] [CrossRef]
- Dashevsky, D.; Rokyta, D.; Frank, N.; Nouwens, A.; Fry, B.G. Electric Blue: Molecular Evolution of Three-Finger Toxins in the Long-Glanded Coral Snake Species Calliophis bivirgatus. Toxins 2021, 13, 124. [Google Scholar] [CrossRef]
- Palasuberniam, P.; Tan, K.Y.; Tan, C.H. De novo venom gland transcriptomics of Calliophis bivirgata flaviceps: Uncovering the complexity of toxins from the Malayan blue coral snake. J. Venom. Anim. Toxins Incl. Trop. Dis. 2021, 27, e20210024. [Google Scholar] [CrossRef]
- Tan, K.Y.; Liew, J.L.; Tan, N.H.; Quah, E.S.H.; Ismail, A.K.; Tan, C.H. Unlocking the secrets of banded coral snake (Calliophis intestinalis, Malaysia): A venom with proteome novelty, low toxicity and distinct antigenicity. J. Proteom. 2019, 192, 246–257. [Google Scholar] [CrossRef]
- Dashevsky, D.; Deuis, J.R.; Vetter, I.; Huynh, T.; Hodgson, W.C.; Tan, C.H.; Nouwens, A.; Fry, B.G. Novel Neurotoxic Activity in Calliophis intestinalis Venom. Neurotox. Res. 2022, 40, 173–178. [Google Scholar] [CrossRef]
- Maljevic, S.; Lerche, H. Potassium channels: A review of broadening therapeutic possibilities for neurological diseases. J. Neurol. 2013, 260, 2201–2211. [Google Scholar] [CrossRef]
- Du, X.; Gamper, N. Potassium channels in peripheral pain pathways: Expression, function and therapeutic potential. Curr. Neuropharmacol. 2013, 11, 621–640. [Google Scholar] [CrossRef]
- Schmitt, N.; Grunnet, M.; Olesen, S.P. Cardiac potassium channel subtypes: New roles in repolarization and arrhythmia. Physiol. Rev. 2014, 94, 609–653. [Google Scholar] [CrossRef]
- Shah, N.H.; Aizenman, E. Voltage-gated potassium channels at the crossroads of neuronal function, ischemic tolerance, and neurodegeneration. Transl. Stroke Res. 2014, 5, 38–58. [Google Scholar] [PubMed]
- Huang, X.; Jan, L.Y. Targeting potassium channels in cancer. J. Cell Biol. 2014, 206, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, T.; Zagotta, W.N.; Aldrich, R.W. Two types of inactivation in Shaker K+ channels: Effects of alterations in the carboxy-terminal region. Neuron 1991, 7, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Kurata, H.T.; Fedida, D. A structural interpretation of voltage-gated potassium channel inactivation. Prog. Biophys. Mol. Biol. 2006, 92, 185–208. [Google Scholar] [CrossRef]
- Doyle, D.A.; Morais Cabral, J.; Pfuetzner, R.A.; Kuo, A.; Gulbis, J.M.; Cohen, S.L.; Chait, B.T.; MacKinnon, R. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science 1998, 280, 69–77. [Google Scholar] [CrossRef]
- Thompson, A.N.; Posson, D.J.; Parsa, P.V.; Nimigean, C.M. Molecular mechanism of pH sensing in KcsA potassium channels. Proc. Natl. Acad. Sci. USA 2008, 105, 6900–6905. [Google Scholar] [CrossRef]
- Cuello, L.G.; Jogini, V.; Cortes, D.M.; Perozo, E. Structural mechanism of C-type inactivation in K(+) channels. Nature 2010, 466, 203–208. [Google Scholar]
- Rivera-Torres, I.O.; Jin, T.B.; Cadene, M.; Chait, B.T.; Poget, S.F. Discovery and characterisation of a novel toxin from Dendroaspis angusticeps, named Tx7335, that activates the potassium channel KcsA. Sci. Rep. 2016, 6, 23904. [Google Scholar] [CrossRef]
- Utkin, Y.N.; Kuch, U.; Kasheverov, I.E.; Lebedev, D.S.; Cederlund, E.; Molles, B.E.; Polyak, I.; Ivanov, I.A.; Prokopev, N.A.; Ziganshin, R.H.; et al. Novel long-chain neurotoxins from Bungarus candidus distinguish the two binding sites in muscle-type nicotinic acetylcholine receptors. Biochem. J. 2019, 476, 1285–1302. [Google Scholar] [CrossRef]
- Kini, R.M. Toxins for decoding interface selectivity in nicotinic acetylcholine receptors. Biochem. J. 2019, 476, 1515–1520. [Google Scholar] [CrossRef]
- Ackermann, E.J.; Taylor, P. Nonidentity of the alpha-neurotoxin binding sites on the nicotinic acetylcholine receptor revealed by modification in alpha-neurotoxin and receptor structures. Biochemistry 1997, 36, 12836–12844. [Google Scholar] [CrossRef]
- Chan, C.Y.L.; Tae, H.S.; Koh, C.Y.; Adams, D.J.; Kini, R.M. Erabutoxin mutants demonstrate interface selectivity at human fetal and adult muscle-type nicotinic acetylcholine receptors. Biochem. Pharmacol. 2025, 241, 117189. [Google Scholar] [CrossRef]
- Dellisanti, C.D.; Yao, Y.; Stroud, J.C.; Wang, Z.Z.; Chen, L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nat. Neurosci. 2007, 10, 953–962. [Google Scholar] [CrossRef] [PubMed]
- Karlin, A. Emerging structure of the nicotinic acetylcholine receptors. Nat. Rev. Neurosci. 2002, 3, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Patrick, J.; Sequela, P.; Vernino, S.; Amador, M.; Luetje, C.; Dani, J.A. Functional diversity of neuronal nicotinic acetylcholine receptors. Prog. Brain Res. 1993, 98, 113–120. [Google Scholar] [PubMed]
- Tremeau, O.; Lemaire, C.; Drevet, P.; Pinkasfeld, S.; Ducancel, F.; Boulain, J.C.; Menez, A. Genetic engineering of snake toxins. The functional site of Erabutoxin a, as delineated by site-directed mutagenesis, includes variant residues. J. Biol. Chem. 1995, 270, 9362–9369. [Google Scholar]
- Modahl, C.M.; Brahma, R.K.; Koh, C.Y.; Shioi, N.; Kini, R.M. Omics Technologies for Profiling Toxin Diversity and Evolution in Snake Venom: Impacts on the Discovery of Therapeutic and Diagnostic Agents. Annu. Rev. Anim. Biosci. 2020, 8, 91–116. [Google Scholar] [CrossRef]
- Hill, R.E.; Mackessy, S.P. Venom yields from several species of colubrid snakes and differential effects of ketamine. Toxicon 1997, 35, 671–678. [Google Scholar] [CrossRef]
- Neri-Castro, E.; Zarzosa, V.; Benard-Valle, M.; Rodriguez-Solis, A.M.; Hernandez-Orihuela, L.; Ortiz-Medina, J.A.; Alagon, A. Quantifying venom production: A study on Micrurus snakes in Mexico. Toxicon 2024, 240, 107658. [Google Scholar] [CrossRef]
- Durban, J.; Juarez, P.; Angulo, Y.; Lomonte, B.; Flores-Diaz, M.; Alape-Giron, A.; Sasa, M.; Sanz, L.; Gutierrez, J.M.; Dopazo, J.; et al. Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genom. 2011, 12, 259. [Google Scholar] [CrossRef]
- Correa-Netto, C.; Junqueira-de-Azevedo Ide, L.; Silva, D.A.; Ho, P.L.; Leitao-de-Araujo, M.; Alves, M.L.; Sanz, L.; Foguel, D.; Zingali, R.B.; Calvete, J.J. Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J. Proteom. 2011, 74, 1795–1809. [Google Scholar] [CrossRef]
- Aird, S.D.; da Silva, N.J.; Qiu, L.; Villar-Briones, A.; Saddi, V.A.; Pires de Campos Telles, M.; Grau, M.L.; Mikheyev, A.S. Coralsnake Venomics: Analyses of Venom Gland Transcriptomes and Proteomes of Six Brazilian Taxa. Toxins 2017, 9, 187. [Google Scholar] [CrossRef]
- Modahl, C.M.; Saviola, A.J.; Mackessy, S.P. Integration of transcriptomic and proteomic approaches for snake venom profiling. Expert. Rev. Proteom. 2021, 18, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Heyborne, W.H.; Mackessy, S.P. Venoms of New World Vinesnakes (Oxybelis aeneus and O. fulgidus). Toxicon 2021, 190, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Srodawa, K.; Cerda, P.A.; Davis Rabosky, A.R.; Crowe-Riddell, J.M. Evolution of Three-Finger Toxin Genes in Neotropical Colubrine Snakes (Colubridae). Toxins 2023, 15, 523. [Google Scholar] [CrossRef]
- Luddecke, T.; Avella, I.; Damm, M.; Schulte, L.; Eichberg, J.; Hardes, K.; Schiffmann, S.; Henke, M.; Timm, T.; Lochnit, G.; et al. The Toxin Diversity, Cytotoxicity, and Enzymatic Activity of Cape Cobra (Naja nivea) Venom. Toxins 2024, 16, 438. [Google Scholar] [CrossRef]
- Yang, C.; Ding, L.; He, Q.; Chen, X.; Zhu, H.; Chen, F.; Yang, W.; Pan, Y.; Tai, Z.; Zhang, W.; et al. Proteomic Profiling of Venoms from Bungarus suzhenae and B. bungaroides: Enzymatic Activities and Toxicity Assessment. Toxins 2024, 16, 494. [Google Scholar] [CrossRef]
- Redureau, D.; Amorim, F.G.; Crasset, T.; Berger, I.; Schaffitzel, C.; Menzies, S.K.; Casewell, N.R.; Quinton, L. Dual Proteomics Strategies to Dissect and Quantify the Components of Nine Medically Important African Snake Venoms. Toxins 2025, 17, 243. [Google Scholar] [CrossRef]
- Li, Y.; Dai, H.; Zhao, Z.; Ye, R.; Han, S.; Yang, D.; Pan, H.; Lin, J.; Dang, X.; Cheng, Y.; et al. Deciphering the venom components of Sea snakes: Integrating dual proteomic techniques to uncover the bioactive potential of Hydrophis curtus. Toxicon 2025, 264, 108451. [Google Scholar] [CrossRef]
- Zarzosa, V.; Neri-Castro, E.; Lomonte, B.; Fernandez, J.; Rodriguez-Barrera, G.; Rodriguez-Lopez, B.; Rodriguez-Solis, A.M.; Olvera-Rodriguez, A.; Benard-Valle, M.; Saviola, A.; et al. Integrative transcriptomic, proteomic, biochemical and neutralization studies on the venom of Micrurus ephippifer. J. Proteom. 2025, 316, 105416. [Google Scholar] [CrossRef]
- Quinton, L.; Demeure, K.; Dobson, R.; Gilles, N.; Gabelica, V.; De Pauw, E. New method for characterizing highly disulfide-bridged peptides in complex mixtures: Application to toxin identification from crude venoms. J. Proteome Res. 2007, 6, 3216–3223. [Google Scholar] [CrossRef]
- Petras, D.; Heiss, P.; Sussmuth, R.D.; Calvete, J.J. Venom Proteomics of Indonesian King Cobra, Ophiophagus hannah: Integrating Top-Down and Bottom-Up Approaches. J. Proteome Res. 2015, 14, 2539–2556. [Google Scholar] [CrossRef] [PubMed]
- Melani, R.D.; Skinner, O.S.; Fornelli, L.; Domont, G.B.; Compton, P.D.; Kelleher, N.L. Mapping Proteoforms and Protein Complexes From King Cobra Venom Using Both Denaturing and Native Top-down Proteomics. Mol. Cell Proteom. 2016, 15, 2423–2434. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, S.; Petras, D.; Engmark, M.; Sussmuth, R.D.; Whiteley, G.; Albulescu, L.O.; Kazandjian, T.D.; Wagstaff, S.C.; Rowley, P.; Wuster, W.; et al. The medical threat of mamba envenoming in sub-Saharan Africa revealed by genus-wide analysis of venom composition, toxicity and antivenomics profiling of available antivenoms. J. Proteom. 2018, 172, 173–189. [Google Scholar]
- Chapeaurouge, A.; Silva, A.; Carvalho, P.; McCleary, R.J.R.; Modahl, C.M.; Perales, J.; Kini, R.M.; Mackessy, S.P. Proteomic Deep Mining the Venom of the Red-Headed Krait, Bungarus flaviceps. Toxins 2018, 10, 373. [Google Scholar] [CrossRef]
- Hempel, B.F.; Damm, M.; Mrinalini; Gocmen, B.; Karis, M.; Nalbantsoy, A.; Kini, R.M.; Sussmuth, R.D. Extended Snake Venomics by Top-Down In-Source Decay: Investigating the Newly Discovered Anatolian Meadow Viper Subspecies, Vipera anatolica senliki. J. Proteome Res. 2020, 19, 1731–1749. [Google Scholar] [CrossRef]
- Ghezellou, P.; Heiles, S.; Kadesch, P.; Ghassempour, A.; Spengler, B. Venom Gland Mass Spectrometry Imaging of Saw-Scaled Viper, Echis carinatus sochureki, at High Lateral Resolution. J. Am. Soc. Mass. Spectrom. 2021, 32, 1105–1115. [Google Scholar] [CrossRef]
- Kleiz-Ferreira, J.M.; Cirauqui, N.; Trajano, E.A.; Almeida, M.S.; Zingali, R.B. Three-Finger Toxins from Brazilian Coral Snakes: From Molecular Framework to Insights in Biological Function. Toxins 2021, 13, 328. [Google Scholar] [CrossRef]
- Wang, C.R.; Zenaidee, M.A.; Snel, M.F.; Pukala, T.L. Exploring Top-Down Mass Spectrometric Approaches To Probe Forest Cobra (Naja melanoleuca) Venom Proteoforms. J. Proteome Res. 2024, 23, 4601–4613. [Google Scholar] [CrossRef]
- Fernandez, J.; Chaves, W.; Vargas-Diaz, D.; Petras, D.; Lomonte, B. Top-down proteomics of venoms from five Micrurus species from Costa Rica: Comparative composition of phospholipase A(2)-rich vs three-finger toxin-rich phenotypes. Toxicon 2024, 252, 108187. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Tyler, T.J.; Durek, T.; Craik, D.J. Native and Engineered Cyclic Disulfide-Rich Peptides as Drug Leads. Molecules 2023, 28, 3189. [Google Scholar] [CrossRef] [PubMed]
- Rink, R.; Arkema-Meter, A.; Baudoin, I.; Post, E.; Kuipers, A.; Nelemans, S.A.; Akanbi, M.H.; Moll, G.N. To protect peptide pharmaceuticals against peptidases. J. Pharmacol. Toxicol. Methods 2010, 61, 210–218. [Google Scholar] [CrossRef]
- Vanhoof, G.; Goossens, F.; De Meester, I.; Hendriks, D.; Scharpe, S. Proline motifs in peptides and their biological processing. FASEB J. 1995, 9, 736–744. [Google Scholar] [CrossRef]
- Fox, J.W.; Elzinga, M.; Tu, A.T. Amino acid sequence of a snake neurotoxin from the venom of Lapemis hardwickii and the detection of a sulfhydryl group by laser Raman spectroscopy. FEBS Lett. 1977, 80, 217–220. [Google Scholar] [CrossRef]
- Kim, H.S.; Tamiya, N. The amino acid sequence and position of the free thiol group of a short-chain neurotoxin from common-death-adder (Acanthophis antarcticus) venom. Biochem. J. 1981, 199, 211–218. [Google Scholar] [CrossRef]
- Aoki-Shioi, N.; Jobichen, C.; Sivaraman, J.; Kini, R.M. Unusual quaternary structure of a homodimeric synergistic-type toxin from mamba snake venom defines its molecular evolution. Biochem. J. 2020, 477, 3951–3962. [Google Scholar] [CrossRef]
- Gunasekaran, K.; Ramakrishnan, C.; Balaram, P. Beta-hairpins in proteins revisited: Lessons for de novo design. Protein Eng. 1997, 10, 1131–1141. [Google Scholar] [CrossRef]
- Minuchehr, Z.; Goliaei, B. Propensity of amino acids in loop regions connecting beta-strands. Protein Pept. Lett. 2005, 12, 379–382. [Google Scholar] [CrossRef]
- Krieger, F.; Moglich, A.; Kiefhaber, T. Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J. Am. Chem. Soc. 2005, 127, 3346–3352. [Google Scholar] [CrossRef]
- Rahman, S.; Lu, X.; Kakkar, V.V.; Authi, K.S. The integrin alpha IIb beta 3 contains distinct and interacting binding sites for snake-venom RGD (Arg-Gly-Asp) proteins. Evidence that the receptor-binding characteristics of snake-venom RGD proteins are related to the amino acid environment flanking the sequence RGD. Biochem. J. 1995, 312 Pt 1, 223–232. [Google Scholar]
- Kini, R.M.; Caldwell, R.A.; Wu, Q.Y.; Baumgarten, C.M.; Feher, J.J.; Evans, H.J. Flanking proline residues identify the L-type Ca2+ channel binding site of calciseptine and FS2. Biochemistry 1998, 37, 9058–9063. [Google Scholar]
- Kini, R.M. Toxinology provides multidirectional and multidimensional opportunities: A personal perspective. Toxicon X 2020, 6, 100039. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Utkin, Y.N. Specific Amino Acid Residues in the Three Loops of Snake Cytotoxins Determine Their Membrane Activity and Provide a Rationale for a New Classification of These Toxins. Toxins 2024, 16, 262. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lei, X.; Li, A.; Li, J.; Li, S.; Chen, L. Scalable production of recombinant three-finger proteins: From inclusion bodies to high quality molecular probes. Microb. Cell Fact. 2024, 23, 48. [Google Scholar] [PubMed]
- Schroeder, K.S.; Neagle, B.D. FLIPR: A New Instrument for Accurate, High Throughput Optical Screening. J. Biomol. Screen. 1996, 1, 75–80. [Google Scholar]
- Obergrussberger, A.; Rinke-Weiss, I.; Goetze, T.A.; Rapedius, M.; Brinkwirth, N.; Becker, N.; Rotordam, M.G.; Hutchison, L.; Madau, P.; Pau, D.; et al. The suitability of high throughput automated patch clamp for physiological applications. J. Physiol. 2022, 600, 277–297. [Google Scholar] [CrossRef]
- Kaushansky, A.; Allen, J.E.; Gordus, A.; Stiffler, M.A.; Karp, E.S.; Chang, B.H.; MacBeath, G. Quantifying protein-protein interactions in high throughput using protein domain microarrays. Nat. Protoc. 2010, 5, 773–790. [Google Scholar] [CrossRef]
- Ramani, S.R.; Tom, I.; Lewin-Koh, N.; Wranik, B.; Depalatis, L.; Zhang, J.; Eaton, D.; Gonzalez, L.C. A secreted protein microarray platform for extracellular protein interaction discovery. Anal. Biochem. 2012, 420, 127–138. [Google Scholar] [CrossRef]
- Duenas, M.E.; Peltier-Heap, R.E.; Leveridge, M.; Annan, R.S.; Buttner, F.H.; Trost, M. Advances in high-throughput mass spectrometry in drug discovery. EMBO Mol. Med. 2023, 15, e14850. [Google Scholar] [CrossRef]
- Henderson, M.J.; Holbert, M.A.; Simeonov, A.; Kallal, L.A. High-Throughput Cellular Thermal Shift Assays in Research and Drug Discovery. SLAS Discov. Adv. Sci. Drug Discov. 2020, 25, 137–147. [Google Scholar] [CrossRef] [PubMed]

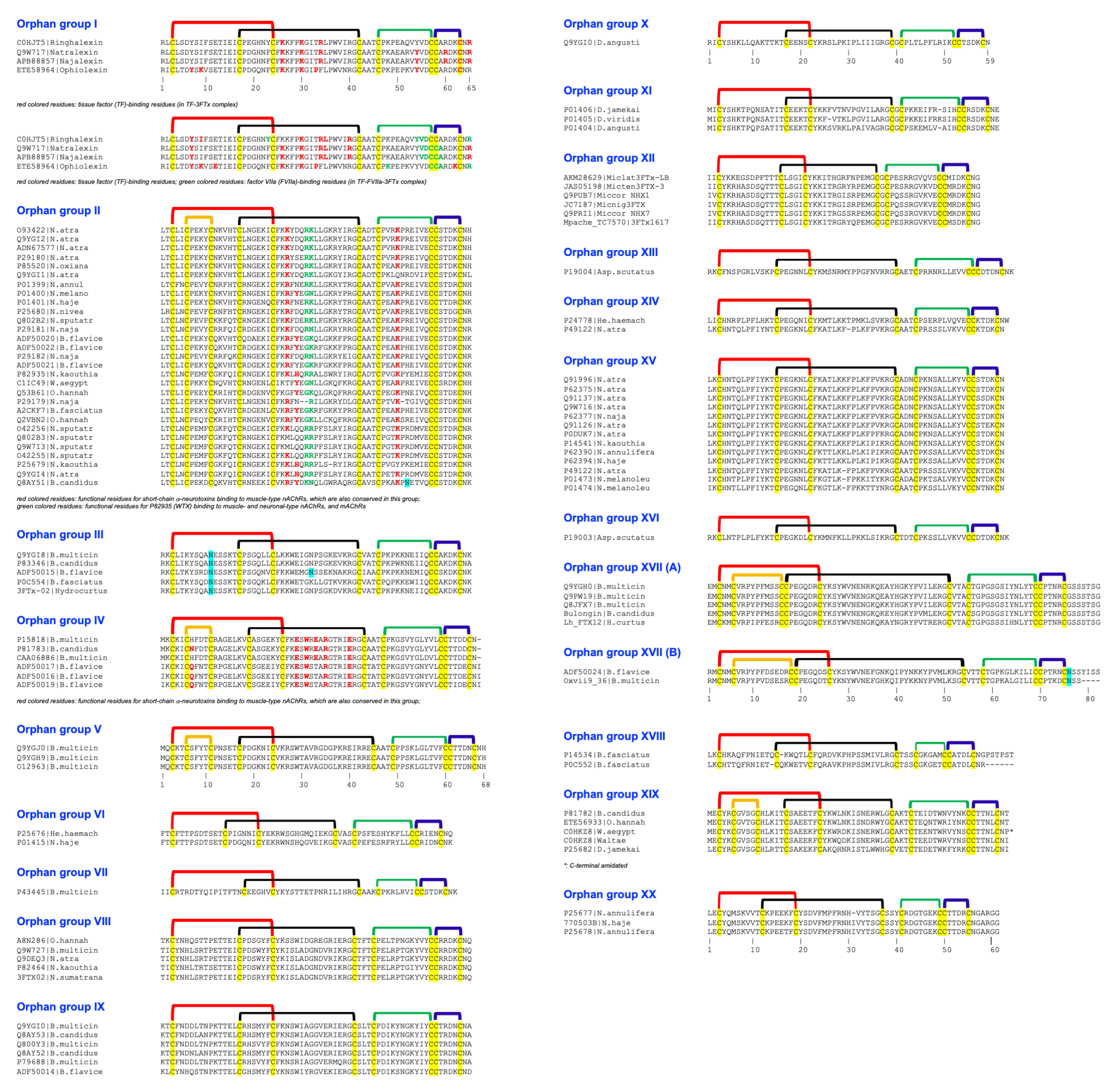



| Group | Toxins |
|---|---|
| Orphan group I | Q9W717 (natralexin); C0HJT5 (ringhalexin); ETE58964 (ophiolexin), APB88857 (najalexin) |
| Orphan group II | O93422, Q9YGI2, Q9YGI1, P29180, P01401, P01400, P29179, P01399, P25680, P29181, P29182, Q9YGI4, Q9W7I3, O42256, O42255, P25679, P82935, ADN67577, P85520, Q802B2, ADF50020, ADF50022, ADF50021, C1IC49, Q53B61, A2CKF7, Q2VBN2, Q802B3, Q8AY51 (bungatoxin) |
| Orphan group III | Q9YGI8, P83346 (bucain), ADF50015, P0C554 |
| Orphan group IV | P15818 (Q9YGI9), P81783 (candoxin), CAA06886, ADF50017, ADF50016, ADF50019 |
| Orphan group V | Q9YGJ0 (γ-bungarotoxin; AAD41806), O12963, Q9YGH9 |
| Orphan group VI | P25676, P01415 |
| Orphan group VII | P43445 |
| Orphan group VIII | Q9W727, Q9DEQ3, P82464, A8N286 (haditoxin) |
| Orphan group IX | Q9YGI0, P79688, Q8AY53, Q800Y3, Q8AY52, ADN67594 |
| Orphan group X | P18329 |
| Orphan group XI | P01406, P01405, P01404 |
| Orphan group XII | Q9PRI1, Q9PUB7, AKM28629, JAS05198, JC7187, Mpache_TC7570|3FTx1617 |
| Orphan group XIII | P19004 |
| Orphan group XIV | P24778, P49122 |
| Orphan group XV | Q91996, P62375 *, Q91126, Q91137, Q9W716, P14541, P62394 *, P49122, P01473, P01474, P62377, P0DUK7, P62390 |
| Orphan group XVI | P19003 |
| Orphan group XVII | Q9YGH0, Q9PW19, bulongin, Q8JFX7 |
| Orphan group XVIII | P14534, P0C552 |
| Orphan group XIX | P81782 (bucandin), P25682, ETE56933, C0HKZ8 (actiflagelin) |
| Orphan group XX | P25677, P25678, 770503B |
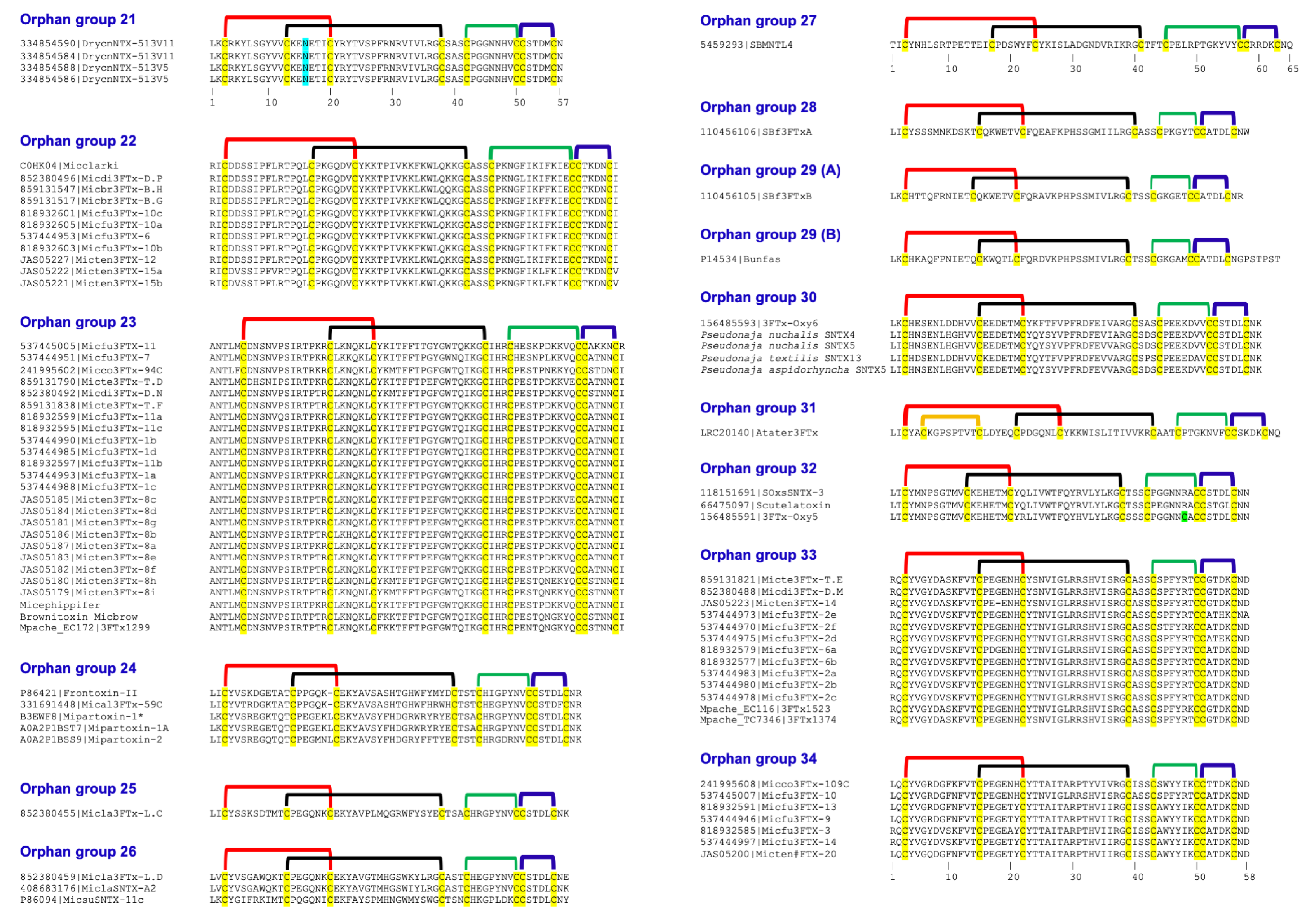
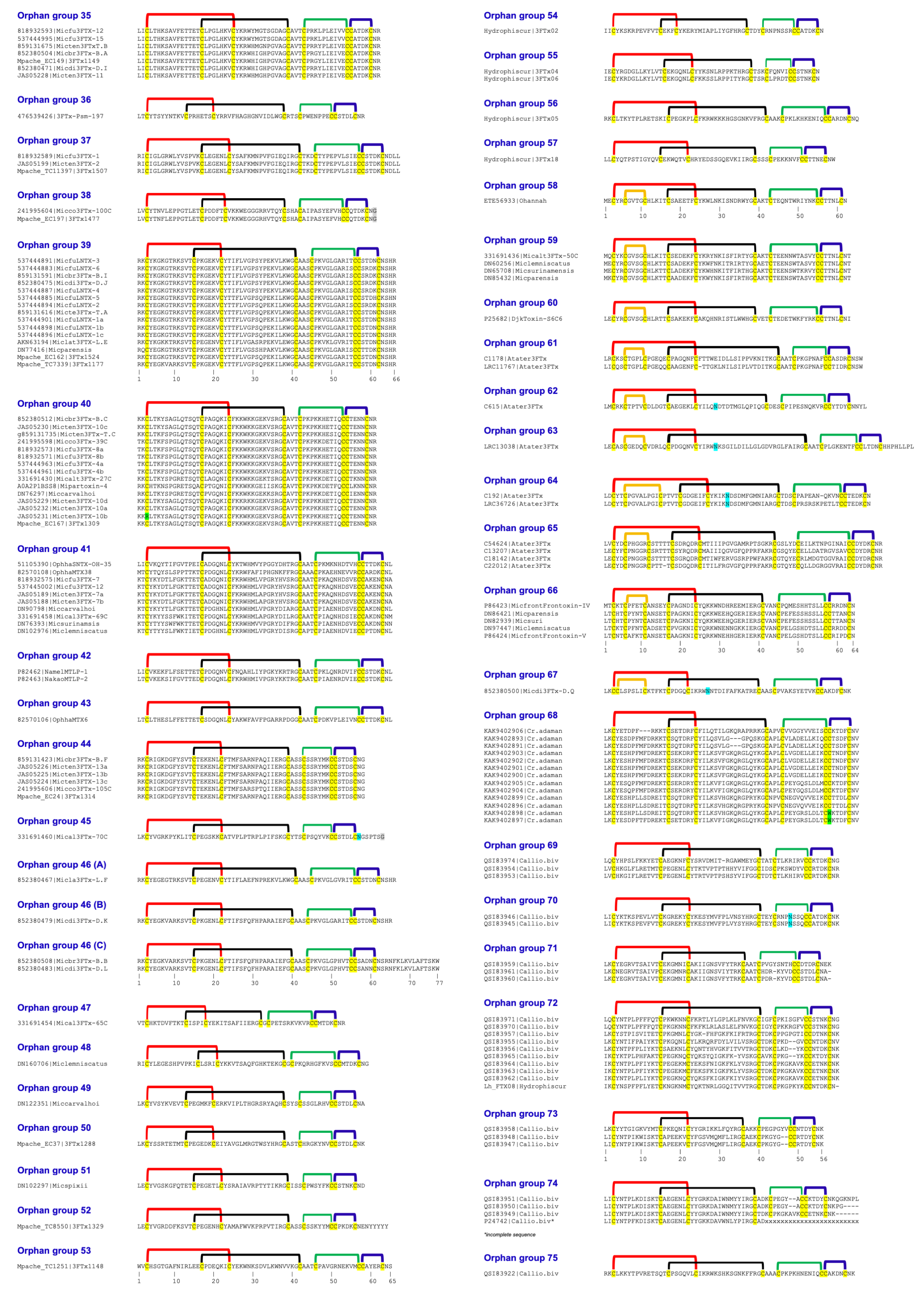
| Orphan 3FTx Group | N-term | First S-S (Fifth S-S) | Second S-S | Gap Residues | Third S-S | Fourth S-S | C-Term |
|---|---|---|---|---|---|---|---|
| 20 groups of orphan 3FTxs initially classified in 2003 [2] | |||||||
| I | 2 | 22 | 26 | 3 | 12 | 6 | 2 |
| II | 2 | 22 (6 *) | 24–26 | 3 | 12 | 6 | 2 |
| III | 2 | 22 | 26 | 3 | 13 | 6 | 2 |
| IV | 2 | 24 ^ (6 *) | 25 | 3 | 13 | 6 | 1–2 |
| V | 2 | 22 (6 *) | 29 | 3 | 12 | 6 | 2 |
| VI | 2 | 19 | 24 | 3 | 13 | 6 | 2 |
| VII | 2 | 22 | 25 | 3 | 9 | 6 | 2 |
| VIII | 2 | 22 | 25 | 3 | 13 | 6 | 2 |
| IX | 2 | 22 | 25 | 3 | 13 | 6 | 2 |
| X | 2 | 22 | 25 | 3 | 13 | 6 | 2 |
| XI | 2 | 20 | 23 | 1 | 12 | 6 | 1 |
| XII | 2 | 20 | 23 | 1 | 12–13 | 6 | 2 |
| XIII | 2 | 19^ | 21 | 1 | 12 | 6 | 2 |
| XIV | 2 | 20 | 26 | 3 | 12 | 6 | 1–2 |
| XV | 2 | 20 | 26 | 3 | 12 | 6 | 1 |
| XVI | 2 | 20 | 26 | 3 | 12 | 6 | 2 |
| XVII (A) | 2 | 22 (11) | 35 | 3 | 15 | 6 | 7 |
| XVII (B) | 2 | 24 (13) | 36 | 3 | 12 | 6 | 3–7 |
| XVIII | 2 | 21 | 25–26 | 3 | 7 * | 6 | 2–8 |
| XIX | 2 | 22 ^ (6 *) | 23 | 3 | 13 | 6 | 2 |
| XX | 2 | 17 * | 25–26 | 3 | 9 | 6 | 6 |
| Further classification of groups of orphan 3FTxs | |||||||
| 21 | 2 | 18 | 25 | 3 | 9 | 6 | 1 |
| 22 | 2 | 22 | 26 | 3 | 14 | 6 | 1 |
| 23 | 2 | 22 | 26 | 3 | 12 | 6 | 1 |
| 24 | 2 | 18–19 | 26–27 | 3 | 9 | 6 | 2 |
| 25 | 2 | 18–19 | 26 | 3 | 9 | 6 | 2–4 |
| 26 | 2 | 18 | 26 | 3 | 9 | 6 | 2 |
| 27 | 2 | 22 | 25 | 3 | 13 | 6 | 2 |
| 28 | 2 | 20 | 26 | 3 | 7 * | 6 | 2 |
| 29A | 2 | 19 | 26 | 3 | 7 * | 6 | 2 |
| 29B | 2 | 19 | 25 | 3 | 7 * | 6 | 8 |
| 30 | 2 | 20 | 26 | 3 | 9 | 6 | 2 |
| 31 | 2 | 26 (10) | 23 | 3 | 9 | 6 | 2 |
| 32 | 2 | 18 | 26 | 3 | 9 | 6 | 2 |
| 33 | 2 | 20 | 25 | 3 | 8 * | 6 | 2 |
| 34 | 2 | 20 | 25 | 3 | 8 * | 6 | 2 |
| 35 | 2 | 22 ^ | 23 | 3 | 12 | 6 | 2 |
| 36 | 2 | 18 | 26 | 3 | 9 | 6 | 2 |
| 37 | 2 | 22 | 26 | 3 | 12 | 6 | 4 |
| 38 | 2 | 21 ^ | 22 | 3 | 12 | 6 | 1–2 |
| 39 | 2 | 20 | 27 | 3 | 12 | 6 | 4 |
| 40 | 2 | 22 ^ | 22 | 3 | 12 | 6 | 2 |
| 41 | 2 | 22 | 26 | 3 | 12 | 6 | 2 |
| 42 | 2 | 22 | 26 | 3 | 12 | 6 | 2 |
| 43 | 2 | 22 | 26 | 3 | 12 | 6 | 2 |
| 44 | 2 | 20 | 25 | 3 | 8 * | 6 | 2 |
| 45 | 2 | 20 | 25 | 3 | 8 * | 6 | 6–7 |
| 46A | 2 | 20 | 26 | 3 | 12 | 6 | 4 |
| 46B | 2 | 20 | 26 | 3 | 12 | 6 | 4 |
| 46C | 2 | 20 | 26 | 3 | 12 | 6 | 16 # |
| 47 | 2 | 16 * | 20 * | 1 | 12 | 6 | 2 |
| 48 | 2 | 19 | 23 | 1 | 12 | 6 | 2 |
| 49 | 2 | 18 | 26 | 3 | 9 | 6 | 2 |
| 50 | 2 | 18 | 26 | 3 | 9 | 6 | 2 |
| 51 | 2 | 20 | 25 | 3 | 8 * | 6 | 2 |
| 52 | 2 | 20 | 25 | 3 | 8 * | 6 | 8 |
| 53 | 2 | 22 | 26 | 3 | 12 | 6 | 2 |
| 54 | 2 | 17 * | 23 | 3 | 9 | 6 | 1 |
| 55 | 2 | 21 | 23 | 3 | 7 * | 6 | 1 |
| 56 | 2 | 22 | 26 | 3 | 12 | 6 | 2 |
| 57 | 2 | 21 | 25 | 3 | 9 | 6 | 2 |
| 58 | 2 | 22 ^ (6 *) | 23 | 3 | 11 | 6 | 1 |
| 59 | 2 | 22 ^ (6 *) | 23 | 3 | 13 | 6 | 2 |
| 60 | 2 | 22 ^ (6 *) | 23 | 3 | 13 | 6 | 1–2 |
| 61 | 2 | 22 (6 *) | 28–29 | 3 | 9 | 6 | 3 |
| 62 | 2 | 22 (6 *) | 26 | 3 | 12 | 6 | 4 |
| 63 | 2 | 22 (6 *) | 36 | 3 | 10 | 6 | 8 |
| 64 | 2 | 24 ^–25 ^ (10) | 24–26 | 3 | 9–13 | 6 | 1–2 |
| 65 | 2 | 22–23 (7) | 28 | 5 | 14 | 6 | 2 |
| 66 | 2 | 22 (6 *) | 25 | 3 | 13 | 6 | 1 |
| 67 | 2 | 20 (8) | 24 | 3 | 12 | 6 | 2 |
| 68 | 2 | 19–22 ^ | 23–26 | 3 | 12 | 6 | 2 |
| 69 | 2 | 20 | 25–26 | 3 | 9 | 6 | 2 |
| 70 | 2 | 20 | 26 | 3 | 9 | 6 | 2 |
| 71 | 2 | 20 ^ | 22 | 3 | 9–10 | 6 | 2–3 |
| 72 | 2 | 20 | 26 | 3 | 7 *–9 | 6 | 2 |
| 73 | 2 | 20 ^ | 22 | 3 | 7 *–9 | 6 | 2 |
| 74 | 2 | 20 | 25 | 3 | 7 *–9 | 6 | 2–8 |
| 75 | 2 | 22 | 25 | 3 | 12 | 6 | 2 |
| 76 | 2 | 22 | 25 | 3 | 12 | 6 | 1 |
| 77 | 2 | 21 ^ | 22 | 3 | 12 | 6 | 2 |
| 78 | 2 | 20 | 27 | 3 | 7 * | 6 | 2 |
| 79 | 2 | 20 | 23 | 3 | 7 * | 6 | 2 |
| 80 | 2 | 20 | 23 | 3 | 7 * | 6 | 2 |
| 81 | 2 | 20 ^ | 21 | 3 | 7 * | 6 | 2–3 |
| 82 | 2 | 20 | 23 | 3 | 7 * | 6 | 2 |
| 83 | 2 | 22 | 22 | 3 | 12 | 6 | 2 |
| 84 | 2 | 19 | 26 | 3 | 9 | 6 | 2–4 |
| 85 | 2 | 19 | 27 | 3 | 9 | 6 | 7 |
| 86 | 2 | 22 | 26 | 3 | 12 | 6 | 1 |
| 87 | 2 | 22 | 26 | 3 | 12 | 6 | 2 |
| 88 | 2 | 23 (7) | 27 | 5 | 14–16 | 6 | 2 |
| 89 | 2 | 20–25 ^ (5 *–6 *) | 23–26 | 3 | 12 | 6 | 2–7 |
| 90 | 2 | 22 ^ (6 *) | 22–26 | 3 | 11–12 | 6 | 1–3 |
| 91 | 2 | 24 ^ (8) | 25 | 3 | 12 | 6 | 3 |
| 92 | 2 | 25 ^ (9) | 26 | 3 | 12 | 6 | 8 |
| 93 | 2 | 25 ^ (9) | 23 | 3 | 12 | 6 | 3 |
| 94 | 2 | 27 ^ (12) | 24 | 3 | 9 | 6 | 1 |
| 95 | 2 | 22 (6 *) | 26 | 3 | 13 | 6 | 1 |
| 96 | 2 | 22 (6 *) | 28 | 3 | 9 | 6 | 2–3 |
| 97 | 2 | 28 ^ (13) | 28 | 3 | 9 | 6 | 11 |
| 98 | 2 | 28 ^ (13) | 24 | 3 | 9 | 6 | 3 |
| 99 | 2 | 24 ^ (6 *) | 23–26 | 3 | 9 | 6 | 3 |
| 100 | 2 | 22 ^ (6 *) | 22 | 3 | 12 | 6 | 2 |
| 101 | 2 | 22–24 (6 *–8) | 26–27 | 3 | 12 | 6 | 1–6 |
| 102 | 2 | 22 (6 *) | 26 | 3 | 12 | 6 | 1 |
| 103 | 2 | 22 (6 *) | 26 | 3 | 12 | 6 | 4 |
| 104 | 2 | 22 (6 *) | 32 | 3 | 12 | 6 | 17 # |
| 105 | 2 | 22 (6 *) | 26 | 3 | 12 | 6 | 2 |
| 106 | 2 | 22 (6 *) | 26 | 3 | 12 | 6 | 1 |
| 107 | 2 | 21 (6 *) | 25 | 3 | 12 | 6 | 2 |
| 108 | 2 | 21 ^ (6 *) | 20 * | 1 | 12 | 6 | 1 |
| 109 | 2 | 22 (6 *) | 26 | 3 | 12 | 6 | 4 |
| 110 | 2 | 20 (6 *) | 24–26 | 3 | 12 | 6 | 8 |
| 111 | 2 | 20 (6 *) | 26 | 3 | 12 | 6 | 4 |
| 112 | 2 | 20 (6 *) | 26 | 3 | 12 | 6 | 2 |
| 113 | 2 | 24 (10) | 27 | 3 | 12 | 6 | 2 |
| 114 | 2 | 24 ^ (6 *) | 25 | 3 | 12 | 6 | 1 |
| 115 | 2 | 22 (6 *) | 26 | 3 | 12 | 6 | 2 |
| N-terminal variants | |||||||
| 116 | 1 | 19 (10) | 24 | 3 | 12 | 6 | 3 |
| 117 | 3 | 26 (10) | 27 | 3 | 12 | 6 | 4 |
| 118 | 3 | 25 (9) | 27 | 3 | 12 | 6 | 3 |
| 119 | 4 | 24 ^ (6 *) | 21 | 3 | 12 | 6 | 5 |
| 120 | 4 | 22 (6 *) | 26 | 3 | 12 | 6 | 4 |
| 121 | 4 | 22 (6 *) | 26 | 3 | 12 | 6 | 20 # |
| 122 | 1 | 23 | 23 | 3 | 12 | 6 | 2 |
| Distinct disulfide variants | |||||||
| 123 | 2 | 21 (6 *) | 23 | 3 | 12 | 0 * | 10 |
| 124 | 2 | 21^ (6 *) | 23 | 3 | 12 | 0 * | 13 # |
| 125 | 2 | 22 | 25 | 3 | 13 | 0 * | 8 |
| 126 | 2 | 25 (9) | 19 * | 3 | 12 | 6 | 8 |
| 127 | 2 | 22^ (6 *) | 20 * | 3 | 12 | 6 | 7 |
| 128 | 2 | 26 (9) | 27 | 8 | 16 | 6 | 18 # |
| 129 | 2 | 22 | 26 | 1 | 12 | 6 | 2 |
| 130 | 2 | 22 | 29 | 3 | 12 | 6 | 3 |
| 131 | 2 | 21 | 29 | 3 | 12 | 6 | 6 |
| 132 | 2 | 20 ^ (6 *) | 20 *–22 | 3 | 11 | 6 | 2–4 |
| 133 | 2 | 22 | 29 | 3 | 12 | 6 | 1 |
| 134 | 2 | 18 | 32 | 3 | 12 | 6 | 4 |
| 135 | 2 | 18 | 26 | 3 | 9 | 6 | 2 |
| 136 | 2 | 18 | 26 | 3 | 9 | 6 | 2 |
| 137 | 2 | 18 | 26 | 3 | 9 | 6 | 2 |
| 138 | 1 | 26 ^ (10) | 24 | 3 | 12 | 6 | 4 |
| 139 | 2 | ? | ? | 3 | 12 | 6 | 2 |
| 140 | 2 | ? | ? | 1 | 12 | 6 | 2 |
| 141 | 2 | 24 (6 *) | 26 ? | 3 | ? | 6 | 2 |
| 142 | 2 | 20 (6 *) | 27 ? | 3 | ? | 6 | 2 |
| 143 | 2 | 21 (6 *) | 23 ? | 3 | ? | 6 | 2 |
| 144 | 2 | 24 (6 *) | 26 ? | 3 | ? | 6 | 2 |
| 145 | 2 | 19 * (6 *) | 24 ? | 3 | ? | 6 | 4 |
| 146 | 2 | 21 ^–25 ^ (6 *–10) | 21–24 ? | 3 | ? | 6 | 1–2 |
| 147 | 2 | 22 (6 *) | 26 ? | 3 | ? | 6 | 1 |
| 148 | 2 | 22 (6 *) | 26 ? | 3 | ? | 6 | 2 |
| 149 | 2 | 22 (6 *) | 26 ? | 3 | ? | 6 | 1–2 |
| 150 | 2 | 22 (6 *) | 26 ? | 3 | ? | 6 | 2 |
| 151 | 2 | 21 (6 *) | 25 ? | 3 | ? | 6 | 2 |
| 152 | 2 | 21 (6 *) | 25 ? | 3 | ? | 6 | 0–1 |
| 153 | 2 | 19 * | 21 ? | 3 | ? | 6 | 2 |
| 154 | 2 | 22 | 24 ? | 3 | ? | 6 | 3 |
| 155 | 2 | 19 * | 25 | 3 | 7 *-8 ? | 6 | 2 |
| 156 | 2 | 21 | 25? | 3 | ? | 6 | 1 |
| 157 | 2 | 22 (6 ? *) | 26 | 3 | 12 | 6 | 6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koh, C.Y.; Kini, R.M. Orphan Three-Finger Toxins from Snake Venoms: Unexplored Library of Novel Biological Ligands with Potential New Structures and Functions. Int. J. Mol. Sci. 2025, 26, 8792. https://doi.org/10.3390/ijms26188792
Koh CY, Kini RM. Orphan Three-Finger Toxins from Snake Venoms: Unexplored Library of Novel Biological Ligands with Potential New Structures and Functions. International Journal of Molecular Sciences. 2025; 26(18):8792. https://doi.org/10.3390/ijms26188792
Chicago/Turabian StyleKoh, Cho Yeow, and R. Manjunatha Kini. 2025. "Orphan Three-Finger Toxins from Snake Venoms: Unexplored Library of Novel Biological Ligands with Potential New Structures and Functions" International Journal of Molecular Sciences 26, no. 18: 8792. https://doi.org/10.3390/ijms26188792
APA StyleKoh, C. Y., & Kini, R. M. (2025). Orphan Three-Finger Toxins from Snake Venoms: Unexplored Library of Novel Biological Ligands with Potential New Structures and Functions. International Journal of Molecular Sciences, 26(18), 8792. https://doi.org/10.3390/ijms26188792






