The Roles of SHCBP1 in Cancer Hallmarks: Molecular Mechanisms and Therapeutic Implications
Abstract
1. Introduction
2. Molecular Characteristics and Regulation of SHCBP1
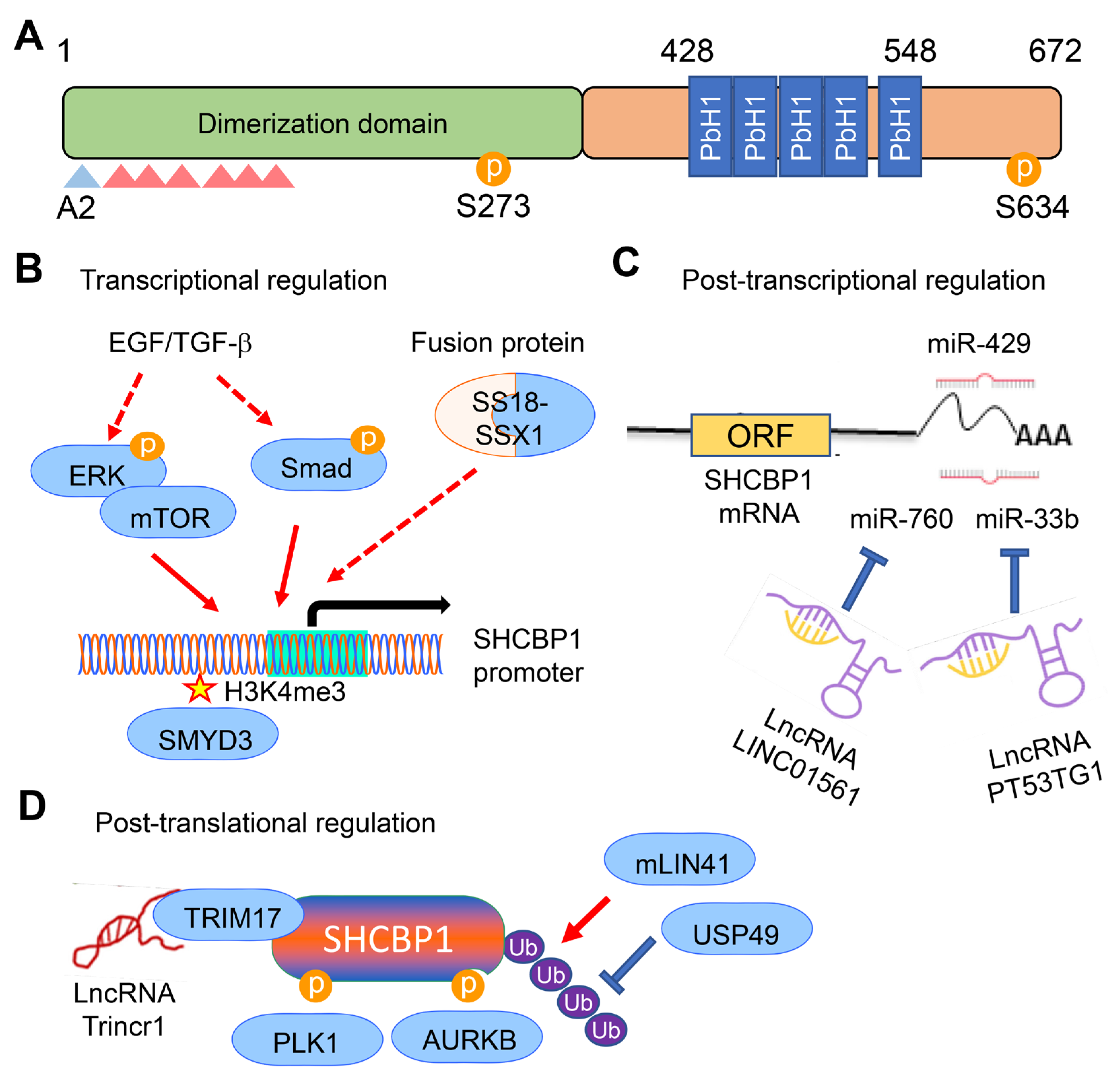
2.1. Genomic Organization and Protein Structure
2.2. Dynamic Subcellular Localization and Cell Cycle Regulation
2.3. Transcriptional and Post-Transcriptional Regulation of SHCBP1
2.4. Post-Translational Modifications of SHCBP1
3. Multifaceted Roles of SHCBP1 in the Hallmarks of Cancer
3.1. Sustaining Proliferative Signaling by SHCBP1
3.2. Evading Growth Suppression by SHCBP1
3.3. Resisting Cell Death by SHCBP1
3.4. Enabling Replicative Immortality by SHCBP1
3.5. Inducing Angiogenesis by SHCBP1
3.6. Activating Invasion and Metastasis by SHCBP1
3.7. Deregulating Cellular Energetics by SHCBP1
3.8. Avoiding Immune Destruction by SHCBP1
4. SHCBP1 as a Diagnostic and Prognostic Biomarker in Cancer
| Cancer Type | Relevant Clinical Characteristics | Prognostic Significance and Reference |
|---|---|---|
| Breast cancer | Tumor size ↑, Grade ↑, Lymph node metastasis ↑, TMB ↑, Stemness index ↑ | OS ↓ p < 0.001 [13], p < 0.01 [72], HR = 1.59 [46], HR = 1.76 [73], HR = 1.06 [74]; RFS ↓ p < 0.001 [13], |
| Pancreatic cancer | Tumor size ↑, Portal vein invasion ↑, AJCC stage ↑, CA19-9 ↑ | OS ↓ p < 0.001 [63], HR = 1.57 [77], HR = 1.6 [78]; DFS ↓ p < 0.001 [63], HR = 1.8 [77]; PFI ↓ HR = 1.57 [77] |
| Lung cancer | TNM stage ↑, Metastasis ↑, Immune evasion ↑, TMB ↑ | OS ↓ p < 0.001 [66], p = 0.0135 [79], p = 0.047 [80], p < 0.001 [81], HR = 1.5 [82] |
| HCC | SHCBP1 level ↑, Immune cell infiltration ↑, TPX2 ↑ | OS ↓ p < 0.001, HR = 2.14 [84], HR = 1.5 [85], HR = 1.5 [86], HR = 1.83 [88] |
| Prostate cancer | PSA ↑, Gleason grade ↑, pT stage ↑, Seminal vesicle invasion ↑ | PFS ↓ p < 0.001 [43]; RFS ↓ p < 0.001 [47] |
| Glioblastoma | pT stage ↑, IDH mutation status ↑, Female tumor ↑ | OS ↓ p < 0.01 [19], p < 0.001 [36], p = 0.039 [90] |
| Pan-cancer | TMB ↑, MSI ↑, TAM ↑, Immune evasion ↑, IC50 ↑ | OS ↓ p < 0.001 [31], HR = 1.29–1.8 [69], HR = 1.764 [93]; DFS ↓ p < 0.05 [69], HR = 1.469 [93]; PFI ↓ p < 0.001 [31], HR = 1.661 [93] |
5. Therapeutic Targeting of SHCBP1 in Cancer
5.1. Genetic Suppression of SHCBP1 for Antitumor Therapy
5.2. Pharmacological Inhibition of SHCBP1 for Anticancer Therapy
5.3. SHCBP1-Targeted Combination Strategies in Cancer Therapy
6. Limitations and Future Directions
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SHCBP1 | SHC SH2-domain binding protein 1 |
| mPAL | Murine protein of activated lymphocytes |
| NSCLC | Non-small cell lung carcinoma |
| HCC | Hepatocellular carcinoma |
| CDK | Cyclin-dependent kinase |
| EMT | Epithelial–mesenchymal transition |
| PbH1 | Parallel beta-helix |
| PLK1 | Polo-like kinase 1 |
| EGF | Epidermal growth factor |
| TGF-β | Transforming growth factor β |
| MiRNA | MicroRNA |
| LncRNA | Long noncoding RNA |
| CeRNA | Competing endogenous RNA |
| AURKB | Aurora kinase B |
| FGF | Fibroblast growth factor |
| USP49 | Ubiquitin-specific peptidase 49 |
| CBP | CREB binding protein |
| HNSCC | Head and neck squamous cell carcinoma |
| VEGF | Vascular endothelial growth factor |
| RTK | Receptor tyrosine kinase |
| PDAC | Pancreatic ductal adenocarcinoma |
| EOGT | EGF domain specific O-linked GlcNActransferase |
| TME | Tumor microenvironment |
| TAM | Tumor-associated macrophage |
| TMB | Tumor mutation burden |
| MSI | Microsatellite instability |
| CAG | Cancer–associated fibroblast |
| ICI | Immune checkpoint inhibitor |
| PPI | Protein–protein interaction |
| SCLC | Small cell lung cancer |
| RNAi | RNA interference |
| SiRNA | Small interfering RNA |
| ShRNA | Short hairpin RNA |
| TFBG | Theaflavin-3,3′-digallate |
References
- Zhang, S.; Xiao, X.; Yi, Y.; Wang, X.; Zhu, L.; Shen, Y.; Lin, D.; Wu, C. Tumor initiation and early tumorigenesis: Molecular mechanisms and interventional targets. Signal Transduct. Target. Ther. 2024, 9, 149. [Google Scholar] [CrossRef]
- de Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Schmandt, R.; Liu, S.K.; McGlade, C.J. Cloning and characterization of mPAL, a novel Shc SH2 domain-binding protein expressed in proliferating cells. Oncogene 1999, 18, 1867–1879. [Google Scholar]
- Asano, E.; Hasegawa, H.; Hyodo, T.; Ito, S.; Maeda, M.; Takahashi, M.; Hamaguchi, M.; Senga, T. The Aurora-B-mediated phosphorylation of SHCBP1 regulates cytokinetic furrow ingression. J. Cell Sci. 2013, 126, 3263–3270. [Google Scholar]
- Zheng, Y.; Zhang, C.; Croucher, D.R.; Soliman, M.A.; St-Denis, N.; Pasculescu, A.; Taylor, L.; Tate, S.A.; Hardy, W.R.; Colwill, K.; et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 2013, 499, 166–171. [Google Scholar] [CrossRef]
- Chen, J.; Lai, F.; Niswander, L. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 2012, 26, 803–815. [Google Scholar] [CrossRef]
- Asano, E.; Hasegawa, H.; Hyodo, T.; Ito, S.; Maeda, M.; Chen, D.; Takahashi, M.; Hamaguchi, M.; Senga, T. SHCBP1 is required for midbody organization and cytokinesis completion. Cell Cycle 2014, 13, 2744–2751. [Google Scholar] [CrossRef]
- Buckley, M.W.; Arandjelovic, S.; Trampont, P.C.; Kim, T.S.; Braciale, T.J.; Ravichandran, K.S. Unexpected phenotype of mice lacking Shcbp1, a protein induced during T cell proliferation. PLoS ONE 2014, 9, e105576. [Google Scholar]
- Montembault, E.; Zhang, W.; Przewloka, M.R.; Archambault, V.; Sevin, E.W.; Laue, E.D.; Glover, D.M.; D’Avino, P.P. Nessun Dorma, a novel centralspindlin partner, is required for cytokinesis in Drosophila spermatocytes. J. Cell Biol. 2010, 191, 1351–1365. [Google Scholar] [CrossRef]
- Ito, M.; Iwasaki, M.; Takeda, M.; Nakamura, T.; Yanagi, Y.; Ohno, S. Measles virus nonstructural C protein modulates viral RNA polymerase activity by interacting with host protein SHCBP1. J. Virol. 2013, 87, 9633–9642. [Google Scholar] [CrossRef]
- Feng, W.; Li, H.C.; Xu, K.; Chen, Y.F.; Pan, L.Y.; Mei, Y.; Cai, H.; Jiang, Y.M.; Chen, T.; Feng, D.X. SHCBP1 is over-expressed in breast cancer and is important in the proliferation and apoptosis of the human malignant breast cancer cell line. Gene 2016, 587, 91–97. [Google Scholar] [CrossRef]
- Wang, F.; Li, Y.; Zhang, Z.; Wang, J.; Wang, J. SHCBP1 regulates apoptosis in lung cancer cells through phosphatase and tensin homolog. Oncol. Lett. 2019, 18, 1888–1894. [Google Scholar] [CrossRef]
- Dong, Y.D.; Yuan, Y.L.; Yu, H.B.; Tian, G.J.; Li, D.Y. SHCBP1 is a novel target and exhibits tumor-promoting effects in gastric cancer. Oncol. Rep. 2019, 41, 1649–1657. [Google Scholar] [PubMed]
- Tao, H.C.; Wang, H.X.; Dai, M.; Gu, C.Y.; Wang, Q.; Han, Z.G.; Cai, B. Targeting SHCBP1 inhibits cell proliferation in human hepatocellular carcinoma cells. Asian Pac. J. Cancer Prev. 2013, 14, 5645–5650. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Zhou, Z.; Wang, X.; Hua, X.; Zou, M.; Zhang, X. SHCBP1 Promotes the Progression of Esophageal Squamous Cell Carcinoma Via the TGFβ Pathway. Appl. Immunohistochem. Mol. Morphol. 2021, 29, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Piccaluga, P.P.; Agostinelli, C.; Califano, A.; Rossi, M.; Basso, K.; Zupo, S.; Went, P.; Klein, U.; Zinzani, P.L.; Baccarani, M.; et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J. Clin. Investig. 2007, 117, 823–834. [Google Scholar] [CrossRef]
- Zhou, Y.; Tan, Z.; Chen, K.; Wu, W.; Zhu, J.; Wu, G.; Cao, L.; Zhang, X.; Zeng, X.; Li, J.; et al. Overexpression of SHCBP1 promotes migration and invasion in gliomas by activating the NF-kappaB signaling pathway. Mol. Carcinog. 2018, 57, 1181–1190. [Google Scholar] [CrossRef]
- Lin, Y.; Cai, H. Biological functions and therapeutic potential of SHCBP1 in human cancer. Biomed. Pharmacother. 2023, 160, 114362. [Google Scholar] [CrossRef]
- Liu, L.; Yang, Y.; Liu, S.; Tao, T.; Cai, J.; Wu, J.; Guan, H.; Zhu, X.; He, Z.; Li, J.; et al. EGF-induced nuclear localization of SHCBP1 activates β-catenin signaling and promotes cancer progression. Oncogene 2019, 38, 747–764. [Google Scholar]
- Peng, C.; Zhao, H.; Song, Y.; Chen, W.; Wang, X.; Liu, X.; Zhang, C.; Zhao, J.; Li, J.; Cheng, G.; et al. SHCBP1 promotes synovial sarcoma cell metastasis via targeting TGF-β1/Smad signaling pathway and is associated with poor prognosis. J. Exp. Clin. Cancer Res. 2017, 36, 141. [Google Scholar] [PubMed]
- Yin, H.; Zhang, C.; Wei, Z.; He, W.; Xu, N.; Xu, Y.; Li, T.; Ren, K.; Kuang, Y.; Zhu, X.; et al. EGF-induced nuclear translocation of SHCBP1 promotes bladder cancer progression through inhibiting RACGAP1-mediated RAC1 inactivation. Cell Death Dis. 2022, 13, 39. [Google Scholar] [PubMed]
- Gu, Q.; Ma, Z.; Wang, Q.; Dai, Y.; Shi, W.; Jiao, Z. Knockout of Shcbp1 sensitizes immunotherapy by regulating alpha-SMA positive cancer-associated fibroblasts. Mol. Carcinog. 2024, 63, 601–616. [Google Scholar] [CrossRef] [PubMed]
- Dephoure, N.; Zhou, C.; Villen, J.; Beausoleil, S.A.; Bakalarski, C.E.; Elledge, S.J.; Gygi, S.P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 10762–10767. [Google Scholar]
- Olsen, J.V.; Vermeulen, M.; Santamaria, A.; Kumar, C.; Miller, M.L.; Jensen, L.J.; Gnad, F.; Cox, J.; Jensen, T.S.; Nigg, E.A.; et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal 2010, 3, ra3. [Google Scholar] [CrossRef]
- Pelicci, G.; Dente, L.; De Giuseppe, A.; Verducci-Galletti, B.; Giuli, S.; Mele, S.; Vetriani, C.; Giorgio, M.; Pandolfi, P.P.; Cesareni, G.; et al. A family of Shc related proteins with conserved PTB, CH1 and SH2 regions. Oncogene 1996, 13, 633–641. [Google Scholar]
- Hutchins, J.R.; Toyoda, Y.; Hegemann, B.; Poser, I.; Heriche, J.K.; Sykora, M.M.; Augsburg, M.; Hudecz, O.; Buschhorn, B.A.; Bulkescher, J.; et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 2010, 328, 593–599. [Google Scholar] [CrossRef]
- Zhou, M.; Duan, L.; Chen, J.; Li, Y.; Yin, Z.; Song, S.; Cao, Y.; Luo, P.; Hu, F.; Yang, G.; et al. The dynamic role of nucleoprotein SHCBP1 in the cancer cell cycle and its potential as a synergistic target for DNA-damaging agents in cancer therapy. Cell Commun. Signal. 2024, 22, 131. [Google Scholar]
- Mo, L.; Deng, M.; Adhav, R.; Chan, Y.; Lei, J.H.; Su, S.M.; Zhang, X.; An, T.; Liu, J.; Li, J.; et al. Oncogenic activation of SMYD3-SHCBP1 promotes breast cancer development and is coupled with resistance to immune therapy. Cell Death Dis. 2025, 16, 220. [Google Scholar] [CrossRef]
- Huang, Y.; You, M.; Wu, Q.; Zhu, W.; Guo, F.; Lin, W. SHCBP1 Is a Prognostic Biomarker Related to the Tumour Immune Microenvironment in Pan-Cancer. Ann. Clin. Lab. Sci. 2022, 52, 904–917. [Google Scholar] [PubMed]
- Peng, C.; Zhao, H.; Chen, W.; Song, Y.; Wang, X.; Li, J.; Qiao, Y.; Wu, D.; Ma, S.; Wang, X.; et al. Identification of SHCBP1 as a novel downstream target gene of SS18-SSX1 and its functional analysis in progression of synovial sarcoma. Oncotarget 2016, 7, 66822–66834. [Google Scholar] [CrossRef] [PubMed]
- Ebegboni, V.J.; Jones, T.L.; Brownmiller, T.; Zhao, P.X.; Pehrsson, E.C.; Rajan, S.S.; Caplen, N.J. ETS1, a target gene of the EWSR1::FLI1 fusion oncoprotein, regulates the expression of the focal adhesion protein TENSIN3. Mol. Cancer Res. 2024, 22, 625–641. [Google Scholar] [CrossRef] [PubMed]
- So, C.L.; Lee, Y.J.; Vokshi, B.H.; Chen, W.; Huang, B.; De Sousa, E.; Gao, Y.; Portuallo, M.E.; Begum, S.; Jagirdar, K.; et al. TFE3 fusion oncoprotein condensates drive transcriptional reprogramming and cancer progression in translocation renal cell carcinoma. Cell Rep. 2025, 44, 115539. [Google Scholar] [CrossRef]
- Zheng, Q.; Wang, T.; Zou, Z.; Ma, W.; Dong, Z.; Zhong, J.; Liu, W.; Xu, Y.; Hu, T.; Sun, W.; et al. Gene Fusions as Potential Therapeutic Targets in Soft Tissue Sarcomas. Biomolecules 2025, 15, 904. [Google Scholar] [CrossRef]
- Xuan, C.; Jin, M.; Wang, L.; Xue, S.; An, Q.; Sun, Q.; Wang, L.; Gao, Y. PART1 and hsa-miR-429-Mediated SHCBP1 Expression Is an Independent Predictor of Poor Prognosis in Glioma Patients. Biomed. Res. Int. 2020, 2020, 1767056. [Google Scholar] [CrossRef]
- Gao, W.; Qi, C.Q.; Feng, M.G.; Yang, P.; Liu, L.; Sun, S.H. SOX2-induced upregulation of lncRNA LINC01561 promotes non-small-cell lung carcinoma progression by sponging miR-760 to modulate SHCBP1 expression. J. Cell Physiol. 2020, 235, 6684–6696. [Google Scholar]
- Wang, H.; Zhang, Z.; Zhang, Y.; Liu, S.; Li, L. Long Non-Coding RNA TP53TG1 Upregulates SHCBP1 to Promote Retinoblastoma Progression by Sponging miR-33b. Cell Transplant. 2021, 30, 9636897211025223. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, G.; Ma, Z.; Li, L.; Liu, M.; Qin, L.; Yu, Z.; Zhao, L.; Liu, Y.; Zhang, X.; et al. Hyperactivation of HER2-SHCBP1-PLK1 axis promotes tumor cell mitosis and impairs trastuzumab sensitivity to gastric cancer. Nat. Commun. 2021, 12, 2812. [Google Scholar] [CrossRef]
- Li, Y.P.; Duan, F.F.; Zhao, Y.T.; Gu, K.L.; Liao, L.Q.; Su, H.B.; Hao, J.; Zhang, K.; Yang, N.; Wang, Y. A TRIM71 binding long noncoding RNA Trincr1 represses FGF/ERK signaling in embryonic stem cells. Nat. Commun. 2019, 10, 1368. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, Z.; Dai, X.; Ruan, R.; Zhong, H.; Wu, Z.; Yao, Y.; Chen, J.; Deng, J.; Xiong, J. Ubiquitin-specific peptidase 49 promotes adenocarcinoma of the esophagogastric junction malignant progression via activating SHCBP1-β-catenin-GPX4 axis. Carcinogenesis 2025, 46, bgae060. [Google Scholar] [CrossRef]
- Lu, H.; Yin, M.; Wang, L.; Cheng, J.; Cheng, W.; An, H.; Zhang, T. FGF13 interaction with SHCBP1 activates AKT-GSK3α/β signaling and promotes the proliferation of A549 cells. Cancer Biol. Ther. 2020, 21, 1014–1024. [Google Scholar]
- Tang, C.; Peng, S.; Chen, Y.; Cheng, B.; Li, S.; Zhou, J.; Wu, Y.; Li, L.; Zhong, H.; Guo, Z.; et al. SHCBP1 is a novel regulator of PLK1 phosphorylation and promotes prostate cancer bone metastasis. MedComm 2025, 6, e70082. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Sun, Z.; Nie, S.; Zhang, T.; Lu, H. Effects of Resveratrol on Mouse B16 Melanoma Cell Proliferation through the SHCBP1-ERK1/2 Signaling Pathway. Molecules 2023, 28, 7614. [Google Scholar] [CrossRef] [PubMed]
- Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Feng, G.; Nian, R.; Han, S.; Ke, M.; Wang, L.; Li, W.; Tian, S.; Lu, H. SHCBP1 Promotes the Proliferation of Breast Cancer Cells by Inhibiting CXCL2. J. Cancer 2023, 14, 3444–3456. [Google Scholar] [CrossRef]
- Xu, N.; Wu, Y.P.; Yin, H.B.; Chen, S.H.; Li, X.D.; Xue, X.Y.; Gou, X. SHCBP1 promotes tumor cell proliferation, migration, and invasion, and is associated with poor prostate cancer prognosis. J. Cancer Res. Clin. Oncol. 2020, 146, 1953–1969. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Chen, G.; Li, W.; Ge, R.; Guo, T.; Zhang, Y.; Zhou, C.; Lin, M. NUSAP1 Promotes Immunity and Apoptosis by the SHCBP1/JAK2/STAT3 Phosphorylation Pathway to Induce Dendritic Cell Generation in Hepatocellular Carcinoma. J. Immunother. 2025, 48, 46–57. [Google Scholar]
- Sun, Y.; Pan, H.; He, Y.; Hu, C.; Gu, Y. Functional roles of the SHCBP1 and KIF23 interaction in modulating the cell-cycle and cisplatin resistance of head and neck squamous cell carcinoma. Head Neck 2022, 44, 591–605. [Google Scholar]
- Zou, A.; Wu, A.; Luo, M.; Zhou, C.; Lu, Y.; Yu, X. SHCBP1 promotes cisplatin induced apoptosis resistance, migration and invasion through activating Wnt pathway. Life Sci. 2019, 235, 116798. [Google Scholar] [CrossRef]
- Qi, G.; Ma, H.; Teng, K.; Gai, P.; Gong, Y.; Chen, J.; Luo, X.; Kong, B. SHCBP1 promotes cisplatin resistance of ovarian cancer through AKT/mTOR/Autophagy pathway. Apoptosis 2025, 30, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kroemer, G.; Kang, R. Targeting cuproplasia and cuproptosis in cancer. Nat. Rev. Clin. Oncol. 2024, 21, 370–388. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Hu, K.; Chen, Q.; Wu, J. The function and immune role of cuproptosis associated hub gene in Barrett’s esophagus and esophageal adenocarcinoma. Biosci. Trends 2023, 17, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.J.; Ma, S. Hallmarks of cancer stemness. Cell Stem Cell 2024, 31, 617–639. [Google Scholar] [CrossRef]
- Deng, B.; Li, A.; Zhu, Y.; Zhou, Y.; Fei, J.; Miao, Y. SHCBP1 contributes to the proliferation and self-renewal of cervical cancer cells and activation of the NF-kappaB signaling pathway through EIF5A. Oncol. Lett. 2023, 25, 246. [Google Scholar] [CrossRef]
- Liu, Z.L.; Chen, H.H.; Zheng, L.L.; Sun, L.P.; Shi, L. Angiogenic signaling pathways and anti-angiogenic therapy for cancer. Signal Transduct. Target. Ther. 2023, 8, 198. [Google Scholar] [CrossRef]
- Mo, M.; Tong, S.; Yin, H.; Jin, Z.; Zu, X.; Hu, X. SHCBP1 regulates STAT3/c-Myc signaling activation to promote tumor progression in penile cancer. Am. J. Cancer Res. 2020, 10, 3138–3156. [Google Scholar]
- Saucier, C.; Khoury, H.; Lai, K.M.; Peschard, P.; Dankort, D.; Naujokas, M.A.; Holash, J.; Yancopoulos, G.D.; Muller, W.J.; Pawson, T.; et al. The Shc adaptor protein is critical for VEGF induction by Met/HGF and ErbB2 receptors and for early onset of tumor angiogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 2345–2350. [Google Scholar] [CrossRef]
- Mani, K.; Deng, D.; Lin, C.; Wang, M.; Hsu, M.L.; Zaorsky, N.G. Causes of death among people living with metastatic cancer. Nat. Commun. 2024, 15, 1519. [Google Scholar] [CrossRef]
- Zhang, G.Y.; Ma, Z.J.; Wang, L.; Sun, R.F.; Jiang, X.Y.; Yang, X.J.; Long, B.; Ye, H.L.; Zhang, S.Z.; Yu, Z.Y.; et al. The Role of Shcbp1 in Signaling and Disease. Curr. Cancer Drug Targets 2019, 19, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Li, L.; Zhao, H.; Li, Z.; Ma, Z.; Gu, Q.; Ye, H.; Jiang, X.; Dong, Y.; Qin, L.; et al. Targeting SHCBP1 Inhibits Tumor Progression by Restoring Ciliogenesis in Ductal Carcinoma. Cancer Res. 2024, 84, 4156–4172. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hu, J.F.; Zhan, Q.; Wang, Z.W.; Li, G.; Pan, J.J.; Huang, L.; Liao, C.Y.; Huang, Y.; Tian, Y.F.; et al. SHCBP1 interacting with EOGT enhances O-GlcNAcylation of NOTCH1 and promotes the development of pancreatic cancer. Genomics 2021, 113, 827–842. [Google Scholar] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef]
- Im, Y.K.; Najyb, O.; Gravel, S.P.; McGuirk, S.; Ahn, R.; Avizonis, D.Z.; Chenard, V.; Sabourin, V.; Hudson, J.; Pawson, T.; et al. Interplay between ShcA Signaling and PGC-1alpha Triggers Targetable Metabolic Vulnerabilities in Breast Cancer. Cancer Res. 2018, 78, 4826–4838. [Google Scholar] [CrossRef]
- Zeng, L.; Wu, S.; Li, Z.; Tang, Y.; Tan, Y.; Liang, R.; Li, Y. A novel mitochondrial quality regulation gene signature for anticipating prognosis, TME, and therapeutic response in LUAD by multi-omics analysis and experimental verification. Cancer Cell Int. 2025, 25, 138. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Wen, Y.; Zhu, Y.; Zhang, C.; Yang, X.; Gao, Y.; Li, M.; Yang, H.; Liu, T.; Tang, H. Chronic inflammation, cancer development and immunotherapy. Front. Pharmacol. 2022, 13, 1040163. [Google Scholar] [CrossRef]
- Jiang, F.; Shi, Y.; Wang, Y.; Ge, C.; Zhu, J.; Fang, H.; Zhang, Y.; Zhang, Y.; Jian, H.; Lei, T.; et al. Characterization of SHCBP1 to prognosis and immunological landscape in pan-cancer: Novel insights to biomarker and therapeutic targets. Aging 2023, 15, 2066–2081. [Google Scholar] [CrossRef]
- Oshi, M.; Katsuta, E.; Yan, L.; Ebos, J.M.L.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. A Novel 4-Gene Score to Predict Survival, Distant Metastasis and Response to Neoadjuvant Therapy in Breast Cancer. Cancers 2020, 12, 1148. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Z.; Zhang, C.; Ouyang, J.; Zhang, G.; Wu, C. A four-gene signature in the tumor microenvironment that significantly associates with the prognosis of patients with breast cancer. Gene 2020, 761, 145049. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, X.; Lv, M.; Sun, E.; Lu, X.; Lu, C. Genes That Predict Poor Prognosis in Breast Cancer via Bioinformatical Analysis. Biomed. Res. Int. 2021, 2021, 6649660. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, G.D.; Bai, P.; Su, L.; Tian, H.; He, M. Eight hub genes as potential biomarkers for breast cancer diagnosis and prognosis: A TCGA-based study. World J. Clin. Oncol. 2022, 13, 675–688. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, J. Construction and Validation of a Prognostic Model Based on mRNAsi-Related Genes in Breast Cancer. Comput. Math. Methods Med. 2022, 2022, 6532591. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Lu, L.; Dai, T.; Li, A.; Yu, Y.; Li, Y.; Xu, Z.; Chen, Y. Construction of a lncRNA-mediated ceRNA network and a genomic-clinicopathologic nomogram to predict survival for breast cancer patients. Cancer Biomark. 2023, 36, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, L.; Wang, Y.; Huo, J.; Wang, J.; Xue, H.; Cai, Y. Identification of gene signatures associated with lactation for predicting prognosis and treatment response in breast cancer patients through machine learning. Sci. Rep. 2025, 15, 13575. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Gu, Q.; Dai, Y.; Wang, Q.; Shi, W.; Jiao, Z. Therapeutic potential of SHCBP1 inhibitor AZD5582 in pancreatic cancer treatment. Cancer Sci. 2024, 115, 820–835. [Google Scholar] [CrossRef]
- Shi, H.; Xu, H.; Chai, C.; Qin, Z.; Zhou, W. Integrated bioinformatics analysis of potential biomarkers for pancreatic cancer. J. Clin. Lab. Anal. 2022, 36, e24381. [Google Scholar] [CrossRef]
- Cao, J.; Yu, C. Identification of Immune Infiltration and Prognostic Biomarkers in Small Cell Lung Cancer Based on Bioinformatic Methods from 3 Studies. Comb. Chem. High. Throughput Screen. 2023, 26, 507–516. [Google Scholar]
- Gong, Z.; Yu, F.; Li, C.; Zhao, B.; Wen, M.; Zhang, S.; Xu, Z.; Wu, A.; Zang, R.; Li, Y.; et al. Four-gene Prognostic Signature and Risk of Brain Metastasis of Lung Adenocarcinoma. Mol. Carcinog. 2025, 64, 1209–1221. [Google Scholar] [CrossRef]
- Dai, Y.; Hu, C.; Zhou, H.; Liu, W.; Lai, W.; Xu, R.; Liao, J.; Wang, J.; Li, G.; Zhang, R. Rucaparib inhibits lung adenocarcinoma cell proliferation and migration via the SHCBP1/CDK1 pathway. FEBS J. 2023, 290, 5720–5743. [Google Scholar] [CrossRef]
- Li, Y.; Shi, R.; Zhu, G.; Chen, C.; Huang, H.; Gao, M.; Xu, S.; Cao, P.; Zhang, Z.; Wu, D.; et al. Construction of a circular RNA-microRNA-messenger RNA regulatory network of hsa_circ_0043256 in lung cancer by integrated analysis. Thorac. Cancer 2022, 13, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhang, S.; Xie, H.; Su, Q.; He, S.; Lei, Z. Prognosis and immunotherapy significances of a cancer-associated fibroblasts-related gene signature in lung adenocarcinoma. Cell. Mol. Biol. 2023, 69, 51–61. [Google Scholar] [PubMed]
- Long, J.; Bai, Y.; Yang, X.; Lin, J.; Yang, X.; Wang, D.; He, L.; Zheng, Y.; Zhao, H. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2019, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Moghimi, A.; Bani Hosseinian, N.; Mahdipour, M.; Ahmadpour, E.; Miranda-Bedate, A.; Ghorbian, S. Deciphering the Molecular Complexity of Hepatocellular Carcinoma: Unveiling Novel Biomarkers and Therapeutic Targets Through Advanced Bioinformatics Analysis. Cancer Rep. 2024, 7, e2152. [Google Scholar] [CrossRef]
- Rahimi-Farsi, N.; Shahbazi, T.; Ghorbani, A.; Mottaghi-Dastjerdi, N.; Yazdani, F.; Mohseni, P.; Guzzi, P.H.; Esmail Nia, G.; Shahbazi, B.; Ahmadi, K. Network-based analysis of candidate oncogenes and pathways in hepatocellular carcinoma. Biochem. Biophys. Rep. 2025, 43, 102086. [Google Scholar] [CrossRef]
- Xie, S.; Jiang, X.; Zhang, J.; Xie, S.; Hua, Y.; Wang, R.; Yang, Y. Identification of significant gene and pathways involved in HBV-related hepatocellular carcinoma by bioinformatics analysis. PeerJ 2019, 7, e7408. [Google Scholar] [CrossRef]
- Li, G.; Wang, Z.; Chen, D.; Yin, J.; Mo, Z.; Sun, B.; Yang, T.; Zhang, X.; Zhai, Z.; Li, Y.; et al. Comprehensive analysis of a TPX2-related TRHDE-AS1/PKIA ceRNA network involving prognostic signatures in Hepatitis B virus-infected hepatocellular carcinoma. Front. Cell. Infect. Microbiol. 2022, 12, 1025900. [Google Scholar] [CrossRef]
- Balraj, A.S.; Muthamilselvan, S.; Raja, R.; Palaniappan, A. PRADclass: Hybrid Gleason Grade-Informed Computational Strategy Identifies Consensus Biomarker Features Predictive of Aggressive Prostate Adenocarcinoma. Technol. Cancer Res. Treat. 2024, 23, 1–20. [Google Scholar] [CrossRef]
- Li, C.; Pu, B.; Gu, L.; Zhang, M.; Shen, H.; Yuan, Y.; Liao, L. Identification of key modules and hub genes in glioblastoma multiforme based on co-expression network analysis. FEBS Open Bio 2021, 11, 833–850. [Google Scholar] [CrossRef]
- Qin, S.; Yuan, Y.; Liu, H.; Pu, Y.; Chen, K.; Wu, Y.; Su, Z. Identification and characterization of sex-dependent gene expression profile in glioblastoma. Neuropathology 2023, 43, 72–83. [Google Scholar] [CrossRef]
- Zhang, X.; Mo, Y.; Feng, L. Ginsenoside Rh7 affects β-catenin nuclear translocation by inhibiting SHCBP1 expression, thereby inhibiting epithelial-mesenchymal transition in gastric cancer cells. Int. J. Med. Sci. 2025, 22, 3053–3069. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhu, L.; Wang, L.; Shen, Z.; Huang, X. Identification of SHCBP1 as a potential biomarker involving diagnosis, prognosis, and tumor immune microenvironment across multiple cancers. Comput. Struct. Biotechnol. J. 2022, 20, 3106–3119. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Rastogi, A.; Jain, M.; Niveriya, K. RNA-based Therapeutics: Past, Present and Future Prospects, Challenges in Cancer Treatment. Curr. Pharm. Biotechnol. 2024, 25, 2125–2137. [Google Scholar] [CrossRef] [PubMed]
- Kara, G.; Calin, G.A.; Ozpolat, B. RNAi-based therapeutics and tumor targeted delivery in cancer. Adv. Drug Deliv. Rev. 2022, 182, 114113. [Google Scholar] [CrossRef]
- Kalaimani, K.; Balachandran, S.; Boopathy, L.K.; Roy, A.; Jayachandran, B.; Sankaranarayanan, S.; Arumugam, M.K. Recent advancements in small interfering RNA based therapeutic approach on breast cancer. Eur. J. Pharmacol. 2024, 981, 176877. [Google Scholar] [CrossRef]
- Kim, W.; Ye, Z.; Simonenko, V.; Shahi, A.; Malikzay, A.; Long, S.Z.; Xu, J.J.; Lu, A.; Horng, J.H.; Wu, C.R.; et al. Codelivery of TGFβ and Cox2 siRNA inhibits HCC by promoting T-cell penetration into the tumor and improves response to Immune Checkpoint Inhibitors. NAR Cancer 2024, 6, zcad059. [Google Scholar]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Chunarkar-Patil, P.; Kaleem, M.; Mishra, R.; Ray, S.; Ahmad, A.; Verma, D.; Bhayye, S.; Dubey, R.; Singh, H.N.; Kumar, S. Anticancer Drug Discovery Based on Natural Products: From Computational Approaches to Clinical Studies. Biomedicines 2024, 12, 201. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; International Natural Product Sciences Taskforce; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Camps-Fajol, C.; Cavero, D.; Minguillon, J.; Surralles, J. Targeting protein-protein interactions in drug discovery: Modulators approved or in clinical trials for cancer treatment. Pharmacol. Res. 2025, 211, 107544. [Google Scholar]
- Bian, H.; Liang, X.; Lu, D.; Lin, J.; Lu, X.; Jin, J.; Zhang, L.; Wu, Y.; Chen, H.; Zhang, W.; et al. In Silico Discovery of Stapled Peptide Inhibitor Targeting the Nur77-PPARgamma Interaction and Its Anti-Breast-Cancer Efficacy. Adv. Sci. 2024, 11, e2308435. [Google Scholar]
- Bonazzi, S.; d’Hennezel, E.; Beckwith, R.E.J.; Xu, L.; Fazal, A.; Magracheva, A.; Ramesh, R.; Cernijenko, A.; Antonakos, B.; Bhang, H.C.; et al. Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy. Cell Chem. Biol. 2023, 30, 235–247 e212. [Google Scholar] [CrossRef]
- Ryan, M.B.; Quade, B.; Schenk, N.; Fang, Z.; Zingg, M.; Cohen, S.E.; Swalm, B.M.; Li, C.; Ozen, A.; Ye, C.; et al. The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers. Cancer Discov. 2024, 14, 1190–1205. [Google Scholar]
- Chirnomas, D.; Hornberger, K.R.; Crews, C.M. Protein degraders enter the clinic—A new approach to cancer therapy. Nat. Rev. Clin. Oncol. 2023, 20, 265–278. [Google Scholar] [PubMed]
- Hinterndorfer, M.; Spiteri, V.A.; Ciulli, A.; Winter, G.E. Targeted protein degradation for cancer therapy. Nat. Rev. Cancer 2025, 25, 493–516. [Google Scholar] [CrossRef] [PubMed]
- Rossari, F.; Minutolo, F.; Orciuolo, E. Past, present, and future of Bcr-Abl inhibitors: From chemical development to clinical efficacy. J. Hematol. Oncol. 2018, 11, 84. [Google Scholar] [CrossRef] [PubMed]
- Batista-Silva, J.P.; Gomes, D.; Sousa, S.F.; Sousa, A.; Passarinha, L.A. Advances in structure-based drug design targeting membrane protein markers in prostate cancer. Drug Discov. Today 2024, 29, 104130. [Google Scholar] [CrossRef]
- Medvedev, K.E.; Schaeffer, R.D.; Grishin, N.V. Leveraging AI to explore structural contexts of post-translational modifications in drug binding. J. Cheminform. 2025, 17, 67. [Google Scholar] [CrossRef]
- Krokidis, M.G.; Koumadorakis, D.E.; Lazaros, K.; Ivantsik, O.; Exarchos, T.P.; Vrahatis, A.G.; Kotsiantis, S.; Vlamos, P. AlphaFold3: An Overview of Applications and Performance Insights. Int. J. Mol. Sci. 2025, 26, 3671. [Google Scholar] [CrossRef]
- Ferrari, V.; Mograbi, B.; Gal, J.; Milano, G. Companion Tests and Personalized Cancer Therapy: Reaching a Glass Ceiling. Int. J. Mol. Sci. 2024, 25, 9991. [Google Scholar] [CrossRef]
- Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. The role of companion diagnostics in the development and use of mutation-targeted cancer therapies. Nat. Biotechnol. 2006, 24, 985–995. [Google Scholar] [CrossRef]

| Targeting Strategy | Therapeutic Agent | Mode of Action | Experimental Model | Therapeutic Effect | Reference |
|---|---|---|---|---|---|
| Genetic suppression | siRNA/ shRNA | Transient gene silencing | Cancer cell lines, Xenograft | Inhibition of cellular proliferation, colony formation, self-renewal, and metastasis Induction of apoptosis | [14,15,43,46,50,51,56,63] |
| CRISPR-Cas9 | Genomic disruption | Cancer cell lines, Xenograft | Decreased tumor progression, suppressed colony formation, and reduced lung metastasis | [13,77] | |
| Genetically engineered knockout mouse | Attenuated tumor growth and diminished lung metastasis Restoration of ciliogenesis or enhancement of T cell infiltration | [24,62] | |||
| Low-molecular-weight compound | AZD5582 | SHCBP1-specific inhibitor | Pancreatic cancer cell, organoid model, Xenograft model | Suppression of cellular proliferation and metastatic potential, Induction of apoptotic pathways | [77] |
| Rucaparib | PARP enzyme inhibitor | Lung cancer cell line, Xenograft model | Decreased cellular proliferation, inhibition of EMT progression, and suppressed metastasis | [81] | |
| Natural product | TFBG | Inhibitor targeting SHCBP1-PLK1 interaction | Gastric cancer cell, Xenograft model | Suppressed colony formation and tumor development, Enhanced drug responsiveness | [39] |
| Resveratrol | SHCBP1 transcriptional repressor | Melanoma cell | Suppression of cellular proliferation, colony formation, and invasive capacity | [44] | |
| Ginsenoside Rh7 | SHCBP1 transcriptional repressor | Gastric cancer cell, Xenograft | Diminished cellular proliferation, colony formation, EMT, and invasiveness | [92] | |
| Combination therapy | siRNA/ ICG-001 | Inhibitor of CBP- β-catenin | Lung cancer cell, Xenograft model | Inhibition of sphere formation, stem cell properties, tumor progression, and cell viability | [21] |
| siRNA/ Chloroquine | Inhibitor of autophagy | Ovarian cancer cell xenograft | Decreased cellular proliferation within tumors, Enhanced sensitivity to cisplatin | [52] | |
| shRNA/ Etoposide | DNA-damaging agent | Lung cancer cell, Xenograft model | Suppressed cell cycle progression and diminished tumor growth, Enhanced apoptosis and cellular senescence | [29] | |
| Erdafitinib/ αPD-1 | FGFR inhibitor/ ICI | Conditional SHCBP1 knockout mouse | Suppression of CAF infiltration and breast cancer progression, Enhanced T cell infiltration | [24] | |
| Trametinib/ αPD-1 | MEK inhibitor/ ICI | Breast cancer cell, Xenograft model | Decreased tumor volume Augmented infiltration of CD4/8+ cells | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-Y.; Park, Y.-J.; Ryu, S.; Hong, S. The Roles of SHCBP1 in Cancer Hallmarks: Molecular Mechanisms and Therapeutic Implications. Int. J. Mol. Sci. 2025, 26, 8778. https://doi.org/10.3390/ijms26188778
Kim H-Y, Park Y-J, Ryu S, Hong S. The Roles of SHCBP1 in Cancer Hallmarks: Molecular Mechanisms and Therapeutic Implications. International Journal of Molecular Sciences. 2025; 26(18):8778. https://doi.org/10.3390/ijms26188778
Chicago/Turabian StyleKim, Hye-Youn, Ye-Jin Park, Soyeon Ryu, and Suntaek Hong. 2025. "The Roles of SHCBP1 in Cancer Hallmarks: Molecular Mechanisms and Therapeutic Implications" International Journal of Molecular Sciences 26, no. 18: 8778. https://doi.org/10.3390/ijms26188778
APA StyleKim, H.-Y., Park, Y.-J., Ryu, S., & Hong, S. (2025). The Roles of SHCBP1 in Cancer Hallmarks: Molecular Mechanisms and Therapeutic Implications. International Journal of Molecular Sciences, 26(18), 8778. https://doi.org/10.3390/ijms26188778






