NeuroDecon: A Neural Network-Based Method for Three-Dimensional Deconvolution of Fluorescent Microscopic Images
Abstract
1. Introduction
2. Results
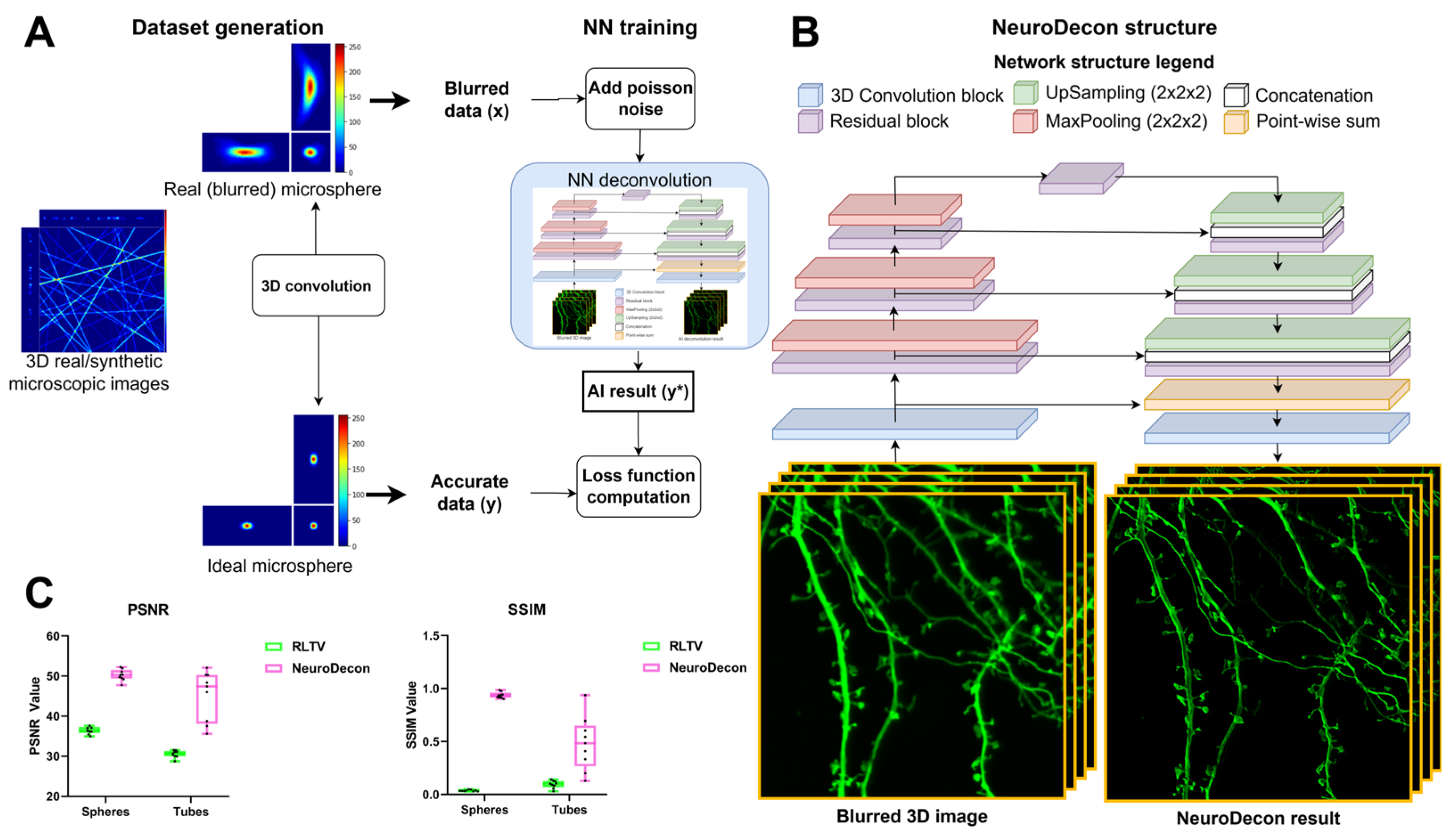
2.1. NeuroDecon Neuronal Network Architecture, Dataset Generation and Training Strategy
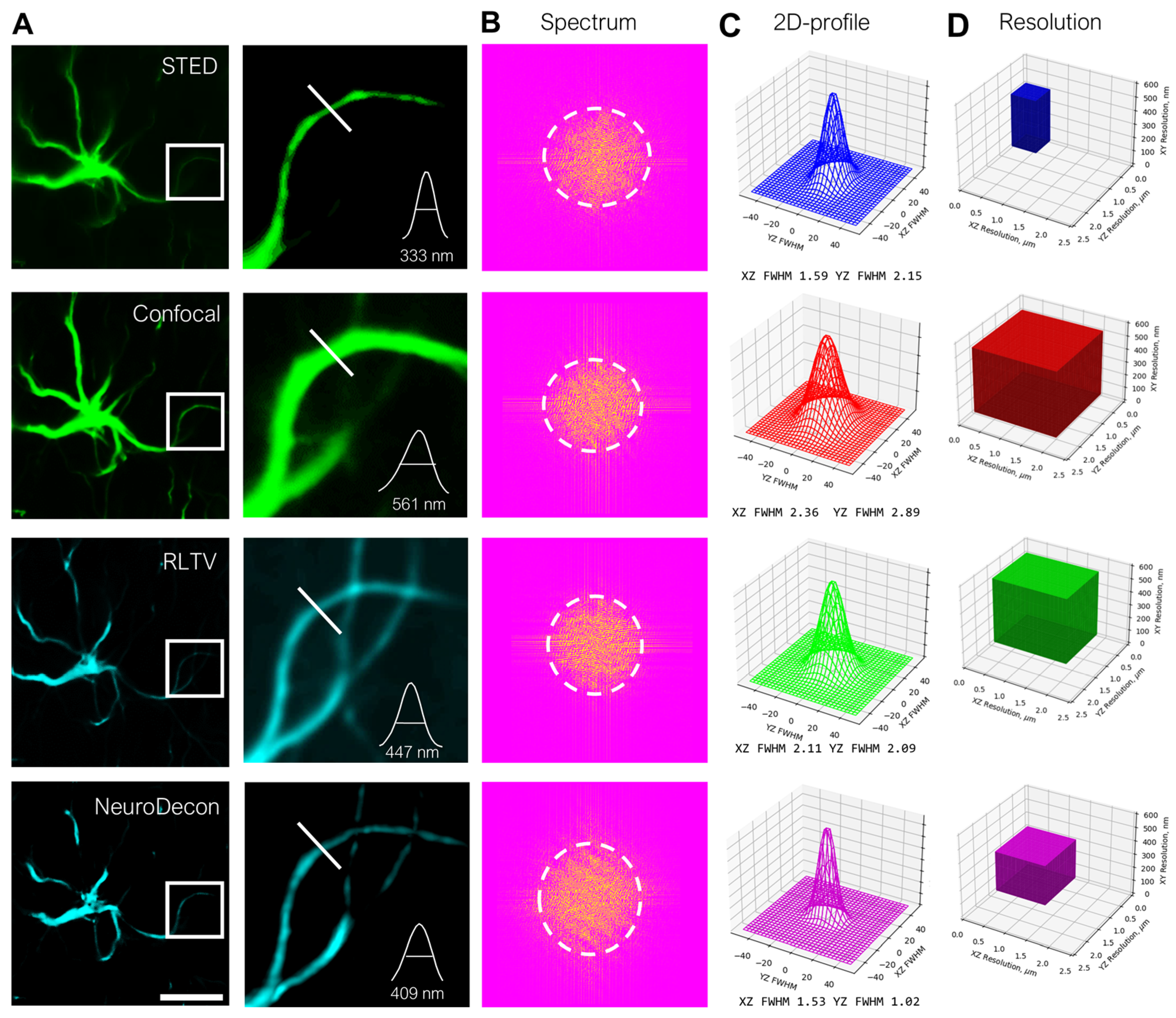
2.2. NeuroDecon Resolution Enhancement of Confocal Images Is Comparable with STED Super Resolution Microscopy
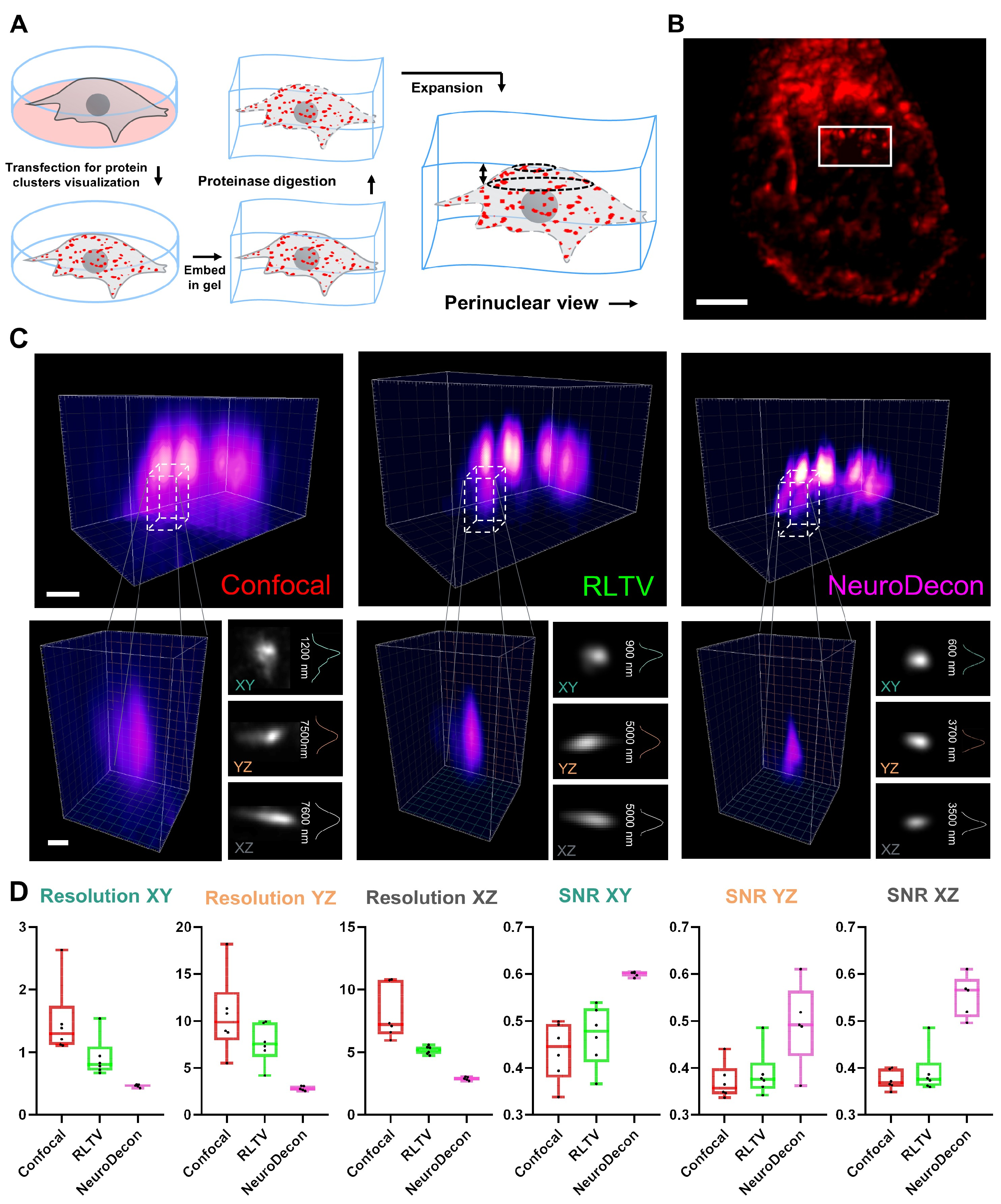
2.3. NeuroDecon Improves Resolution and Reduces Noise in Expansion Microscopy
2.4. NeuroDecon Improves Live- and Fixed- Cell Confocal Image Quality to Reveal Intricate Organelle Structure
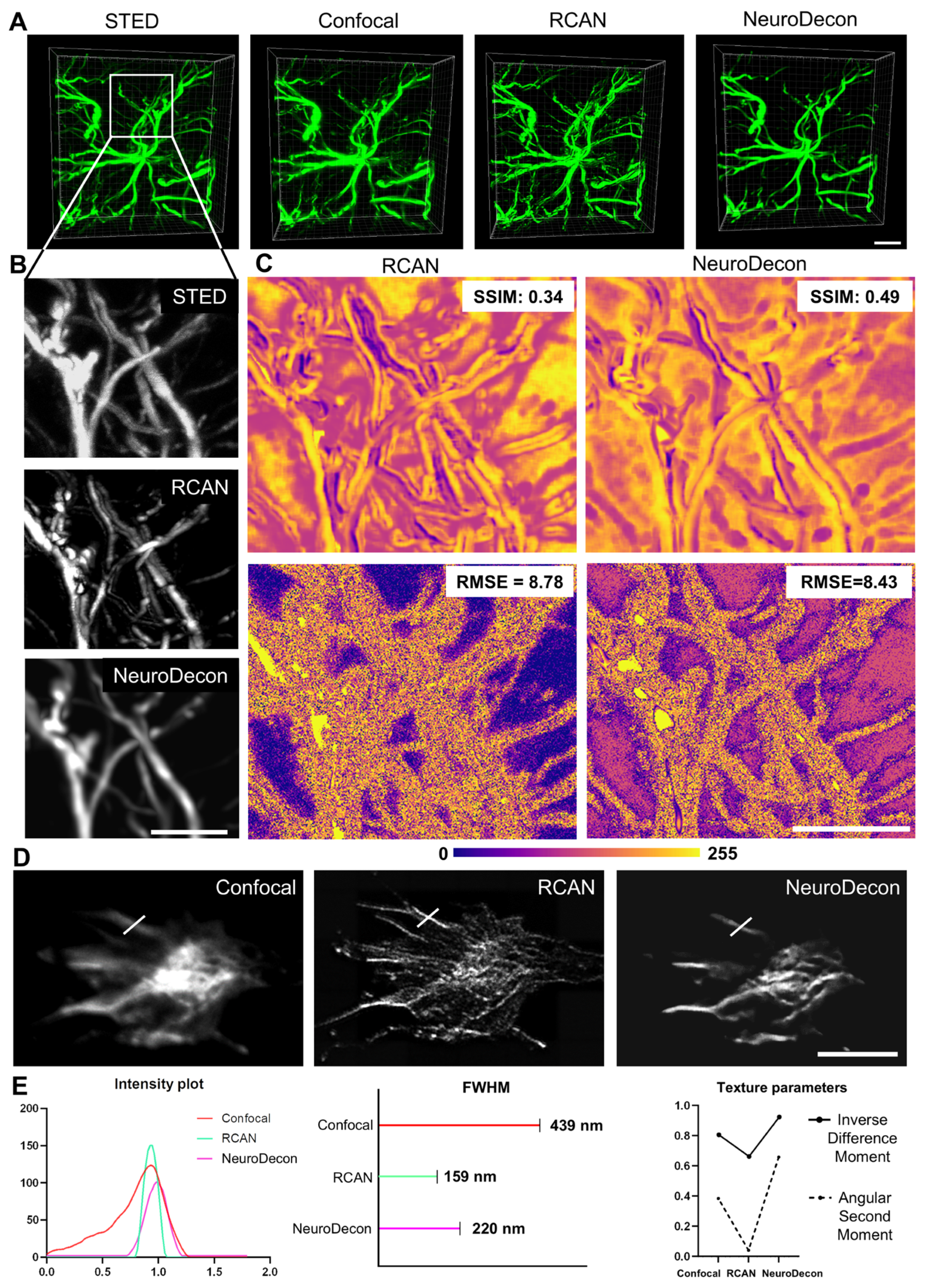
2.5. NeuroDecon and Similar Existing Deep Learning Method for Image Restoration
3. Discussion
4. Materials and Methods
4.1. Image Synthesis
4.1.1. Spheres Synthesis
4.1.2. Tubes Synthesis
4.1.3. Accurate Bead Calculation
4.1.4. Blurred Bead Calculation
4.2. Model Architecture and Training
4.2.1. Model Description
4.2.2. Dataset Generation Mathematical Justification
4.2.3. Dataset Generation Procedure
4.2.4. Blurred Bead Calculation
4.3. Other Deconvolution Methods
4.3.1. Richardson–Lucy Total Variation
4.3.2. 3D-RCAN
4.4. Sample Preparation
4.5. Image Quality Assessment
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PSF | Point Spread Function |
| RLN | Richardson–Lucy Network |
| RLTV | Richardson–Lucy Total Variation |
| PSNR | Peak Signal-to-Noise Ratio |
| SSIM | Structure Similarity |
| STED | Stimulated Emission Depletion Microscopy |
| ExM | Expansion Microscopy |
| ER | Endoplasmic Reticulum |
| ASM | Angular Second Moment |
| ANOVA | Analysis of Variance |
References
- Sibarita, J.B. Deconvolution microscopy. Adv. Biochem. Eng. Biotechnol. 2005, 95, 201–243. [Google Scholar]
- Lucy, L.B. An iterative technique for the rectification of observed distributions. Astron. J. 1974, 79, 745. [Google Scholar] [CrossRef]
- Richardson, W.H. Bayesian-Based Iterative Method of Image Restoration. J. Opt. Soc. Am. 1972, 62, 55–59. [Google Scholar] [CrossRef]
- Kirshner, H.; Sage, D.; Unser, M. 3D PSF Models for Fluorescence Microscopy in ImageJ. In Proceedings of the Twelfth International Conference on Methods and Applications of Fluorescence Spectroscopy, Imaging and Probes, Strasbourg, French, 11 September 2011. [Google Scholar]
- Laasmaa, M.; Vendelin, M.; Peterson, P. Application of regularized Richardson–Lucy algorithm for deconvolution of confocal microscopy images. J. Microsc. 2011, 243, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Su, Y.; Guo, M.; Han, X.; Liu, J.; Vishwasrao, H.D.; Li, X.; Christensen, R.; Sengupta, T.; Moyle, M.W.; et al. Incorporating the image formation process into deep learning improves network performance. Nat. Methods 2022, 19, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Lecture Notes in Computer Science, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; Volume 9351. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tillberg, P.W.; Boyden, E.S. Expansion microscopy. Science 2015, 347, 543–548. [Google Scholar] [CrossRef]
- Chozinski, T.J.; Halpern, A.R.; Okawa, H.; Kim, H.-J.; Tremel, G.J.; Wong, R.O.L.; Vaughan, J.C. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 2016, 13, 485–488. [Google Scholar] [CrossRef]
- Chang, J.-B.; Chen, F.; Yoon, Y.-G.; Jung, E.E.; Babcock, H.; Kang, J.S.; Asano, S.; Suk, H.-J.; Pak, N.; Tillberg, P.W.; et al. Iterative expansion microscopy. Nat. Methods 2017, 14, 593–599. [Google Scholar] [CrossRef]
- Jensen, E.C. Overview of Live-Cell Imaging: Requirements and Methods Used. Anat. Rec. 2013, 296, 1–8. [Google Scholar] [CrossRef]
- Rakovskaya, A.; Volkova, E.; Bezprozvanny, I.; Pchitskaya, E. Hippocampal dendritic spines store-operated calcium entry and endoplasmic reticulum content is dynamic microtubule dependent. Sci. Rep. 2025, 15, 1314. [Google Scholar] [CrossRef] [PubMed]
- Pain, C.; Kriechbaumer, V.; Kittelmann, M.; Hawes, C.; Fricker, M. Quantitative analysis of plant ER architecture and dynamics. Nat. Commun. 2019, 10, 984. [Google Scholar] [CrossRef] [PubMed]
- Culley, S.; Albrecht, D.; Jacobs, C.; Pereira, P.M.; Leterrier, C.; Mercer, J.; Henriques, R. Quantitative mapping and minimization of super-resolution optical imaging artifacts. Nat. Methods 2018, 15, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Laine, R.F.; Tosheva, K.L.; Gustafsson, N.; Gray, R.D.M.; Almada, P.; Albrecht, D.; Risa, G.T.; Hurtig, F.; Lindås, A.-C.; Baum, B.; et al. NanoJ: A high-performance open-source super-resolution microscopy toolbox. J. Phys. D Appl. Phys. 2019, 52, 163001. [Google Scholar]
- Pchitskaya, E.; Vasiliev, P.; Smirnova, D.; Chukanov, V.; Bezprozvanny, I. SpineTool is an open-source software for analysis of morphology of dendritic spines. Sci. Rep. 2023, 13, 10561. [Google Scholar] [CrossRef]
- Chen, J.; Sasaki, H.; Lai, H.; Su, Y.; Liu, J.; Wu, Y.; Zhovmer, A.; Combs, C.A.; Rey-Suarez, I.; Chang, H.-Y.; et al. Three-dimensional residual channel attention networks denoise and sharpen fluorescence microscopy image volumes. Nat. Methods 2021, 18, 678–687. [Google Scholar] [CrossRef]
- Wang, H.; Rivenson, Y.; Jin, Y.; Wei, Z.; Gao, R.; Günaydın, H.; Bentolila, L.A.; Kural, C.; Ozcan, A. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 2019, 16, 103–110. [Google Scholar]
- Qiao, C.; Li, D.; Guo, Y.; Liu, C.; Jiang, T.; Dai, Q.; Li, D. Evaluation and development of deep neural networks for image super-resolution in optical microscopy. Nat. Methods 2021, 18, 194–202. [Google Scholar] [CrossRef]
- Weigert, M.; Schmidt, U.; Boothe, T.; Müller, A.; Dibrov, A.; Jain, A.; Wilhelm, B.; Schmidt, D.; Broaddus, C.; Culley, S.; et al. Content-aware image restoration: Pushing the limits of fluorescence microscopy. Nat. Methods 2018, 15, 1090–1097. [Google Scholar] [CrossRef]
- Qiao, C.; Zeng, Y.; Meng, Q.; Chen, X.; Chen, H.; Jiang, T.; Wei, R.; Guo, J.; Fu, W.; Lu, H.; et al. Zero-shot learning enables instant denoising and super-resolution in optical fluorescence microscopy. Nat. Commun. 2024, 15, 4180. [Google Scholar]
- Goodwin, P.C. Quantitative deconvolution microscopy. Methods Cell Biol. 2014, 123, 177–192. [Google Scholar] [PubMed]
- Guo, M.; Wu, Y.; Hobson, C.M.; Su, Y.; Qian, S.; Krueger, E.; Christensen, R.; Kroeschell, G.; Bui, J.; Chaw, M.; et al. Deep learning-based aberration compensation improves contrast and resolution in fluorescence microscopy. Nat. Commun. 2025, 16, 313. [Google Scholar] [CrossRef] [PubMed]
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Conference on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Sage, D.; Donati, L.; Soulez, F.; Fortun, D.; Schmit, G.; Seitz, A.; Guiet, R.; Vonesch, C.; Unser, M. DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods 2017, 115, 28–41. [Google Scholar] [CrossRef]
- Rakovskaya, A.; Chigriai, M.; Bezprozvanny, I.; Pchitskaya, E. Expansion Microscopy Application for Calcium Protein Clustering Imaging in Cells and Brain Tissues. Curr. Protoc. 2023, 3, e789. [Google Scholar] [CrossRef]
- Ustinova, A.; Volkova, E.; Rakovskaya, A.; Smirnova, D.; Korovina, O.; Pchitskaya, E. Generate and Analyze Three-Dimensional Dendritic Spine Morphology Datasets with SpineTool Software. Curr. Protoc. 2024, 4, e70061. [Google Scholar] [CrossRef]
- Descloux, A.; Grußmayer, K.S.; Radenovic, A. Parameter-free image resolution estimation based on decorrelation analysis. Nat. Methods 2019, 16, 918–924. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sachuk, A.; Volkova, E.; Rakovskaya, A.; Chukanov, V.; Pchitskaya, E. NeuroDecon: A Neural Network-Based Method for Three-Dimensional Deconvolution of Fluorescent Microscopic Images. Int. J. Mol. Sci. 2025, 26, 8770. https://doi.org/10.3390/ijms26188770
Sachuk A, Volkova E, Rakovskaya A, Chukanov V, Pchitskaya E. NeuroDecon: A Neural Network-Based Method for Three-Dimensional Deconvolution of Fluorescent Microscopic Images. International Journal of Molecular Sciences. 2025; 26(18):8770. https://doi.org/10.3390/ijms26188770
Chicago/Turabian StyleSachuk, Alexander, Ekaterina Volkova, Anastasiya Rakovskaya, Vyacheslav Chukanov, and Ekaterina Pchitskaya. 2025. "NeuroDecon: A Neural Network-Based Method for Three-Dimensional Deconvolution of Fluorescent Microscopic Images" International Journal of Molecular Sciences 26, no. 18: 8770. https://doi.org/10.3390/ijms26188770
APA StyleSachuk, A., Volkova, E., Rakovskaya, A., Chukanov, V., & Pchitskaya, E. (2025). NeuroDecon: A Neural Network-Based Method for Three-Dimensional Deconvolution of Fluorescent Microscopic Images. International Journal of Molecular Sciences, 26(18), 8770. https://doi.org/10.3390/ijms26188770






