Abstract
Liquid biopsy has emerged as a valuable tool for the detection and monitoring of colorectal cancer (CRC), providing minimally invasive insights into tumor biology through circulating biomarkers such as circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). Additional biomarkers, including tumor-educated platelets (TEPs) and exosomal RNAs, offer further potential for early detection and prognostic role, although ongoing clinical validation is still needed. This review summarizes the current evidence on the diagnostic, prognostic, and predictive capabilities of liquid biopsy in both metastatic and non-metastatic CRC. In the non-metastatic setting, liquid biopsy is gaining traction in early detection through screening and in identifying minimal residual disease (MRD), potentially guiding adjuvant treatment and reducing overtreatment. In contrast, liquid biopsy is more established in metastatic CRC for monitoring treatment responses, clonal evolution, and mechanisms of resistance. The integration of ctDNA-guided treatment algorithms into clinical practice could optimize therapeutic strategies and minimize unnecessary interventions. Despite promising advances, challenges remain in assay standardization, early-stage sensitivity, and the integration of multi-omic data for comprehensive tumor profiling. Future efforts should focus on enhancing the sensitivity of liquid biopsy platforms, validating emerging biomarkers, and expanding multi-omic approaches to support more targeted and personalized treatment strategies across CRC stages.
1. Introduction
Colorectal cancer (CRC) is among the most prevalent gastrointestinal malignancies and represents the second leading cause of cancer-related mortality worldwide [1,2,3]. Lifestyle factors such as poor diet and physical inactivity significantly contribute to its global burden [4]. CRC typically arises through the stepwise progression of colorectal adenomas to invasive carcinoma, often resulting in late-stage diagnosis and limited treatment options [5]. Prognosis is stage-dependent, with five-year survival rates decreasing from over 90% in stage I to 14% in stage IV [6,7,8], emphasizing the need for effective early detection strategies.
Conventional diagnostic methods, such as colonoscopy, imaging, biopsy with histological examination, and tumor markers, face limitations including invasiveness, limited sensitivity in early disease, and challenges related to tumor heterogeneity. Non-invasive tools such as the fecal immunochemical test (FIT) offer greater accessibility but lack adequate sensitivity for early-stage CRC [9]. Novel stool-based biomarkers, including DNA [10], mRNA [11], proteins [12], and microbiota profiles [13], are under development, yet their clinical utility remains constrained by high costs, variable adherence, and limited sensitivity for precancerous lesions.
Liquid biopsy has emerged as a promising non-invasive modality for tumor detection and monitoring. It enables the analysis of circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), tumor-educated platelets (TEPs), exosomes, and tumor-associated proteins from blood or other body fluids [14,15]. This approach supports repeated sampling, real-time molecular profiling, and longitudinal assessment of tumor dynamics, aligning with the principles of precision oncology. In CRC, liquid biopsy holds significant potential for early detection, post-surgical surveillance, and individualized treatment planning, especially in patients with unresectable disease. It also offers valuable insights into therapeutic response, resistance mechanisms, and disease progression (Figure 1).
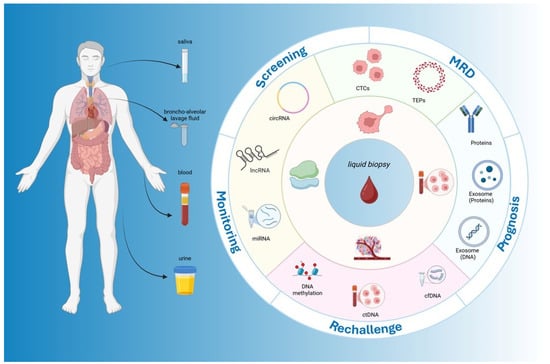
Figure 1.
Clinical applications of liquid biopsy in colorectal cancer. The figure illustrates the sources, analytes, and clinical applications of liquid biopsy in CRC. On the left, different body fluids (blood, urine, saliva, and bronchoalveolar lavage fluid) are depicted as collection matrices. The central circle highlights the key analytes, including circulating tumor DNA (ctDNA), cell-free DNA (cfDNA), microRNAs (miRNAs), long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), proteins, tumor-educated platelets (TEPs), and exosomes. The outer ring summarizes the major clinical applications across the CRC continuum: screening, minimal residual disease (MRD) detection, prognosis, rechallenge and treatment selection, and monitoring. This schematic figure underscores the multidimensional utility of liquid biopsy by integrating diverse analytes and specimen types at various decision-making points throughout CRC management.
This review explores the application of liquid biopsy in CRC, focusing on its biological relevance, detection technologies, areas of applications, current limitations, and future clinical integration.
1.1. Historical Development of Liquid Biopsy: From Discovery to Clinical Translation
The evolution of liquid biopsy can be categorized into four key phases: the exploratory stage (pre-1990s), foundational research (1990s), technological and industrial expansion (2000–2010), and clinical translation and integration (2010–present) [16].
In the exploratory phase, pivotal discoveries laid the groundwork for modern liquid biopsy. In 1869, Ashworth identified malignant cell-like elements in the blood, now recognized as CTC [17]. In 1948, Mandel and Métais detected cell-free DNA (cfDNA) in plasma [18], while Wolf provided the first electron micrographs of extracellular vesicles (EVs) in 1967 [19]. In the early 1980s, Johnstone et al. described exosome biogenesis [20], and in 1977 Leon et al. found elevated cfDNA levels in cancer patients, linking them to tumor burden [21].
Refined methodologies, with polymerase chain reaction (PCR), allowed the detection of KRAS mutations in cfDNA from pancreatic cancer patients in 1994 [22]. In 1996, Raposo et al. demonstrated antigen presentation by EVs from immune cells [23], and in 1998 CTC isolation was correlated with tumor stage and prognosis [24].
The early 2000s brought significant technological advances. In 2005, CTC enumeration predicted survival in metastatic breast cancer [25]. In 2008, Diehl et al. used BEAMing (beads, emulsion, amplification, and magnetics) to track ctDNA mutations in CRC, correlating them with tumor burden and carcinoembryonic antigen (CEA) serum levels [26].
Since 2010, clinical applications have expanded. In 2014, the European Medicines Agency (EMA) approved plasma ctDNA testing for epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) [27], followed by the U.S. Food and Drug Administration (FDA) approval of the Epi proColon® (Berlin, Germany) test for CRC screening in 2016 [28]. The FDA also authorized ctDNA-based PIK3CA testing for hormone receptor-positive/human epidermal growth factor receptor 2-negative (HR+/HER2−) breast cancer [29] and BRCA1/2 mutation detection for poly ADP-ribose polymerase (PARP) inhibitor use in metastatic castration-resistant prostate cancer (mCRPC) [30].
Despite regulatory limitations, clinical use of liquid biopsy continues to grow, driven by robust translational research and ongoing clinical trials.
1.2. Molecular Biomarkers in Liquid Biopsy: Features, Advantages, Limitations, and Detection Techniques
Liquid biopsy provides a minimally invasive method for obtaining molecular data from blood, urine, and saliva in real time, encompassing CTCs, ctDNA, exosomes, TEPs, and non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) [31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97]. A comparative summary of the main circulating biomarkers, including detection methods, sensitivity, specificity, and clinical applications, is provided in Table 1.

Table 1.
Molecular biomarkers in liquid biopsy. CTCs: circulating tumor cells; ctDNA: circulating tumor DNA; ddPCR: digital droplet polymerase chain reaction; qPCR: quantitative polymerase chain reaction; NGS: next-generation sequencing; MRD: minimal residual disease; EpCAM: epithelial cell adhesion molecule; EMT: epithelial–mesenchymal transition; TEPs: tumor-educated platelets; RNA-seq: RNA sequencing; RT-qPCR: reverse transcription quantitative polymerase chain reaction; RNA-FISH: RNA fluorescence in situ hybridization; lncRNAs: long non-coding RNAs; circRNAs: circular RNAs.
CTCs, first identified by Ashworth in 1869 [31], are tumor cells that detach from primary or metastatic lesions and enter the bloodstream, correlating with metastasis and prognosis in cancers such as breast and C [32,33,34,35]. Due to their low concentration (less than 10 cells per 10 mL of blood) and heterogeneity, detection methods involve enrichment and immunofluorescent labeling [36,37]. The CellSearch® system, the only U.S. FDA-approved platform for CTC detection, uses epithelial cell adhesion molecule (EpCAM)-coated beads, but it may miss CTCs undergoing epithelial–mesenchymal transition (EMT), affecting sensitivity [38,39,40,41,42,43,44].
ctDNA, a small fraction of tumor-derived DNA, reflects the tumor’s mutational landscape and constitutes 0.1–1% of cfDNA [45,46,47]. Detection of ctDNA involves techniques such as digital droplet PCR (ddPCR), next-generation sequencing (NGS), and quantitative PCR (qPCR) [48,49,50,51,52]. However, ctDNA detection is challenging in patients with low tumor burden, and its fragmented nature may increase false negatives [53,54,55,56].
Exosomes, extracellular vesicles (30–150 nm), carry proteins, nucleic acids and lipids, reflecting the tumor microenvironment (TME) and protecting cargo from enzymatic degradation [57,58,59,60,61]. Isolation methods include ultracentrifugation, size-exclusion chromatography, and microfluidics, though protocol variability affects consistency [62,63,64,65,66,67,68].
TEPs absorb RNAs, proteins and vesicles from tumor cells, with RNA profiles potentially indicating tumor type, location, and mutation status [69,70]. RNA sequencing (RNA-seq) is the primary detection method but, despite promising accuracy in distinguishing cancer patients from controls, TEPs remain investigational and lack FDA-approved assays [71,72,73,74].
ncRNAs, including miRNAs, lncRNAs, and circRNAs, are emerging biomarkers [75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96]. miRNAs (18–25 nucleotides) regulate gene expression and are detected via qPCR, but hemolysis may impact results [75,76]. lncRNAs (over 200 nucleotides) regulate gene expression and chromatin structure, detectable via RNA fluorescence in situ hybridization (RNA-FISH), reverse transcription qPCR (RT-qPCR), and RNA-seq, though their variability complicates interpretation [77,78,79,80,81,82,83,84,85,86,87,88,89]. circRNAs (200–600 nucleotides), characterized by a covalently closed-loop structure, act as miRNA sponges and transcriptional regulators and are detectable by divergent primer RT-PCR and RNA-seq, though standardization is still needed [90,91,92,93,94,95,96,97].
Despite advancements, liquid biopsy faces challenges in standardization, sensitivity, and cost. Further research is necessary to optimize detection methods, validate biomarker specificity, and establish clinical guidelines to enable broader clinical adoption.
1.3. Clinical Applications of Liquid Biopsy in Colorectal Cancer
In recent years, liquid biopsy has rapidly evolved from a promising research tool to a clinically relevant strategy in the management of CRC [98]. Its minimally invasive nature, combined with the ability to provide dynamic molecular insights, offers significant advantages over traditional tissue-based approaches. In the next chapters, we explore the key clinical applications of liquid biopsy across three major settings of CRC management: screening, where early detection can significantly impact patient outcomes; localized disease, where liquid biopsy can assist in risk stratification, detection of MRD, and guidance of adjuvant therapy; and metastatic disease, where it plays a crucial role in treatment selection, monitoring response, and identifying mechanisms of resistance. Together, these applications underscore the transformative potential of liquid biopsy in achieving more personalized and adaptive care in CRC (Figure 1).
2. Screening
The primary goal of CRC screening is early diagnosis to improve patient outcomes. Commonly used methods include FIT, which has a sensitivity of 74% and specificity of 96%; the high-sensitivity guaiac-based fecal occult blood test (HSgFOBT), with 70% sensitivity and 93% specificity; and colonoscopy which offers 95% sensitivity and 86% specificity [99]. However, these methods have limitations, particularly the lower sensitivity of FIT and HSgFOBT and the low patient compliance with invasive procedures like colonoscopy [99,100,101]. Developing less invasive, more accessible diagnostic tests remains a key clinical need [102]. Liquid biopsy, detecting circulating biomarkers in blood, offers a potential solution to overcome these limitations [101]. The role of CTCs in the early diagnosis of CRC remains limited, due to the low sensitivity observed in several studies [103,104]. Therefore, this chapter will primarily focus on the role of circulating nucleic acids and other biomarkers (Table 2).

Table 2.
Overview of CRC screening methods: conventional and liquid biopsy approaches. ctDNA: circulating tumor DNA; cfDNA: cell-free DNA; miRNA: microRNA; lncRNA: long non-coding RNA; circRNA: circular RNA; FIT: fecal immunochemical test; HSgFOBT: high-sensitivity guaiac-based fecal occult blood test; SEPT9: Septin 9 (gene involved in colorectal cancer); SFRP2: Secreted Frizzled-Related Protein 2; SDC2: Syndecan-2; IGFBP-2: Insulin-like Growth Factor Binding Protein 2; PKM2: Pyruvate Kinase M2; DKK-3: Dickkopf WNT Signaling Pathway Inhibitor 3.
2.1. Circulating Cell-Free DNA and Circulating Tumor DNA
Circulating cfDNA, released into the bloodstream through apoptosis and necrosis, has emerged as a key tool for molecular assessment of CRC and other cancers [135,136,137]. A small fraction of cfDNA, known as ctDNA, carries tumor-specific genetic alterations, including somatic mutations, structural variants, copy-number changes, and microsatellite instability, enabling non-invasive tumor monitoring [138]. The potential of ctDNA testing lies in detecting malignancy-associated genetic signatures, addressing limitations of conventional CRC screening [139]. However, in early-stage CRC, low ctDNA levels increase the risk of false negatives [140]. Mutational heterogeneity further complicates ctDNA analysis, as certain mutations may not be universally present [141]. To enhance sensitivity, emerging approaches focus on DNA methylation markers, fragmentomics, and proteomics to detect even minimal tumor DNA more effectively [140].
The EpiProColon® assay, FDA-approved in 2016, targets SEPT9 gene methylation for CRC screening, achieving 74% specificity and 96% sensitivity in a meta-analysis of 3202 CRC patients and 7284 controls [105,106,107]. Subsequent studies in younger populations reported 96.3% specificity and 90.8% sensitivity, but limited sample size (27 CRC patients, 87 controls) constrains broader applicability [108]. Beyond SEPT9, the SpecColon test targets SFRP2 and SDC2 methylation, showing promise in identifying CRC-associated epigenetic changes [109]. Other potential markers include IKZF1, BCAT1, NTRK3, SRFP1-2, EHD3, TMEM240, SMAD3, and VIM, though further validation is necessary to confirm clinical utility and integration into routine CRC screening workflows [142,143].
The available data show that approximately three out of four FIT+ patients are negative for diagnostic endoscopic evaluation. A recent Italian study evaluated the role of cfDNA in identifying FIT+ patients at higher risk of having a positive colonoscopy. This trial enrolled 711 FIT+ patients, who subsequently underwent both the QuantiDNA™ (Pleasanton, CA, USA) test and colonoscopy. It was found that the QuantiDNA™ (Pleasanton, CA, USA) test is effective in reducing the need for colonoscopies by approximately 33.4%, proving to be non-inferior to the standard of care in the early identification of colorectal lesions [110].
2.2. Non-Coding RNAs
Recent studies highlight the potential of miRNAs as non-invasive biomarkers for CRC due to their dual role as oncogenes and tumor suppressors in processes like proliferation, apoptosis, angiogenesis, and metastasis [111,144]. Synthesized in the nucleus and present in extracellular fluids like blood and serum, miRNAs are released via secretory mechanisms, mainly in extracellular vesicles (exosomes) or passively from damaged cells [112]. Their stability in bodily fluids enhances their diagnostic and prognostic value, enabling early cancer detection and monitoring [111,112]. Techniques such as qRT-PCR, microarray analysis, and NGS facilitate the identification of tumor-specific miRNA expression profiles relevant to CRC diagnosis [101].
The growing interest in miRNAs as early CRC biomarkers has led to studies on miR-21, which showed promising diagnostic accuracy with 77% sensitivity and 83% specificity [113,114,115,116]. Other miRNAs, including miR-210, miR-144-3p, and miR-1246, have also been identified as potential CRC biomarkers, though further validation is needed [115,117,118,119,120].
Research indicates that miRNA panels combining multiple markers often outperform single miRNAs in diagnostic accuracy. For instance, a panel comprising miR-193a-5p, miR-210, miR-513a-5p, and miR-628-3p achieved an AUC of 0.92, with 90% sensitivity and 80% specificity [118]. However, due to the heterogeneous expression of miRNAs, single-marker approaches are less reliable, and comprehensive panels may more accurately reflect tumor-specific profiles [121,145].
LncRNAs, detectable in blood, plasma, serum, and urine, exhibit significant diagnostic potential due to their stability in bodily fluids and resistance to RNase degradation [122,123,124,125]. They can circulate within apoptotic bodies, microvesicles, and exosomes, facilitating their transport and protecting them from degradation [126]. Aberrant lncRNA expression can influence key oncogenic pathways such as WNT/β-catenin, PI3K/Akt, EGFR, NOTCH, mTOR, and TP53, linking them to CRC progression [127]. Several circulating lncRNAs have been investigated as potential CRC biomarkers. Notably, CCAT1 and HOTAIR are consistently reported as elevated in CRC patients compared to healthy controls, suggesting their potential role in early diagnosis [128]. Additionally, other lncRNAs such as BLACAT1, CRNDE, NEAT1, and UCA1 have also demonstrated diagnostic potential, with some identified within exosomes, indicating a role in intercellular communication and metastasis [146,147]. Exosomal forms of 91H, CRNDE-h, UCA1, TUG1, and multiple LNCV6 transcripts (e.g., LNCV6_116109, LNCV6_98390) are under investigation as potential components of multi-marker panels for improved diagnostic accuracy [146,147].
CircRNAs, characterized by stable covalently closed-loop structures, exhibit resistance to exonuclease degradation [129]. They play roles in gene transcription and splicing regulation and are gaining attention as potential biomarkers for early CRC detection. Exosomal circ_0004771, significantly upregulated in CRC patients and reduced post surgery, suggests potential for early diagnosis and monitoring [130]. Although circRNA research is still emerging, their structural stability positions them as promising diagnostic and prognostic markers. Currently, the only FDA-approved lncRNA-based test, PCA3, is available for prostate cancer, highlighting the potential for similar circRNA-based diagnostics in CRC [131].
2.3. Other Biomarkers
Insulin-like growth factors (IGF-1 and IGF-2) promote tumor growth and act as mitogens for colon mucosa, advancing cancer progression [132]. Insulin-like growth factor-binding protein 2 (IGFBP-2), which binds IGF-2, is linked to cancer metastasis through its interaction with HSP27 [133]. Elevated serum IGFBP-2 levels in CRC patients correlate with neoplastic changes and increased CEA concentrations, indicating potential for early diagnosis and monitoring, though its standalone sensitivity and specificity are limited [134].
Pyruvate Kinase M2 (PKM2), a glycolytic enzyme, is overexpressed in CRC, gastric cancer, and adenomas [148]. While it shows high sensitivity as a blood and fecal biomarker, its specificity remains suboptimal when used alone [149].
Dickkopf-3 (DKK3), part of the tumor endothelial marker family, is frequently silenced in CRC due to promoter hypermethylation [150]. Although its role is not fully defined, DKK3 is implicated in angiogenesis and tumor progression, suggesting its potential as a CRC biomarker [151,152].
Individually, IGFBP-2, PKM2, and DKK3 exhibit limited diagnostic accuracy, but a combined panel has demonstrated improved performance, with 57% sensitivity for early-stage CRC, 76% for advanced stages, and 95% specificity, underscoring its potential as a non-invasive blood-based diagnostic tool [134].
3. Localized CRC
Following surgical treatment, adjuvant chemotherapy is recommended for patients with stage II-III CRC. However, the existing risk stratification factors primarily rely on clinical and pathological parameters, which, though important, do not fully encompass all the nuances necessary for optimal patient management. Consequently, the possibility of overtreatment or undertreatment in patients with early-stage CRC remains an ongoing concern. Therefore, the identification of supplementary diagnostic tools to refine patient selection and guide subsequent therapeutic decisions remains an unmet clinical need. From this perspective, identifying biomarkers that can detect MRD represents an important goal [99,153,154].
3.1. Detection of MRD in CRC: Plasma-Only vs. Tumor-Informed Assay
The detection of MRD in CRC is a rapidly evolving field with significant implications for post-surgical management and adjuvant treatment decision-making. MRD refers to residual tumor cells that persist after primary treatment and may lead to recurrence. Liquid biopsy, particularly through ctDNA analysis, has emerged as a promising non-invasive method for detecting MRD, providing molecular-level insights not achievable through conventional imaging [155,156,157,158].
Two main methodologies exist for ctDNA-based MRD detection: plasma-only and tumor-informed assays. Plasma-only assays, also known as tumor-agnostic assays, analyze cfDNA from blood samples without prior tissue sequencing [155]. These assays target predefined gene panels commonly mutated in CRC, such as KRAS, TP53, and APC [159,160,161]. Examples of commercial plasma-only assays include Guardant Reveal™ (Redwood City, CA, USA) and AVENIO ctDNA Surveillance Kit V2™, which utilize targeted sequencing and fragmentomics to enhance MRD detection [162,163]. Fragmentomics leverages differences in fragment size and integrity to distinguish ctDNA from normal cfDNA, improving the detection of low-frequency mutations [164,165].
While plasma-only assays offer a simpler workflow without tumor sampling, they may miss mutations specific to a patient’s tumor, increasing the risk of false negatives. Additionally, their sensitivity can be affected by ctDNA shedding rates, which vary by tumor size, location, and biological characteristics, particularly in tumors with low proliferative capacity or poor vascularization [158,160,165].
In contrast, tumor-informed assays adopt a personalized approach by first sequencing the primary tumor to identify patient-specific mutations [166,167]. These mutations are then targeted in subsequent cfDNA samples, allowing more sensitive MRD detection, especially in cases with low ctDNA shedding [155,166,168]. However, these assays require initial tumor tissue sampling, which may not always be feasible, and may miss new mutations arising after the biopsy [158,166].
The choice between plasma-only and tumor-informed assays depends on factors such as tumor biology, disease stage, and clinical context. Plasma-only assays may suit early-stage CRC with high ctDNA shedding, while tumor-informed assays offer greater sensitivity in advanced or low-shedding cases by targeting patient-specific mutations [157,158,165]. The timing of blood collection is crucial for MRD detection accuracy, as ctDNA levels can fluctuate in the first weeks after surgery [169,170]. Blood samples are typically collected five to eight weeks after surgery to minimize the detection of DNA released due to surgical trauma rather than residual disease [169,170,171]. Testing before initiating adjuvant chemotherapy can also provide a baseline for treatment response and recurrence monitoring.
Despite their potential, both plasma-only and tumor-informed assays face challenges, including low ctDNA concentration, high cfDNA background, and false positives due to clonal hematopoiesis. Advances in NGS, fragmentomics, and epigenetic analysis may further refine MRD detection, enhancing both sensitivity and specificity. Integrating multi-omic approaches could provide a more comprehensive assessment of residual disease, supporting personalized treatment strategies in CRC [155,172,173].
3.2. Patient Selection, Treatment Modulation, and Follow-Up in Non-Metastatic Colon Cancer
Several studies have evaluated the prognostic role of ctDNA as a marker of MRD in CRC, aiming to guide adjuvant therapy (Table 3).

Table 3.
Clinical applications of ctDNA in localized colorectal cancer MRD: minimal residual disease; ctDNA: circulating tumor DNA; HR: hazard ratio; OS: overall survival; DFS: disease-free survival; RFS: relapse-free survival; CRC: colorectal cancer; TTR: time to recurrence; ASCO: American Society of Clinical Oncology.
Among these, a prospective study by Reinert et al., involving 130 patients with stage I–III CRC, detected preoperative ctDNA in 88.5% of cases [174]. Post treatment, ctDNA analysis identified 87.5% of relapses. On postoperative day 30, ctDNA-positive patients had a 7-fold increased risk of relapse (HR: 7.2; 95% CI: 2.7–19.0; p < 0.001), which rose to 17-fold following adjuvant chemotherapy (HR: 17.5; 95% CI: 5.4–56.5; p < 0.001). Post-treatment surveillance revealed that ctDNA-positive patients had a 43.5-fold increased recurrence risk (HR: 43.5; 95% CI: 9.8–193.5; p < 0.001). During post-treatment surveillance, patients who tested positive for ctDNA were over 40 times more likely to experience disease recurrence than ctDNA-negative patients (HR: 43.5; 95% CI: 9.8–193.5; p < 0.001). In multivariate analyses, ctDNA status remained an independent predictor of relapse, even after adjusting for established clinicopathologic risk factors. Serial ctDNA assessments detected recurrences up to 16.5 months earlier than radiologic imaging, with a mean lead time of 8.7 months [174].
The CIRCULATE-Japan project, comprising the observational GALAXY study and phase III VEGA and ALTAIR trials, assessed ctDNA-guided therapy in stages II–IV of CRC [175,176]. A 2023 analysis involving 2280 patients showed that MRD-positive (MRD+) patients had worse disease-free survival (DFS) at 24 months (HR: 16.9, p < 0.0001) and benefited from adjuvant chemotherapy (HR: 0.4, p < 0.0001). Among those receiving 6 months of adjuvant chemotherapy, DFS significantly improved (HR: 0.4, p = 0.002) [177]. An update presented at the ASCO 2024 Annual Meeting revealed that ctDNA-positive patients who achieved clearance had a significantly lower relapse risk (HR: 5.4; 95% CI: 3.58–7.67; p < 0.0001). Among 181 ctDNA+ patients treated with adjuvant therapy, 72.9% achieved ctDNA clearance, with 54% maintaining clearance and 46% experiencing re-emergence. Notably, patients with sustained clearance demonstrated markedly better outcomes (HR: 32.57; 95% CI: 9.94–106.76; p < 0.0001) [178].
The DYNAMIC II trial assessed ctDNA-guided therapy in stage II CRC, enrolling 455 patients randomized 2:1 to ctDNA-guided treatment or standard care (SOC). Only 15% of patients in the ctDNA arm received adjuvant chemotherapy compared to 28% in the SOC group, without compromising 3-year relapse-free survival (3 y RFS: 91.7% vs. 92.5%) [179]. However, chemotherapy regimens differed, with oxaliplatin-based therapy predominating in the ctDNA group and single-agent fluoropyrimidine in the control group, representing a study limitation. Another limitation was that ctDNA-negative patients with resected T4 tumor had recurrence rate similar to ctDNA-positive patients, suggesting that ctDNA alone may not be sufficient to assess recurrence risk in stage II colon cancer [179].
At the ASCO 2025 Annual Meeting, the DYNAMIC-III trial confirmed the prognostic value of postoperative ctDNA in stage III colon cancer. Patients with detectable ctDNA showed a high risk of recurrence. However, ctDNA-guided escalation of adjuvant chemotherapy, including FOLFOXIRI, did not improve RFS, underscoring the urgent need for alternative therapeutic strategies in this high-risk subgroup [180].
The PEGASUS trial assessed ctDNA-guided treatment adjustments in stage III and T4N0 stage II CRC patients. Preliminary results showed that 7% of ctDNA-negative patients relapsed after treatment de-escalation, mainly in the peritoneum and lungs, associated with low ctDNA shedding. Among ctDNA-positive patients treated with CAPOX and subsequently switched to FOLFIRI, 46% achieved ctDNA clearance. These findings offer encouraging early evidence for the potential of ctDNA-guided therapeutic modifications [181].
The COBRA trial focused on ctDNA clearance after adjuvant chemotherapy in the ctDNA+ cohort, but phase II was terminated early due to a high rate of false positives in interim analysis [182].
At the ASCO Gastrointestinal Cancers Symposium 2025, results from the CALGB/SWOG 80702 trial confirmed ctDNA as a strong prognostic marker in stage III colon cancer and suggested its potential as a predictive biomarker that would benefit from adding celecoxib to adjuvant FOLFOX chemotherapy [183].
Ongoing trials, including CIRCULATE-US, AFFORD, DYNAMIC-III, and CLAUDIA, are investigating intensified chemotherapy regimens like FOLFOXIRI in MRD-positive patients (NCT05174169, NCT05427669, ACTRN126170015, NCT05534087). Concurrently, phase II trials FANTASTIC and AURORA are exploring FOLFOXIRI and FOLFOXIRI plus bevacizumab in resected oligometastatic CRC [184,185]. Additionally, several studies aim to validate ctDNA for monitoring recurrence and identifying candidates for further intervention in post-adjuvant therapy [186,187,188,189,190].
3.3. Liquid Biopsy in Non-Metastatic Rectal Cancer
Preoperative chemoradiotherapy followed by total mesorectal excision (TME) is the standard treatment for non-metastatic rectal adenocarcinoma [189,191]. Recently, total neoadjuvant treatment (TNT) has shown superiority in pathological complete response (pCR) and overall survival (OS), as demonstrated in the PRODIGE 23 trial [192,193,194]. In this context, ctDNA may serve as a predictive marker for treatment response and guide postoperative therapy adjustments (Table 4).

Table 4.
ctDNA in non-metastatic rectal cancer: predictive and prognostic roles. LARC: locally advanced rectal cancer; ctDNA: circulating tumor DNA; OS: overall survival; RFS: relapse-free survival; TNT: total neoadjuvant therapy; pCR: pathological complete response.
Appelt et al. reported that ctDNA-positive patients had worse outcomes in terms of OS compared to ctDNA-negative patients [195]. In a prospective study by Murahashi et al., ctDNA was detected in 57.6% of baseline samples and 22.3% of samples post treatment in 85 patients with locally advanced rectal cancer (LARC). ctDNA clearance from baseline to post-neoadjuvant treatment was associated with higher pCR rates (p = 0.0276). Postoperative ctDNA positivity was linked to worse RFS, especially when combined with CEA analysis (ctDNA, p = 0.0127; CEA, p = 0.0105) [196].
A meta-analysis by Chang et al. further supported these findings, showing that post-neoadjuvant ctDNA positivity was associated with worse RFS (HR: 9.16; 95% CI: 5.48–15.32), OS (HR: 8.49; 95% CI: 2.20–32.72), and lower pCR rates (OR: 0.40; 95% CI: 0.18–0.89) [197]. Additional studies confirmed ctDNA as a predictive and prognostic marker in this setting [198,199,200].
Tie et al. also demonstrated a significant association between ctDNA positivity, post-neoadjuvant therapy, surgery, and worse RFS (HR: 6.6 and 13.0, respectively) [201].
Building on these data, the ongoing DYNAMIC RECTAL trial is assessing the role of ctDNA in guiding adjuvant chemotherapy decisions in LARC. Patients are randomized to SOC or ctDNA-guided chemotherapy, with adjuvant chemotherapy use as the primary endpoint and 3-year RFS as a key secondary endpoint. Preliminary ASCO 2024 data from 230 LARC patients showed that 46% of the ctDNA-guided group received adjuvant chemotherapy, compared to 77% in the control group. The 3-year RFS was 76% in the ctDNA-guided group versus 82% in the control group, suggesting potential for reducing overtreatment. Additionally, among ctDNA-negative patients, lung-only recurrences accounted for 80% of relapses, indicating a potential limitation in detecting pulmonary metastases [202]. However, the small sample size and lack of TNT inclusion limit definitive conclusions regarding the non-inferiority of the ctDNA-guided approach.
4. Advanced Disease: Target and Response Evaluation in Metastatic Colorectal Cancer
The use of liquid biopsy in metastatic CRC (mCRC) has transformed the clinical management of this disease, providing a non-invasive means of continuously monitoring tumor dynamics. The decision-making process for first-line treatment in mCRC is highly dependent on the molecular characterization of the tumor. Key biomarkers, including RAS, BRAF, HER2, and mismatch repair/microsatellite instability (MMR/MSI) status, are routinely assessed to guide therapeutic strategies [203]. Traditionally, molecular profiling is conducted through tissue biopsy, but liquid biopsy has emerged as a crucial complementary tool, particularly when tissue samples are unavailable or inadequate. ctDNA, CTCs, exosomal RNA, and extracellular vesicle DNA (evDNA) offer real-time molecular insights, addressing the limitations of spatial and temporal tumor heterogeneity [204].
4.1. Molecular Concordance, Dynamic Profiling, and Prognostic Insights
The primary challenge in integrating liquid biopsy into clinical practice is ensuring its reliability and concordance with tissue-based molecular analyses. Several studies report high concordance rates for RAS and BRAF mutations between liquid and tissue biopsies (80–100%), particularly when NGS and ddPCR techniques are used [205,206,207,208,209]. However, ctDNA detectability and concordance can be influenced by factors such as tumor burden and metastatic site. Patients with liver-dominant disease often show higher ctDNA levels, while those with lung or peritoneal metastases exhibit lower ctDNA shedding, resulting in reduced sensitivity (Se) and specificity (Sp), sometimes as low as 60% [206,210,211,212,213,214].
Chemotherapy prior to ctDNA extraction can lower the variant allele fraction (VAF): higher VAFs are noted in liver metastases and lower in cancer with mucinous differentiation, contributing to discordance [210,213,214]. Additionally, a long interval between tissue and blood sample collection may reduce concordance due to tumor evolution and emerging genetic alterations.
Unlike single-timepoint tissue biopsies, liquid biopsy can capture dynamic molecular changes over time. Some mCRC patients initially classified as RAS-mutant in tissue may later present as RAS wild-type (wt) in ctDNA, a phenomenon known as “RAS conversion” [215,216,217]. Pesola et al. reported RAS conversion rates ranging from 5.5% to 78%, influenced by methodological differences [218]. This shift, potentially due to chemotherapy-induced clonal selection, may allow patients to regain sensitivity to anti-EGFR therapies, underscoring the value of serial ctDNA monitoring for real-time molecular reassessment [219,220,221,222,223,224,225,226,227,228].
High baseline ctDNA levels have been strongly associated with worse prognosis, a trend that persists even in later treatment lines [229,230,231]. Elevated RAS VAF at baseline correlates with poor prognosis, even in cases where tissue biopsy indicates RAS-wt status [232,233,234]. Conversely, patients with RAS mutations in tissue but not in plasma tend to have better survival outcomes [233].
Monitoring ctDNA during treatment can provide early indications of therapeutic response, often detectable as early as two weeks after chemotherapy initiation, much earlier than radiological assessments [230,235,236,237]. Lower ctDNA levels after induction are linked to longer PFS, with ctDNA dynamics reflecting those of traditional tumor markers like CEA [230,238,239,240].
While ctDNA is the most widely used plasma biomarker, CTCs provide comprehensive tumor information and can serve as a platform for cell culture. CTC presence at CRC diagnosis is associated with poor prognosis, especially when expressing mesenchymal markers like vimentin, linked to metastatic disease and decreased DFS [241,242,243,244].
Tumor-derived extracellular vesicles (EVs) also offer a promising source of circulating DNA (evDNA), carrying driver mutations with acceptable sensitivity and high specificity [245,246]. Among epigenetic markers, ctDNA hypermethylation, such as NPY promoter methylation detected via ddPCR, is gaining interest. Higher baseline methylation correlates with poor prognosis, and its decline during treatment mirrors radiological response [247,248].
Exosomes, present in most body fluids, carry diverse nucleic acids, including miRNA, lncRNA, and circRNA [249]. These RNAs are not only useful in distinguishing metastatic from non-metastatic CRC but also hold potential in detecting chemoresistance, as demonstrated for miRNAs [250,251]. Predictive models are being developed, incorporating specific lncRNAs and circRNAs [252,253,254,255,256].
4.2. Key Molecular and Immunohistochemical Targets in mCRC Identifiable Through Liquid Biopsy
4.2.1. Anti-EGFR Therapy: Treatment Selection, Resistance Mechanisms, and Rechallenge Strategies
In RAS-wt, mismatch repair-proficient/microsatellite stable (pMMR/MSS) mCRC, first-line treatment typically involves chemotherapy (fluoropyrimidine plus oxaliplatin or irinotecan) combined with epidermal growth factor receptor inhibitors (EGFRis) like cetuximab or panitumumab [203]. Due to primary resistance to EGFRi conferred by RAS mutations, comprehensive molecular testing is essential prior to treatment initiation.
Liquid biopsy enhances RAS mutation detection by overcoming the spatial and temporal limitations of tissue biopsy. Several studies have shown that ctDNA analysis can more accurately identify RAS mutations in patients initially considered RAS wt [212,234,237,257]. Table 5 summarizes the main studies evaluating the role of liquid biopsy in treatment selection and anti-EGFR rechallenge in mCRC.

Table 5.
Overview of liquid biopsy studies in anti-EGFR therapy for metastatic colorectal cancer. EGFRi: epidermal growth factor receptor inhibitors; ctDNA: circulating tumor DNA; RAS-wt: RAS wild-type; PFS: progression-free survival; OS: overall survival; ORR: objective response rate; BRAF: B-Raf Proto-Oncogene, Serine/Threonine Kinase; MAP2K1: Mitogen-Activated Protein Kinase Kinase 1; VEGFi: Vascular Endothelial Growth Factor Inhibitors; PI3K: Phosphoinositide 3-Kinase; MET: Mesenchymal–Epithelial Transition Factor; BRAF/EGFR: B-Raf Proto-Oncogene, Serine/Threonine Kinase/Epidermal Growth Factor Receptor.
The LIBImAb study presented at ASCO 2023 reported RAS mutations in 9.5% of patients initially deemed RAS-wt by tissue biopsy [258], while the FIRE-4 trial found a 13% discordance, underscoring the importance of baseline ctDNA testing to refine EGFRi therapy [259,260].
Beyond RAS, liquid biopsy enables broader molecular profiling, identifying resistance mechanisms involving ERBB2, EGFR, FGFR1, PIK3CA, PTEN, AKT1, MAP2K1, and gene amplifications in KRAS, ERBB2, MET, and fusions in ALK, ROS1, NTRK1-3, and RET [275,276,277]. The VALENTINO trial identified these alterations as predictors of poor PFS and OS during panitumumab maintenance [261]. Real-time ctDNA monitoring is key to capturing emerging resistance mutations, particularly secondary RAS mutations often missed by tissue biopsy [234,257,278,279]. However, chemotherapy plus EGFRi appears to reduce resistance mechanisms involving ERBB2, BRAF, PIK3CA, MET, FLT3, and MAP2K1 [280,281,282,283,284,285,286].
Adaptive or switch maintenance therapy is being investigated in patients developing resistance mutations during first-line EGFRi treatment, through trials such as MODUL (NCT02291289), Rapid 1 (NCT04786600), and MoLiMoR (NCT04554836). The COPERNIC trial (NCT05487248) is assessing ctDNA dynamics to guide third-line therapy. It features an initial pilot phase to define optimal tumor fraction (TF) thresholds and timing, followed by a randomized phase to evaluate the clinical impact of ctDNA-driven treatment adjustments.
Rechallenge with EGFR inhibitors is also being explored based on the hypothesis that resistant clones may regress after a drug-free interval, potentially restoring drug sensitivity [286,287]. Early rechallenge strategies relied solely on baseline tissue RAS-wt status, disregarding molecular evolution over time [288,289].
In the phase II CRICKET trial, cetuximab plus irinotecan met its objective response rate (ORR) endpoint; however, ctDNA analysis revealed that nearly 50% of patients harbored RAS mutations at rechallenge, limiting responses to RAS-wt cases [262]. The Japanese REMARRY trial refined this approach by requiring ctDNA-confirmed RAS-wt status before rechallenge with panitumumab plus irinotecan. Although the ORR endpoint was not met, longer EGFRi-free intervals were associated with better outcomes [263,264].
The VELO trial showed that panitumumab plus trifluridine/tipiracil provided prolonged benefit in patients with ctDNA-confirmed RAS/BRAF-wt [265]. The CAVE study demonstrated favorable OS with cetuximab plus avelumab, although patient selection was not ctDNA-driven [266].
The CHRONOS trial confirmed the utility of ctDNA-based ultra-selection for panitumumab rechallenge [267]. Similarly, the REMARRY-PURSUIT trial evaluated ctDNA RAS dynamics to guide anti-EGFR rechallenge, reporting a modest ORR of 14% due to the exclusion of BRAF- and EGFR-mutated cases [263].
Mariani et al. highlighted the importance of the EGFRi-free interval, showing that rechallenge after ≥14–16 months was associated with better PFS and OS, supporting the use of both clinical and molecular criteria for patient selection [268].
The CAVE-2 GOIM trial, presented at ASCO GI 2025, employed comprehensive genomic profiling (CGP) via the FoundationOne Liquid CDx platform, identifying acquired RAS or BRAF mutations in 31.4% of patients and resistance alterations in 42%. Notably, patients who were wt for KRAS, NRAS, BRAFV600E, EGFR, ERBB2, MAP2K1, and PIK3CA experienced significantly longer PFS [269].
Recently, the PARERE phase II trial, presented at ESMO GI 2025, evaluated the optimal sequencing of panitumumab and regorafenib in RAS/BRAF-wt mCRC patients selected by ctDNA. Administering panitumumab before regorafenib resulted in improved outcomes, supporting ctDNA-guided rechallenge strategies in chemorefractory settings [272].
Novel strategies to overcome resistance are under investigation. For example, trametinib plus panitumumab was tested in patients with ctDNA-detected RAS, BRAF, or MAP2K1 mutations but was discontinued due to poor tolerability [270]. Similarly, targeting MET amplification with tepotinib plus cetuximab showed promise but faced accrual limitations (NCT04515394) [271].
Interestingly, early studies suggest that VEGF inhibitors (VEGFis) may promote reversion to RAS-wt clones in RAS-mut patients, potentially restoring sensitivity to EGFR inhibitors [290], a hypothesis currently under further investigation.
Efforts are also underway to target actionable mutations beyond RAS. The C-PRECISE-01 trial is selecting patients with ct-DNA detected PIK3CA mutations for treatment with cetuximab plus the PI3K inhibitor MEN1611 [273]. Meanwhile, the OrigAMI-1 trial showed that ctDNA-guided rechallenge with amivantamab in left-sided RAS/BRAF/EGFR-wt mCRC patients resulted in greater clinical benefit when administered after a longer EGFRi-free interval (>8.8 months), highlighting the importance of timing and molecular selection in anti-EGFR re-treatment [274].
Rechallenge strategies are increasingly reliant on ctDNA-based assessment of RAS mutational status. Trials such as PARERE (NCT04787341) [A], CAVE-2 (NCT05291156) [269], CITRIC [291], PULSE (NCT03992456), and CAPRI 2 GOIM (NCT05312398) incorporate NGS-based ctDNA profiling to guide treatment across various lines.
4.2.2. Targeting BRAF-Mutant: Applications and Overcoming Resistance
BRAF-V600E mutations, detected in approximately 10% of mCRC cases, are associated with poor prognosis and resistance to standard chemotherapy [292,293]. Liquid biopsy provides a sensitive, non-invasive method for detecting this alteration, particularly in patients with liver metastases. Recent advancements in NGS have achieved detection rates exceeding 90%, outperforming conventional tissue biopsy [206,212,213,294,295,296].
In BRAF-V600E-mutant mCRC, treatment after progression on standard chemotherapy plus bevacizumab, or after immune checkpoint inhibitors in dMMR/MSI-H tumors, involves dual inhibition of the BRAF and EGFR pathways. The encorafenib–cetuximab regimen is the current standard in this setting [297,298,299,300]. Investigational strategies are exploring BRAF-targeted therapy in earlier lines, either as monotherapy or combined with chemotherapy or immunotherapy for dMMR/MSI-H cases [295,301,302].
Despite these efforts, response rates remain modest, with ORR around 20% and median duration of response under six months. Primary resistance remains a challenge [298]. Identifying resistance mechanisms, both primary and acquired, is crucial to refine patient selection and optimize combination strategies. Liquid biopsy enables real-time monitoring of clonal evolution and emerging resistance [303,304]. Table 6 provides an overview of studies assessing the impact of ctDNA monitoring on treatment outcomes in BRAF-mutant mCRC.

Table 6.
Overview of liquid biopsy on BRAF-targeted therapies in metastatic colorectal cancer. BRAF: serine/threonine-protein kinase B-Raf; BRAF-mut: BRAF-mutated; ctDNA: circulating tumor DNA; CNV: copy number variation; MEK: mitogen-activated protein kinase kinase; MET: mesenchymal epithelial transition factor; MSS: microsatellite stable; OS: overall survival; PFS: progression-free survival; RNF43: ring finger protein 43; VAF: variant allele frequency.
The BEACON trial provided key insights into resistance mechanisms. ctDNA analysis at baseline and progression confirmed BRAF-V600E in nearly 90% of cases and identified acquired alterations in KRAS, NRAS, MAP2K1, and MET amplification [305]. These alterations were observed at similar rates in both the BRAF-EGFR doublet and BRAF-EGFR-MEK triplet arms, with the triplet arm showing slightly fewer MAPK mutations but more pathway-related amplifications. At least one resistance-associated alteration was identified in over 60% of progressing patients, often with multiple concurrent mutations [308,309].
Alterations in DNA repair and PI3K signaling genes also emerged as key resistance mechanisms, with baseline mutations linked to early progression and poor outcomes [308]. RNF43 mutations, particularly in MSS tumors, were associated with better responses to BRAF-targeted therapy, potentially due to WNT pathway inhibition mitigating MAPK-driven progression [305,308,309,310].
Efforts to overcome resistance have led to small-scale rechallenge studies. Ji et al. reported three cases where rechallenge strategies led to responses, including one patient with a rising BRAF-V600E variant allele fraction (VAF) but no new MAPK mutations who achieved an eight-cycle response to encorafenib–cetuximab [306]. Another patient with prior MET amplification responded to vemurafenib–cetuximab–irinotecan following MET amplification decrease post-intervening therapy [306]. Oddo et al. also reported a rechallenge using vemurafenib and crizotinib in a patient with BRAF-V600E and MET amplification, with ctDNA dynamics predicting radiologic progression by eight weeks [307].
Plasma BRAF-V600E VAF has emerged as a key surrogate for tumor burden and prognosis. High baseline VAF (≥2%) correlates with increased metastatic load, liver involvement, and poorer clinical outcomes but also predicts greater benefit from triplet therapy including MEK inhibition [304,309]. In the BEACON trial, lower VAF was associated with longer OS across all treatment arms [293,306]. Ye et al. confirmed that patients with low or undetectable baseline VAF (<5%) had superior PFS and OS, with a >50% decline in VAF during treatment correlating with partial response or disease stabilization [308].
4.2.3. Emerging Biomarkers: HER2, MSI, Exosomal RNA, and Methylation Analysis
The detection of additional driver alterations through liquid biopsy remains underexplored, but the growing use of NGS assays has enhanced the capacity to assess multiple genetic alterations simultaneously, offering a more comprehensive view of tumor heterogeneity. Despite this, concordance between plasma-based and tissue-based analyses varies depending on the specific alteration.
HER2/ERBB2 amplification, typically assessed by immunohistochemistry (IHC) or in situ hybridization (ISH), shows variable concordance with plasma-based copy number variation (CNV) analysis, with agreement rates ranging from 66.7% to 97.9% [311,312]. Nevertheless, ctDNA-based CNV has proven predictive of response to HER2-targeted therapies [313]. Siravegna et al. established a CNV threshold of ≥2.4 copies adjusted for ctDNA fraction, correlating with PFS and objective response in HER2-amplified patients identified by tissue analysis. The DESTINY-CRC01 trial similarly reported that higher HER2 CNV or serum HER2 extracellular domain levels predicted better responses to trastuzumab deruxtecan [314].
ctDNA profiling has also demonstrated predictive value in assessing response to trastuzumab and pertuzumab, with similar accuracy when compared to tissue analysis [315]. A decline in ctDNA levels within three weeks of treatment initiation was associated with favorable outcomes. Meanwhile, ctDNA identified receptor tyrosine kinase (RTK), RAS, or PI3K pathway mutations in 67% of non-responders, compared to 19% in matched tissue samples, underscoring the utility of liquid biopsy in detecting resistance mechanisms.
ERBB2 mutations, primarily linked to resistance to EGFRi, may also emerge during HER2-targeted therapy, though their predictive role remains unclear [315,316,317]. Ongoing phase II studies are investigating this phenomenon [317].
In dMMR/MSI-H mCRC, ctDNA-based MSI detection using ddPCR or NGS demonstrated high accuracy [318,319,320], suggesting its use in clinical practice, particularly when tissue samples are unavailable or when discordant results occur between MSI status and clinical features [321]. Despite lower tumor heterogeneity in this subgroup, discordance between tissue and plasma remains a concern [322,323,324,325].
Moreover, ctDNA enables dynamic monitoring of tumor mutational burden (TMB) and MSI, providing insights into treatment response and resistance in patients receiving immune checkpoint inhibitors (ICIs) [326,327]. This is especially relevant as up to 30% of dMMR/MSI-H patients exhibit primary resistance to immunotherapy [328]. In such cases, liquid biopsy may help identify non-responders earlier, potentially guiding timely treatment adjustments, such as switching to dual checkpoint blockade or ICI–chemotherapy combinations.
A case series by Kasi et al. described three dMMR/MSI-H mCRC patients who progressed on pembrolizumab but responded to nivolumab plus ipilimumab, with ctDNA becoming undetectable shortly after dual ICI therapy initiation [329].
In MSS mCRC, ctDNA has provided insights into immunotherapy mechanisms. In the AVETUX trial, which evaluated FOLFOX plus cetuximab and avelumab in RAS/BRAFwt MSS patients, early ctDNA reduction at four weeks correlated with high response rates. Additionally, the clonality and diversity of peripheral and tumor-infiltrating lymphocytes after three treatment cycles predicted disease control [330,331].
Interestingly, PD-L1 mutations emerged in a subset of responders, likely driven by selective pressure from avelumab, resulting in reduced binding affinity but enhanced T cell-mediated killing [332]. These findings illustrate the potential of liquid biopsy to monitor adaptive changes during immunotherapy and refine biomarker-driven strategies in mCRC.
5. Conclusions
Liquid biopsy has emerged as a promising non-invasive approach for detecting and monitoring CRC, offering molecular insights through ctDNA, CTCs, ncRNAs, and other biomarkers. While its clinical utility is well-established in metastatic CRC, its application in non-metastatic disease, particularly for screening and MRD detection, remains in the exploratory phase.
Based on current evidence, we propose a stage-specific conceptual algorithm for integrating liquid biopsy into CRC management, encompassing screening, MRD detection in localized disease, and real-time monitoring in metastatic settings (Figure 2). This flowchart summarizes the most promising applications reported in the literature and may serve as a reference for future clinical validation.
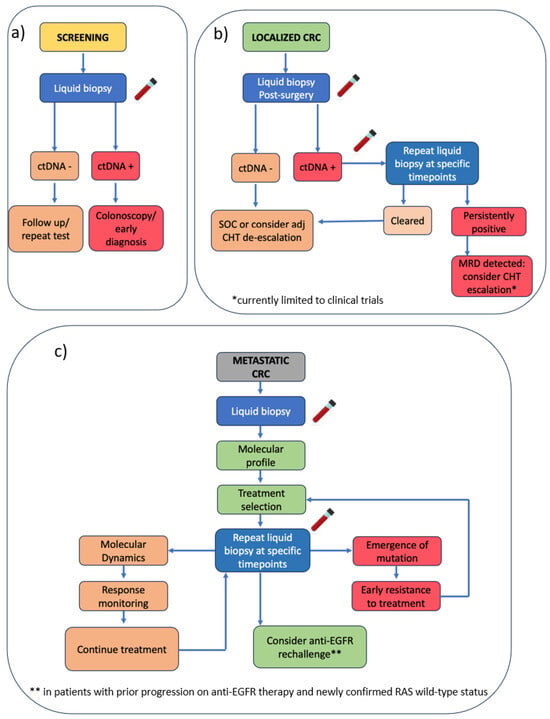
Figure 2.
Proposed stage-specific algorithm for integrating liquid biopsy in CRC management. (a) Screening: liquid biopsy used for early detection; a positive ctDNA result leads to colonoscopy and potential early diagnosis, while a negative test prompts follow-up or repeat testing. (b) Localized CRC: post-surgical ctDNA assessment to guide adjuvant chemotherapy decisions. (c) mCRC: liquid biopsy enables molecular profiling to support treatment selection and monitoring response. CRC = colorectal cancer; ctDNA = circulating tumor DNA; MRD = minimal residual disease; CHT = chemotherapy; adj = adjuvant; EGFR = epidermal growth factor receptor.
In metastatic CRC, liquid biopsy supports real-time treatment monitoring, resistance detection, and tracking of tumor evolution. Technologies such as NGS, dPCR, and methylation-based assays enable the identification of actionable alterations (e.g., RAS, BRAF, HER2), facilitating targeted therapy selection and rechallenge decisions. ctDNA profiling also captures emerging resistance mechanisms, supporting timely therapeutic adaptation. However, implementation is challenged by variability in assay sensitivity, particularly in low-shedding tumors, as well as occasional discordance between tissue and plasma-based results.
In non-metastatic CRC, the role of liquid biopsy is less defined but gaining traction, particularly for MRD detection and adjuvant therapy tailoring. Trials like DYNAMIC II and CIRCULATE-Japan have demonstrated its potential to identify patients at high risk of relapse and to spare low-risk individuals from unnecessary chemotherapy. Nonetheless, most studies to date are single-arm, with limited follow-up and lacking randomized validation. Technical issues, including inconsistent ctDNA shedding and sampling intervals, as well as confounding from clonal hematopoiesis and tumor heterogeneity, further complicate interpretation, and clinical adoption.
Across all stages, the lack of standardized methodologies limits cross-study comparability and slows the development of uniform clinical recommendations. Many rechallenge trials in metastatic CRC, despite being prospective, are constrained by small sample sizes, heterogeneous inclusion criteria (e.g., EGFRi-free interval, molecular profiling strategies), and variable ctDNA platforms.
Despite these limitations, the integration of molecular profiling, adaptive trial designs, and ctDNA monitoring marks a clear shift toward personalized treatment. To fully realize the potential of liquid biopsy in clinical practice, several hurdles must be addressed: the development of analytically and clinically validated assays with high sensitivity and specificity; consensus on sampling intervals and result interpretation; and robust cost-effectiveness analyses. Expanding biomarker panels to include additional resistance pathways and adopting multi-omic frameworks, combining ctDNA with transcriptomic and proteomic data, may further enhance patient stratification and therapy personalization.
Harmonization of protocols for serial sampling, timing, and thresholds is essential to improve reproducibility and facilitate meaningful comparisons across studies. Large, prospective randomized trials using standardized ctDNA methodologies remain the cornerstone for establishing definitive clinical utility.
In parallel, practical and ethical aspects must be addressed. Cost-effectiveness will determine feasibility in resource-limited settings, while inequitable access to advanced diagnostics may widen disparities in care. Additionally, incidental findings, such as germline mutations, raise ethical concerns around patient counseling and data interpretation. Addressing these challenges is critical to ensuring responsible and equitable adoption.
From a regulatory perspective, liquid biopsy is gaining recognition. The U.S. FDA has granted Breakthrough Device designation to multiple ctDNA assays (e.g., Signatera™ by Natera, Austin, TX, USA) for MRD detection in CRC and other cancers [333]. Moreover, both the NCCN and ESMO have begun incorporating ctDNA into clinical guidelines for applications such as molecular profiling in metastatic CRC and MRD-driven therapy decisions [334,335]. However, broader clinical integration depends on the generation of high-quality prospective data and methodological standardization.
In conclusion, liquid biopsy holds great promise as a minimally invasive, dynamic tool across all CRC stages. Overcoming current limitations will be key to making it a cornerstone of precision oncology, enabling more refined, data-driven treatment decisions, and ultimately improving patient outcomes.
Author Contributions
P.Z., D.S., G.S., A.P. and M.S. collectively conceived and designed this comprehensive review. P.Z., A.P., G.S., D.S., C.D., P.A.F., F.C. (Flaviana Cau), A.P.D., M.P. (Monica Piras), S.M., M.P. (Marco Puzzoni), V.P., F.C. (Ferdinando Coghe), G.F., M.S. contributed to the initial draft of the manuscript. P.Z., D.S., G.S., A.P. and M.S. provided supervision, graphics support, editing, and finalization of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data analyzed during this study are included in this published article.
Acknowledgments
We thank all the investigators and patients for participating in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal Cancer Statistics, 2020. Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Chan, A.T.; Sun, J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 2020, 158, 322–340. [Google Scholar] [CrossRef]
- Simon, K. Colorectal Cancer Development and Advances in Screening. Clin. Interv. Aging 2016, 11, 967–976. [Google Scholar] [CrossRef]
- Chetroiu, D.; Pop, C.S.; Filip, P.V.; Beuran, M. How and Why Do We Screen for Colorectal Cancer? J. Med. Life 2021, 14, 462–467. [Google Scholar] [CrossRef]
- Levin, T.R.; Corley, D.A.; Jensen, C.D.; Schottinger, J.E.; Quinn, V.P.; Zauber, A.G.; Lee, J.K.; Zhao, W.K.; Udaltsova, N.; Ghai, N.R.; et al. Effects of Organized Colorectal Cancer Screening on Cancer Incidence and Mortality in a Large Community-Based Population. Gastroenterology 2018, 155, 1383–1391.e5. [Google Scholar] [CrossRef]
- Shaukat, A.; Levin, T.R. Current and future colorectal cancer screening strategies. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 521–531. [Google Scholar] [CrossRef]
- Hanna, M.; Dey, N.; Grady, W.M. Emerging Tests for Noninvasive Colorectal Cancer Screening. Clin. Gastroenterol. Hepatol. 2023, 21, 604–616. [Google Scholar] [CrossRef]
- Imperiale, T.F.; Ransohoff, D.F.; Itzkowitz, S.H. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef]
- Barnell, E.K.; Wurtzler, E.M.; La Rocca, J.; Fitzgerald, T.; Petrone, J.; Hao, Y.; Kang, Y.; Holmes, F.L.; Lieberman, D.A. Multitarget Stool RNA Test for Colorectal Cancer Screening. JAMA 2023, 330, 1760–1768. [Google Scholar] [CrossRef]
- Bosch, L.J.W.; de Wit, M.; Pham, T.V.; Coupé, V.M.H.; Hiemstra, A.C.; Piersma, S.R.; Oudgenoeg, G.; Scheffer, G.L.; Mongera, S.; Sive Droste, J.T. Novel Stool-Based Protein Biomarkers for Improved Colorectal Cancer Screening: A Case-Control Study. Ann. Intern. Med. 2017, 167, 855–866. [Google Scholar] [CrossRef]
- Iadsee, N.; Chuaypen, N.; Techawiwattanaboon, T.; Jinato, T.; Patcharatrakul, T.; Malakorn, S.; Petchlorlian, A.; Praditpornsilpa, K.; Patarakul, K. Identification of a novel gut microbiota signature associated with colorectal cancer in Thai population. Sci. Rep. 2023, 13, 6702. [Google Scholar] [CrossRef]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Marcuello, M.; Vymetalkova, V.; Neves, R.P.L.; Duran-Sanchon, S.; Vedeld, H.M.; Tham, E.; van Dalum, G.; Flügen, G.; Garcia-Barberan, V.; Fijneman, R.J.; et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Asp. Med. 2019, 69, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid biopsy in cancer current: Status, challenges and future prospects. Signal Transduct. Target. Ther. 2024, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Galvis, M.M.; Romero, C.S.; Bueno, T.O.; Teng, Y. Toward a New Era for the Management of Circulating Tumor Cells. Adv. Exp. Med. Biol. 2021, 1286, 125–134. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear acids in human blood plasma. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Wolf, P. The Nature and Significance of Platelet Products in Human Plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Pan, B.T.; Johnstone, R.M. Fate of the Transferrin Receptor During Maturation of Sheep Reticulocytes In Vitro: Selective Externalization of the Receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble Normal and Mutated DNA Sequences from Single-Copy Genes in Human Blood. Cancer Epidemiol. Biomark. Prev. 1994, 3, 67–71. [Google Scholar]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.M.; Geuze, H.J. B Lymphocytes Secrete Antigen-Presenting Vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Smirnov, D.A.; Zweitzig, D.R.; Foulk, B.W.; Miller, M.C.; Doyle, G.V.; Pienta, K.J.; Meropol, N.J.; Weiner, L.M.; Cohen, S.J.; Moreno, J.G.; et al. Global Gene Expression Profiling of Circulating Tumor Cells. Cancer Res. 2005, 65, 4993–4997. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Szabo, S.A.; Holdhoff, M.; Leary, R.J.; Kinzler, K.W.; et al. Circulating Mutant DNA to Assess Tumor Dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Cardona, A.F.; Casadio, C.; Chae, Y.K.; Chouaid, C.; et al. Liquid Biopsy for Advanced Non–Small Cell Lung Cancer: A Consensus Statement from the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Song, L.; Peng, X.; Li, Y.; Xiao, W.; Jia, J.; Dong, C.; Gong, Y.; Zhou, G.; Han, X. The SEPT9 gene methylation assay is capable of detecting colorectal adenoma in opportunistic screening. Epigenomics 2017, 9, 599–610. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor–Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [Google Scholar] [CrossRef]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating Tumor Cells: Approaches to Isolation and Characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef]
- Lianidou, E.S.; Strati, A.; Markou, A. Circulating Tumor Cells as Promising Novel Biomarkers in Solid Cancers. Crit. Rev. Clin. Lab. Sci. 2014, 51, 160–171. [Google Scholar] [CrossRef]
- Agashe, R.; Kurzrock, R. Circulating tumor cells: From the laboratory to the cancer clinic. Cancers 2020, 12, 2361. [Google Scholar] [CrossRef]
- Cohen, S.J.A.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients with Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Vidlarova, M.; Vrbkova, J.; Kucerova, L.; Minarik, M. Recent advances in methods for circulating tumor cell detection. Int. J. Mol. Sci. 2023, 24, 3902. [Google Scholar] [CrossRef] [PubMed]
- Kojima, T.; Hashimoto, Y.; Watanabe, Y.; Kagawa, S.; Uno, F.; Kuroda, S.; Tazawa, H.; Kyo, S.; Mizuguchi, H.; Urata, Y.; et al. A simple biological imaging system for detecting viable human circulating tumor cells. J. Clin. Investig. 2009, 119, 3172–3181. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.I.; Musso, N.; Romano, A.; Caruso, G.; Petralia, S.; Lanzanò, L.; Broggi, G.; Camarda, M. The role of dielectrophoresis for cancer diagnosis and prognosis. Cancers 2022, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Lozar, T.; Jesenko, T.; Kloboves Prevodnik, V.; Cemazar, M.; Hosta, V.; Jericevic, A.; Nolde, N.; Grasic Kuhar, C. Preclinical and clinical evaluation of magnetic-activated cell separation technology for CTC isolation in breast cancer. Front. Oncol. 2020, 10, 554554. [Google Scholar] [CrossRef] [PubMed]
- Petrik, J.; Zemanova, M.; Dolezelova, H.; Krystof, V.; Kral, M. Circulating tumor cells in colorectal cancer: Detection systems and clinical utility. Int. J. Mol. Sci. 2022, 23, 13582. [Google Scholar] [CrossRef]
- Rushton, A.J.; Nteliopoulos, G.; Shaw, J.A.; Coombes, R.C. A review of circulating tumour cell enrichment technologies. Cancers 2021, 13, 970. [Google Scholar] [CrossRef] [PubMed]
- Andree, K.C.; van Dalum, G.; Terstappen, L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016, 10, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Cui, Y.; Jiang, H.; Sui, D.; Wang, Y.; Jiang, Z.; Lin, S. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget 2016, 7, 71330–71340. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Gong, Y.; Wang, Y.; Xie, J.; Cheng, J.; Huang, Q. Comprehensive atlas of circulating rare cells detected by SE-iFISH and image scanning platform in patients with various diseases. Front. Oncol. 2022, 12, 821454. [Google Scholar] [CrossRef]
- Li, Y.Z.; Kong, S.N.; Liu, Y.P.; Yang, Y.; Zhang, H.M. Can liquid biopsy based on ctDNA/cfDNA replace tissue biopsy for the precision treatment of EGFR-mutated NSCLC? J. Clin. Med. 2023, 12, 1438. [Google Scholar] [CrossRef]
- Campos-Carrillo, A.; Weitzel, J.N.; Sahoo, P.; Rockne, R.; Mokhnatkin, J.V.; Murtaza, M.; Gray, S.W.; Goetz, L.; Goel, A.; Schork, N.; et al. Circulating tumor DNA as an early cancer detection tool. Pharmacol. Ther. 2020, 207, 107458. [Google Scholar] [CrossRef]
- Underhill, H.R.; Kitzman, J.O.; Hellwig, S.; Welker, N.C.; Daza, R.; Baker, D.N.; Gligorich, K.M.; Rostomily, R.C.; Bronner, M.P.; Shendure, J. Fragment length of circulating tumor DNA. PLoS Genet. 2016, 12, e1006162. [Google Scholar] [CrossRef]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef]
- Jia, N.; Chang, L.; Gao, X.; Shi, X.; Dou, X.; Guan, M.; Shao, Y.; Li, N.; Cheng, Y.; Ying, H.; et al. Association of emergence of new mutations in circulating tumor DNA during chemotherapy with clinical outcome in metastatic colorectal cancer. BMC Cancer 2021, 21, 845. [Google Scholar] [CrossRef]
- Lin, C.; Liu, X.; Zheng, B.; Ke, R.; Tzeng, C.M. Liquid biopsy, ctDNA diagnosis through NGS. Life 2021, 11, 890. [Google Scholar] [CrossRef]
- Mandlik, J.S.; Patil, A.S.; Singh, S. Next-generation sequencing (NGS): Platforms and applications. J. Pharm. Bioall. Sci. 2024, 16 (Suppl. S1), S41–S45. [Google Scholar] [CrossRef] [PubMed]
- Kojabad, A.A.; Farzanehpour, M.; Hejazi, M.S.; Ramezani, A.; Hashemi, M.; Ghaedi, K. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J. Med. Virol. 2021, 93, 4182–4197. [Google Scholar] [CrossRef]
- Chen, Z.; Li, C.; Zhou, Y.; Yao, Y.; Liu, J.; Wu, M.; Su, J. Liquid biopsies for cancer: From bench to clinic. MedComm 2023, 4, e329. [Google Scholar] [CrossRef] [PubMed]
- Hannigan, B.; Ye, W.; Mehrotra, M.; Lam, V.; Bolivar, A.; Zalles, S.; Barkoh, B.A.; Duose, D.; Hu, P.C.; Broaddus, R.; et al. Liquid biopsy assay for lung carcinoma using centrifuged supernatants from fine-needle aspiration specimens. Ann. Oncol. 2019, 30, 963–969. [Google Scholar] [CrossRef]
- Olmedillas-López, S.; Olivera-Salazar, R.; García-Arranz, M.; García-Olmo, D. Current and emerging applications of droplet digital PCR in oncology: An updated review. Mol. Diagn. Ther. 2022, 26, 61–87. [Google Scholar] [CrossRef]
- Ståhlberg, A.; Krzyzanowski, P.M.; Jackson, J.B.; Egyud, M.; Stein, L.; Godfrey, T.E. Simple multiplexed PCR-based barcoding of DNA for ultrasensitive mutation detection by next-generation sequencing. Nat. Protoc. 2017, 12, 664–682. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Witwer, K.W.; Théry, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles 2019, 8, 1648167. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Challenges and directions in studying cell–cell communication by extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2022, 23, 369–382. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Gong, H.; Luo, S.; Cui, Y. The role of exosomes and their applications in cancer. Int. J. Mol. Sci. 2021, 22, 12204. [Google Scholar] [CrossRef]
- Han, Q.F.; Li, W.J.; Hu, K.S.; Gao, J.; Zhai, W.L.; Yang, J.H.; Zhang, S.J. Exosome biogenesis: Machinery, regulation, and therapeutic implications in cancer. Mol. Cancer 2022, 21, 207. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A review of exosomal isolation methods: Is size exclusion chromatography the best option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Wang, J.; Ni, J.; Beretov, J.; Thompson, J.; Graham, P.; Li, Y. Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit. Rev. Oncol. Hematol. 2020, 145, 102860. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, K.; Yuan, W.; Gao, Z. Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med. Sci. Monit. 2018, 24, 9307–9316. [Google Scholar] [CrossRef] [PubMed]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef]
- Mu, Y.; Huang, X.; Yang, Y.; Huang, Z.; Chen, J.; Li, S.; Dong, Y.; Chen, F.; Zhang, R.; Chen, J.; et al. Study of serum exosome miRNA as a biomarker for early onset adult ocular myasthenia gravis. Gene 2024, 896, 148034. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, L.; Zhu, S.; Yang, H.; Ye, Z.; Wang, H.; Wu, H.; Wu, Y.; Sun, Q.; Liu, X.; et al. Exosomal derived miR-1246 from hydroquinone-transformed cells drives S phase accumulation arrest by targeting cyclin G2 in TK6 cells. Chem. Biol. Interact. 2024, 387, 110809. [Google Scholar] [CrossRef]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X. Review on strategies and technologies for exosome isolation and purification. Front. Bioeng. Biotechnol. 2021, 9, 811971. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Li, H.; Xiong, J.; Wang, J.; Huang, Y. Application of tumor-educated platelets as new fluid biopsy markers in various tumors. Clin. Transl. Oncol. 2023, 25, 114–125. [Google Scholar] [CrossRef]
- Best, M.G.; Wesseling, P.; Wurdinger, T. Tumor-educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res. 2018, 78, 3407–3412. [Google Scholar] [CrossRef]
- Nilsson, R.J.; Balaj, L.; Hulleman, E.; van Rijn, S.; Pegtel, D.M.; Walraven, M.; Widmark, A.; Gerritsen, W.R.; Verheul, H.M.; Vandertop, W.P.; et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011, 118, 3680–3683. [Google Scholar] [CrossRef]
- Stratz, C.; Nuhrenberg, T.G.; Binder, H.; Valina, C.M.; Trenk, D.; Hochholzer, W.; Neumann, F.J.; Fiebich, B. Micro-array profiling exhibits remarkable intra-individual stability of human platelet micro-RNA. Thromb. Haemost. 2012, 107, 634–641. [Google Scholar] [CrossRef]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef]
- Ye, B.; Li, F.; Chen, M.; Weng, Y.; Qi, C.; Xie, Y.; Zhang, Q.; Ding, H.; Zhang, J.; Gao, X. A panel of platelet-associated circulating long non-coding RNAs as potential biomarkers for colorectal cancer. Genomics 2022, 114, 31–37. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Toden, S.; Goel, A. Non-coding RNAs as liquid biopsy biomarkers in cancer. Br. J. Cancer 2022, 126, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Jaé, N.; Holdt, L.; Dimmeler, S. Long noncoding RNAs: From clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 2016, 67, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Chen, Q.; Su, Y.; He, X.; Zhao, W.; Wu, C.; Zhang, W.; Si, X.; Dong, B.; Zhao, L.; Gao, Y.; et al. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol. Lett. 2016, 12, 1361–1366. [Google Scholar] [CrossRef]
- Lin, Q.; Guan, W.; Ren, W.; Zhang, L.; Zhang, J.; Xu, G. MALAT1 affects ovarian cancer cell behavior and patient survival. Oncol. Rep. 2018, 39, 2644–2652. [Google Scholar] [CrossRef]
- Takahashi, K.; Yan, I.; Haga, H.; Patel, T. Long non-coding RNAs in epithelial-mesenchymal transition of pancreatic cancer. Front. Mol. Biosci. 2021, 8, 717890. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Lou, X.; Meng, N.; Li, Z.; Teng, Y.; Zou, Y.; Wang, F. Peripheral blood-based DNA methylation of long non-coding RNA H19 and metastasis-associated lung adenocarcinoma transcript 1 promoters are potential non-invasive biomarkers for gastric cancer detection. Cancer Control 2021, 28, 10732748211043667. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Ramón, Y.C.; Segura, M.F.; Hümmer, S. Interplay between ncRNAs and cellular communication: A proposal for understanding cell-specific signaling pathways. Front. Genet. 2019, 10, 281. [Google Scholar] [CrossRef]
- Meyer, C.; Garzia, A.; Tuschl, T. Simultaneous detection of the subcellular localization of RNAs and proteins in cultured cells by combined multicolor RNA FISH and IF. Methods 2017, 118–119, 101–110. [Google Scholar] [CrossRef]
- Freeman, W.M.; Walker, S.J.; Vrana, K.E. Quantitative RT-PCR: Pitfalls and potential. Biotechniques 1999, 26, 112–122, 124–125. [Google Scholar] [CrossRef]
- He, S.L.; Green, R. Northern blotting. Methods Enzymol. 2013, 530, 75–87. [Google Scholar] [CrossRef]
- Urbanek, M.O.; Nawrocka, A.U.; Krzyzosiak, W.J. Small RNA detection by in situ hybridization methods. Int. J. Mol. Sci. 2015, 16, 13259–13286. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA sequencing: New technologies and applications in cancer research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef]
- Diener, T.O. Potato spindle tuber “virus”. IV. A replicating, low molecular weight RNA. Virology 1971, 45, 411–428. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Hentze, M.W.; Preiss, T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013, 32, 923–925. [Google Scholar] [CrossRef]
- Peng, D.; Luo, L.; Zhang, X.; Wei, C.; Zhang, Z.; Han, L. CircRNA: An emerging star in the progression of glioma. Biomed. Pharmacother. 2022, 151, 113150. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Q.; Liao, Q. CircHIPK3: A promising cancer-related circular RNA. Am. J. Transl. Res. 2020, 12, 6694–6704. [Google Scholar] [PubMed]
- Liu, T.; Huang, T.; Shang, M.; Han, G. CircRNA ITCH: Insight into its role and clinical application prospect in tumor and non-tumor diseases. Front. Genet. 2022, 13, 927541. [Google Scholar] [CrossRef]
- Su, K.; Yi, Q.; Dai, X.; Liu, O. Circular RNA ITCH: An emerging multifunctional regulator. Biomolecules 2022, 12, 359. [Google Scholar] [CrossRef]
- Liang, Y.; Liu, N.; Yang, L.; Tang, J.; Wang, Y.; Mei, M. A brief review of circRNA biogenesis, detection, and function. Curr. Genom. 2021, 22, 485–495. [Google Scholar] [CrossRef]
- Pretta, A.; Lai, E.; Donisi, C.; Spanu, D.; Ziranu, P.; Pusceddu, V.; Puzzoni, M.; Massa, E.; Scartozzi, M. Circulating tumour DNA in gastrointestinal cancer in clinical practice: Just a dream or maybe not? World J. Clin. Oncol. 2022, 13, 980–983. [Google Scholar] [CrossRef]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef]
- Inadomi, J.M.; Vijan, S.; Janz, N.K.; Fagerlin, A.; Thomas, J.P.; Lin, Y.V.; Muñoz, R.; Lau, C.; Somsouk, M.; El-Nachef, N.; et al. Adherence to colorectal cancer screening: A randomized clinical trial of competing strategies. Arch. Intern. Med. 2012, 172, 575–582. [Google Scholar] [CrossRef]
- Zhou, H.; Zhu, L.; Song, J.; Wang, G.; Li, P.; Li, W.; Luo, P.; Sun, X.; Wu, J.; Liu, Y.; et al. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol. Cancer 2022, 21, 86. [Google Scholar] [CrossRef] [PubMed]
- Vega-Benedetti, A.F.; Loi, E.; Moi, L.; Orrù, S.; Ziranu, P.; Pretta, A.; Lai, E.; Puzzoni, M.; Ciccone, L.; Casadei-Gardini, A.; et al. Colorectal cancer early detection in stool samples tracing CpG islands methylation alterations affecting gene expression. Int. J. Mol. Sci. 2020, 21, 4494. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.J.; Hong, H.J.; Sun, J.; Yu, C.R.; Liu, H.S.; Li, P.Y.; Zheng, M.H. Detection and clinical value of circulating tumor cells as an assisted prognostic marker in colorectal cancer patients. Cancer Manag. Res. 2021, 13, 4567–4578. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, T.S.A.; Meiners, J.; Riethdorf, S.; König, A.; Melling, N.; Gorges, T.; Karstens, K.F.; Izbicki, J.R.; Pantel, K.; Reeh, M. Prognostic value of preoperative circulating tumor cells counts in patients with UICC stage I-IV colorectal cancer. PLoS ONE 2021, 16, e0252897. [Google Scholar] [CrossRef]
- Church, T.R.; Wandell, M.; Lofton-Day, C.; Mongin, S.J.; Burger, M.; Payne, S.R.; Castaños-Vélez, E.; Blumenstein, B.A.; Rösch, T.; Osborn, N.; et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014, 63, 317–325. [Google Scholar] [CrossRef]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef]
- Sun, G.; Meng, J.; Duan, H.; Zhang, D.; Tang, Y. Diagnostic assessment of septin9 DNA methylation for colorectal cancer using blood detection: A meta-analysis. Pathol. Oncol. Res. 2019, 25, 1525–1534. [Google Scholar] [CrossRef]
- Loomans-Kropp, H.A.; Song, Y.; Gala, M.; Parikh, A.R.; Van Seventer, E.E.; Alvarez, R.; Hitchins, M.P.; Shoemaker, R.H.; Umar, A. Methylated Septin9 (mSEPT9): A promising blood-based biomarker for the detection and screening of early-onset colorectal cancer. Cancer Res. Commun. 2022, 2, 90–98. [Google Scholar] [CrossRef]
- Zhao, G.; Ma, Y.; Li, H.; Li, S.; Zhu, Y.; Liu, X.; Xiong, S.; Liu, Y.; Miao, J.; Fei, S.; et al. A novel plasma-based early colorectal cancer screening assay based on methylated SDC2 and SFRP2. Clin. Chim. Acta 2020, 503, 84–89. [Google Scholar] [CrossRef]
- Scimia, M.; Pepe, F.; Russo, G.; Malapelle, U.; Scimia, S.; Alfieri, A.; Olivieri, V.; Chuang, R.; Tanaka, H.; Sha, M.; et al. Evaluation of cell-free DNA long fragments in the triage of FIT+ patients enrolled in a colorectal cancer screening program: An Italian prospective, cross-sectional study. J. Mol. Pathol. 2024, 5, 533–543. [Google Scholar] [CrossRef]
- Sohel, M.M.H. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci. 2020, 248, 117473. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef]
- AlZaabi, A.; Shalaby, A. A Systematic Review of Diagnostic Performance of Circulating MicroRNAs in Colorectal Cancer Detection with a Focus on Early-Onset Colorectal Cancer. Int. J. Mol. Sci. 2024, 25, 9565. [Google Scholar] [CrossRef]
- Ghareib, A.F.; Mohamed, R.H.; Abd El-Fatah, A.R.; Saadawy, S.F. Assessment of Serum MicroRNA-21 Gene Expression for Diagnosis and Prognosis of Colorectal Cancer. J. Gastrointest. Cancer 2020, 51, 818–823. [Google Scholar] [CrossRef]
- Sabry, D.; El-Deek, S.E.M.; Maher, M.; El-Baz, M.A.H.; El-Bader, H.M.; Amer, E.; Hassan, E.A.; Fathy, W.; El-Deek, H.E.M. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: Impact of HIF-1α-VEGF signaling pathway. Mol. Cell. Biochem. 2019, 454, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, D.; Guan, S.; Dong, M. Diagnostic role of circulating MiR-21 in colorectal cancer: An update meta-analysis. Ann. Med. 2021, 53, 87–102. [Google Scholar] [CrossRef]
- Tan, Y.; Lin, J.J.; Yang, X.; Gou, D.M.; Fu, L.; Li, F.R.; Yu, X.F. A panel of three plasma microRNAs for colorectal cancer diagnosis. Cancer Epidemiol. 2019, 60, 67–76. [Google Scholar] [CrossRef]
- Nakamura, K.; Hernández, G.; Sharma, G.G.; Wada, Y.; Banwait, J.K.; González, N.; Perea, J.; Balaguer, F.; Takamaru, H.; Saito, Y.; et al. A Liquid Biopsy Signature for the Detection of Patients with Early-Onset Colorectal Cancer. Gastroenterology 2022, 163, 1242–1251.e2. [Google Scholar] [CrossRef]
- Bavelloni, A.; Ramazzotti, G.; Poli, A.; Piazzi, M.; Focaccia, E.; Blalock, W.; Faenza, I. MiRNA-210: A Current Overview. Anticancer Res. 2017, 37, 6511–6521. [Google Scholar] [CrossRef]
- Wei, C.; Li, Y.; Huang, K.; Li, G.; He, M. Exosomal miR-1246 in body fluids is a potential biomarker for gastrointestinal cancer. Biomark. Med. 2018, 12, 1185–1196. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhu, A.; Bhardwaj, M.; Schrotz-King, P.; Brenner, H. Fecal microRNAs, Fecal microRNA Panels, or Combinations of Fecal microRNAs with Fecal Hemoglobin for Early Detection of Colorectal Cancer and Its Precursors: A Systematic Review. Cancers 2021, 14, 65. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Hüttenhofer, A.; Vogel, J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006, 34, 635–646. [Google Scholar] [CrossRef]
- Ragusa, M.; Barbagallo, C.; Statello, L.; Condorelli, A.G.; Battaglia, R.; Tamburello, L.; Barbagallo, D.; Di Pietro, C.; Purrello, M. Non-coding landscapes of colorectal cancer. World J. Gastroenterol. 2015, 21, 11709–11739. [Google Scholar] [CrossRef]
- Qi, P.; Zhou, X.Y.; Du, X. Circulating long non-coding RNAs in cancer: Current status and future perspectives. Mol. Cancer 2016, 15, 39. [Google Scholar] [CrossRef]
- Bagheri, R.; Ghorbian, M.; Ghorbian, S. Tumor circulating biomarkers in colorectal cancer. Cancer Treat. Res. Commun. 2024, 38, 100787. [Google Scholar] [CrossRef]
- Yang, P.; Yang, Y.; An, W.; Xu, J.; Zhang, G.; Jie, J.; Zhang, Q. The long noncoding RNA-ROR promotes the resistance of radiotherapy for human colorectal cancer cells by targeting the p53/miR-145 pathway. J. Gastroenterol. Hepatol. 2017, 32, 837–845. [Google Scholar] [CrossRef]
- Zhao, W.; Song, M.; Zhang, J.; Kuerban, M.; Wang, H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 14131–14140. [Google Scholar] [PubMed] [PubMed Central]
- Dong, Y.; He, D.; Peng, Z.; Peng, W.; Shi, W.; Wang, J.; Li, B.; Zhang, C.; Duan, C. Circular RNAs in cancer: An emerging key player. J. Hematol. Oncol. 2017, 10, 2. [Google Scholar] [CrossRef]
- Pan, B.; Qin, J.; Liu, X.; He, B.; Wang, X.; Pan, Y.; Sun, H.; Xu, T.; Xu, M.; Chen, X.; et al. Identification of Serum Exosomal hsa-circ-0004771 as a Novel Diagnostic Biomarker of Colorectal Cancer. Front. Genet. 2019, 10, 1096. [Google Scholar] [CrossRef]
- Mugoni, V.; Ciani, Y.; Nardella, C.; Demichelis, F. Circulating RNAs in prostate cancer patients. Cancer Lett. 2022, 524, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Gligorijević, N.; Dobrijević, Z.; Šunderić, M.; Robajac, D.; Četić, D.; Penezić, A.; Miljuš, G.; Nedić, O. The Insulin-like Growth Factor System and Colorectal Cancer. Life 2022, 12, 1274. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.S.; Huang, C.Y.; Lee, C.H.; Chen, W.Y.; Huang, M.T.; Wei, P.L.; Chang, Y.J. IGFBP2 plays an important role in heat shock protein 27-mediated cancer progression and metastasis. Oncotarget 2017, 8, 54978–54992. [Google Scholar] [CrossRef]
- Fung, K.Y.; Tabor, B.; Buckley, M.J.; Priebe, I.K.; Purins, L.; Pompeia, C.; Brierley, G.V.; Lockett, T.; Gibbs, P.; Tie, J.; et al. Blood-based protein biomarker panel for the detection of colorectal cancer. PLoS ONE 2015, 10, e0120425. [Google Scholar] [CrossRef]
- Wang, X.; Shi, X.Q.; Zeng, P.W.; Mo, F.M.; Chen, Z.H. Circulating cell free DNA as the diagnostic marker for colorectal cancer: A systematic review and meta-analysis. Oncotarget 2018, 9, 24514–24524. [Google Scholar] [CrossRef]
- Lockney, N.A.; Henderson, R.H.; Swarts, S.G.; Zhang, Z.; Zhang, B.; Li, J.; Zlotecki, R.A.; Morris, C.G.; Casey-Sawicki, K.A.; Okunieff, P.G. Measuring radiation toxicity using circulating cell-free DNA in prostate cancer patients. Int. J. Part. Ther. 2021, 8, 28–35. [Google Scholar] [CrossRef]
- Qian, C.; Ju, S.; Qi, J.; Zhao, J.; Shen, X.; Jing, R.; Yu, J.; Li, L.; Shi, Y.; Zhang, L.; et al. Alu-based cell-free DNA: A novel biomarker for screening of gastric cancer. Oncotarget 2016, 8, 54037–54045. [Google Scholar] [CrossRef]
- Krebs, M.G.; Malapelle, U.; André, F.; Paz-Ares, L.; Schuler, M.; Thomas, D.M.; Vainer, G.; Yoshino, T.; Rolfo, C. Practical considerations for the use of circulating tumor DNA in the treatment of patients with cancer: A narrative review. JAMA Oncol. 2022, 8, 1830–1839. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y.; Hu, T.; He, X.; Zou, Y.; Deng, Q.; Ke, J.; Lian, L.; He, X.; Zhao, D.; et al. A novel cell-free DNA methylation-based model improves the early detection of colorectal cancer. Mol. Oncol. 2021, 15, 2702–2714. [Google Scholar] [CrossRef]
- Vojjala, N.; Gibatova, V.; Shah, R.N.; Singal, S.; Prabhu, R.; Krishnamoorthy, G.; Riggins, K.; Moka, N. Integrating Circulating Tumor DNA into Clinical Management of Colorectal Cancer: Practical Implications and Therapeutic Challenges. Cancers 2025, 17, 2520. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Murray, D.H.; Symonds, E.L.; Young, G.P.; Byrne, S.; Rabbitt, P.; Roy, A.; Cornthwaite, K.; Karapetis, C.S.; Pedersen, S.K. Relationship between post-surgery detection of methylated circulating tumor DNA with risk of residual disease and recurrence-free survival. J. Cancer Res. Clin. Oncol. 2018, 144, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Worm Ørntoft, M.B. Review of blood-based colorectal cancer screening: How far are circulating cell-free DNA methylation markers from clinical implementation? Clin. Color. Cancer 2018, 17, e415–e433. [Google Scholar] [CrossRef] [PubMed]
- Vega-Benedetti, A.F.; Loi, E.; Moi, L.; Restivo, A.; Cabras, F.; Deidda, S.; Pretta, A.; Ziranu, P.; Orrù, S.; Scartozzi, M.; et al. Colorectal cancer promoter methylation alteration affects the expression of glutamate ionotropic receptor AMPA type subunit 4 alternative isoforms potentially relevant in colon tissue. Hum. Cell 2022, 35, 310–319. [Google Scholar] [CrossRef]
- Swarbrick, S.; Wragg, N.; Ghosh, S.; Stolzing, A. Systematic Review of miRNA as Biomarkers in Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 6156–6167. [Google Scholar] [CrossRef]
- Galamb, O.; Barták, B.K.; Kalmár, A.; Nagy, Z.B.; Szigeti, K.A.; Tulassay, Z.; Igaz, P.; Molnár, B. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J. Gastroenterol. 2019, 25, 5026–5048. [Google Scholar] [CrossRef]
- Hu, D.; Zhan, Y.; Zhu, K.; Bai, M.; Han, J.; Si, Y.; Zhang, H.; Kong, D. Plasma Exosomal Long Non-Coding RNAs Serve as Biomarkers for Early Detection of Colorectal Cancer. Cell. Physiol. Biochem. 2018, 51, 2704–2715. [Google Scholar] [CrossRef]
- Demır, A.S.; Erdenen, F.; Müderrısoğlu, C.; Toros, A.B.; Bektaş, H.; Gelışgen, R.; Tabak, Ö.; Altunoğlu, E.; Uzun, H.; Erdem Huq, G.E.; et al. Diagnostic and prognostic value of tumor M2-pyruvate kinase levels in patients with colorectal cancer. Turk. J. Gastroenterol. 2013, 24, 36–42. [Google Scholar] [CrossRef]
- Uppara, M.; Adaba, F.; Askari, A.; Clark, S.; Hanna, G.; Athanasiou, T.; Faiz, O. A systematic review and meta-analysis of the diagnostic accuracy of pyruvate kinase M2 isoenzymatic assay in diagnosing colorectal cancer. World J. Surg. Oncol. 2015, 13, 48. [Google Scholar] [CrossRef]
- St Croix, B.; Rago, C.; Velculescu, V.; Traverso, G.; Romans, K.E.; Montgomery, E.; Lal, A.; Riggins, G.J.; Lengauer, C.; Vogelstein, B.; et al. Genes expressed in human tumor endothelium. Science 2000, 289, 1197–1202. [Google Scholar] [CrossRef]
- Kikuchi, A.; Matsumoto, S.; Sada, R. Dickkopf signaling, beyond Wnt-mediated biology. Semin. Cell Dev. 2022, 125, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Zitt, M.; Untergasser, G.; Amberger, A.; Moser, P.; Stadlmann, S.; Zitt, M.; Müller, H.M.; Mühlmann, G.; Perathoner, A.; Margreiter, R.; et al. Dickkopf-3 as a new potential marker for neoangiogenesis in colorectal cancer: Expression in cancer tissue and adjacent non-cancerous tissue. Dis. Markers 2008, 24, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Kneuertz, P.J.; Chang, G.J.; Hu, C.Y.; Rodriguez-Bigas, M.A.; Eng, C.; Vilar, E.; Skibber, J.M.; Feig, B.W.; Cormier, J.N.; You, Y.N. Overtreatment of young adults with colon cancer: More intense treatments with unmatched survival gains. JAMA Surg. 2015, 150, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Gandini, A.; Taieb, J.; Blons, H.; Netter, J.; Laurent-Puig, P.; Gallois, C. Early-Onset colorectal Cancer: From the laboratory to the clinic. Cancer Treat. Rev. 2024, 130, 102821. [Google Scholar] [CrossRef]
- Crisafulli, G. Liquid Biopsy and Challenge of Assay Heterogeneity for Minimal Residual Disease Assessment in Colon Cancer Treatment. Genes 2025, 16, 71. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Thierry, A.R.; El Messaoudi, S.; Mollevi, C.; Raoul, J.L.; Guimbaud, R.; Pezet, D.; Artru, P.; Assenat, E.; Borg, C.; Mathonnet, M.; et al. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment. Ann. Oncol. 2017, 28, 2149–2159. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Parikh, A.R.; Parikh, A.R.; Chee, B.H.; Tsai, J.; Rich, T.A.; Price, K.S.; Patel, S.A.; Zhang, L.; Ibrahim, F.; Esquivel, M.; et al. Minimal Residual Disease using a Plasma-Only Circulating Tumor DNA Assay to Predict Recurrence of Metastatic Colorectal Cancer Following Curative Intent Treatment. Clin. Cancer Res. 2024, 30, 2964–2973. [Google Scholar] [CrossRef]
- Reinert, T.; Schøler, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.; Lamy, P.; Kannerup, A.S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumor DNA to monitor disease burden in patients with colorectal cancer. Gastroenterology 2019, 157, 1082–1092. [Google Scholar] [CrossRef]
- Dasari, A.; Shen, C.; Halperin, D.M.; Zhao, H.; Zhou, S.; Xu, Y.; Shih, T.; Yao, J.C. Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol. 2017, 3, 1335–1342. [Google Scholar] [CrossRef] [PubMed]
- Guardant Health. Guardant Reveal™: ctDNA-Based MRD Detection in Colorectal Cancer. Available online: https://guardantcomplete.com/hcp/solutions/guardant-reveal/ (accessed on 3 July 2025).
- Roche. AVENIO ctDNA Surveillance Kit V2™: Longitudinal Monitoring of Tumor Burden and MRD Detection. Available online: https://sequencing.roche.com/us/en/products/group/avenio-ctdna-surveillance-kits.html (accessed on 3 July 2025).
- Lo, Y.M.D.; Han, D.S.C.; Jiang, P.; Chiu, R.W.K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021, 372, eaaw3616. [Google Scholar] [CrossRef]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaap4921. [Google Scholar] [CrossRef] [PubMed]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.J.; Tsui, D.W.Y.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef]
- Michaud, D.S.; Kelsey, K.T.; Papathanasiou, E.; Genco, C.A.; Giovannucci, E. Periodontal disease and risk of all cancers among male never smokers: An updated analysis of the Health Professionals Follow-up Study. Ann. Oncol. 2016, 27, 941–947. [Google Scholar] [CrossRef]
- Henriksen, T.V.; Demuth, C.; Frydendahl, A.; Nors, J.; Nesic, M.; Rasmussen, M.H.; Larsen, O.H.; Jaensch, C.; Løve, U.S.; Andersen, P.V.; et al. Timing of ctDNA Analysis Aimed at Guiding Adjuvant Treatment in Colorectal Cancer. Clin. Cancer Res. 2025, 31, 1676–1685. [Google Scholar] [CrossRef]
- Coombes, R.C.; Page, K.; Salari, R.; Hastings, R.K.; Armstrong, A.; Ahmed, S.; Ali, S.; Cleator, S.; Kenny, L.; Stebbing, J.; et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 2019, 25, 4255–4263. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Swanton, C. Early stage NSCLC—Challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 2018, 15, 577–586. [Google Scholar] [CrossRef]
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019, 5, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Kotaka, M.; Shirasu, H.; Watanabe, J.; Yamazaki, K.; Hirata, K.; Akazawa, N.; Matsuhashi, N.; Yokota, M.; Ikeda, M.; Kato, K.; et al. Association of circulating tumor DNA dynamics with clinical outcomes in the adjuvant setting for patients with colorectal cancer from an observational GALAXY study in CIRCULATE-Japan. J. Clin. Oncol. 2022, 40 (Suppl. S4), 9. [Google Scholar] [CrossRef]
- Taniguchi, H.; Nakamura, Y.; Kotani, D.; Yukami, H.; Mishima, S.; Sawada, K.; Shirasu, H.; Ebi, H.; Yamanaka, T.; Aleshin, A.; et al. CIRCULATE-Japan: Circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021, 112, 2915–2920. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Kotani, D.; Saori, M.; Yukami, H.; Watanabe, J.; Akazawa, N.; Kataoka, K.; Hirata, K.; Yamazaki, K.; Yeh, K.-H.; et al. Circulating tumor DNA as a prognostic biomarker in patients with resected colorectal cancer: An updated 24-month disease-free survival analysis from GALAXY study (CIRCULATE-Japan). Ann. Oncol. 2023, 34 (Suppl. S2), S415–S416. [Google Scholar] [CrossRef]
- Yukami, H.; Nakamura, Y.; Mishima, S.; Ando, K.; Bando, H.; Watanabe, J.; Hirata, K.; Akazawa, N.; Ikeda, M.; Yokota, M.; et al. Circulating tumor DNA dynamics in patients with colorectal cancer with molecular residual disease: Updated analysis from GALAXY study in the CIRCULATE-Japan. J. Clin. Oncol. 2024, 42 (Suppl. S6), 6. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Loree, J.M.; Cohen, J.D.; Espinosa, D.; Wong, R.; Jay Price, T.; Tebbutt, N.T.; Lee, M.; Burge, M.E.; et al. ctDNA-guided adjuvant chemotherapy escalation in stage III colon cancer: Primary analysis of the ctDNA-positive cohort from the randomized AGITG dynamic-III trial (intergroup study of AGITG and CCTG). J. Clin. Oncol. 2025, 43 (Suppl. S17), 3503. [Google Scholar] [CrossRef]
- Lonardi, S.; Pietrantonio, F.; Llavero, M.; Montagut Viladot, C.; Sartore Bianchi, A.; Zampino, M.G.; Elez Fernandez, M.E.; Santos Vivas, C.; Mandalà, M.; Tamberi, S.; et al. The PEGASUS trial: Post-surgical liquid biopsy-guided treatment of stage III and high-risk stage II colon cancer patients. J. Clin. Oncol. 2020, 38, TPS4124. [Google Scholar] [CrossRef]
- Morris, V.K.; Yothers, G.; Kopetz, S.; Puhalla, S.L.; Lucas, P.C.; Iqbal, A.; Boland, P.M.; Deming, D.A.; Scott, A.J.; Lim, H.J.; et al. Phase II results of ctDNA as a predictive biomarker in adjuvant chemotherapy in patients with stage II colon cancer: NRG-GI005 (COBRA). J. Clin. Oncol. 2024, 42 (Suppl. S3), 5. [Google Scholar] [CrossRef]
- Nowak, J.A.; Shi, Q.; Twombly, T.; Pederson, L.D.; Ma, C.; Väyrynen, J.P.; Zhao, M.M.; Takashima, Y.; Shergill, A.; Kumar, P.; et al. Prognostic and predictive role of circulating tumor DNA (ctDNA) in stage III colon cancer treated with celecoxib: Findings from CALGB (Alliance)/SWOG 80702. J. Clin. Oncol. 2025, 43, LBA14. [Google Scholar] [CrossRef]
- Kataoka, K.; Yamada, T.; Yamazaki, K.; Mori, K.; Matsuhashi, N.; Shiozawa, M.; Iwai, T.; Goto, M.; Yasui, M.; Takii, Y.; et al. Trial protocol of a Phase II study of mFOLFOXIRI after metastasectomy in patients with oligometastatic colorectal cancer (FANTASTIC Study). J. Anus Rectum Colon 2024, 8, 246–252. [Google Scholar] [CrossRef]
- Oki, E.; Nakanishi, R.; Ando, K.; Takemasa, I.; Watanabe, J.; Matsuhashi, N.; Kato, T.; Kagawa, Y.; Kotaka, M.; Hirata, K.; et al. Recurrence monitoring using ctDNA in patients with metastatic colorectal cancer: COSMOS-CRC-03 and AURORA studies. ESMO Gastrointest. Oncol. 2024, 3, 100034. [Google Scholar] [CrossRef]
- Taieb, J.; Taly, V.; Henriques, J.; Bourreau, C.; Mineur, L.; Bennouna, J.; Desrame, J.; Louvet, C.; Lepere, C.; Mabro, M.; et al. Prognostic value and relation with adjuvant treatment duration of ctDNA in stage III colon cancer: A post hoc analysis of the PRODIGE-GERCOR IDEA-France trial. Clin. Cancer Res. 2021, 27, 5638–5646. [Google Scholar] [CrossRef] [PubMed]
- Pappas, L.; Yaeger, R.; Kasi, P.M.; Saha, M.A.; Saba Azad, N.; Horick, N.K.; Corcoran, R.B.; Diaz, L.A.; Parikh, A.R. Early identification and treatment of occult metastatic disease in stage III colon cancer (SU2C ACT3 clinical trial). J. Clin. Oncol. 2024, 42, 148. [Google Scholar] [CrossRef]
- Nors, J.; Henriksen, T.V.; Gotschalck, K.A.; Juul, T.; Søgaard, J.; Iversen, L.H.; Andersen, C.L. IMPROVE-IT2: Implementing noninvasive ctDNA analysis to optimize operative and postoperative treatment for colorectal cancer—Study protocol. Acta Oncol. 2020, 59, 336–341. [Google Scholar] [CrossRef]
- Kasi, P.M.; Sawyer, S.; Guilford, J.; Munro, M.; Ellers, S.; Wulff, J.; Hook, N.; Krinshpun, S.; Koyen Malashevich, A.; Malhotra, M.; et al. BESPOKE study protocol: A multicentre, prospective observational study to evaluate the impact of ctDNA-guided therapy on patients with colorectal cancer. BMJ Open 2021, 11, e047831. [Google Scholar] [CrossRef]
- Kasi, P.M.; Aushev, V.N.; Ensor, J.; Langer, N.; Wang, C.G.; Lewis Cannon, T.; Derby Berim, L.; Feinstein, T.; Grothey, A.; McCollom, J.W.; et al. Circulating tumor DNA for informing adjuvant chemotherapy in stage II/III colorectal cancer: Interim analysis of BESPOKE CRC study. J. Clin. Oncol. 2024, 42, 9. [Google Scholar] [CrossRef]
- Glynne-Jones, R.; Wyrwicz, L.; Tiret, E.; Brown, G.; Rödel, C.; Cervantes, A.; Arnold, D. Rectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28 (Suppl. S4), iv22–iv40. [Google Scholar] [CrossRef]
- Bahadoer, R.R.; Dijkstra, E.A.; van Etten, B.; Marijnen, C.A.M.; Putter, H.; Kranenbarg, E.M.; Roodvoets, A.G.H.; Nagtegaal, I.D.; Beets-Tan, R.G.H.; Blomqvist, L.K.; et al. Short-course radiotherapy followed by chemotherapy before total mesorectal excision versus preoperative chemoradiotherapy in locally advanced rectal cancer (RAPIDO): A randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 29–42. [Google Scholar] [CrossRef]
- Conroy, T.; Bosset, J.F.; Etienne, P.L.; Rio, E.; François, É.; Mesgouez-Nebout, N.; Vendrely, V.; Artignan, X.; Bouché, O.; Gargot, D.; et al. Neoadjuvant chemotherapy with FOLFIRINOX and preoperative chemoradiotherapy for patients with locally advanced rectal cancer (UNICANCER-PRODIGE 23): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 702–715. [Google Scholar] [CrossRef]
- Conroy, T.; Castan, F.; Etienne, P.L.; Rio, E.; Mesgouez-Nebout, N.; Evesque, L.; Vendrely, V.; Artignan, X.; Bouché, O.; Gargot, D.; et al. Total neoadjuvant therapy with mFOLFIRINOX versus preoperative chemoradiotherapy in patients with locally advanced rectal cancer: Long-term results of the UNICANCER-PRODIGE 23 trial. Ann. Oncol. 2024, 35, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Appelt, A.L.; Andersen, R.F.; Lindebjerg, J.; Jakobsen, A. Prognostic value of serum NPY hypermethylation in neoadjuvant chemoradiotherapy for rectal cancer: Secondary analysis of a randomized trial. Am. J. Clin. Oncol. 2020, 43, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Murahashi, S.; Akiyoshi, T.; Sano, T.; Fukunaga, Y.; Noda, T.; Ueno, M.; Zembutsu, H. Serial circulating tumour DNA analysis for locally advanced rectal cancer treated with preoperative therapy: Prediction of pathological response and postoperative recurrence. Br. J. Cancer 2020, 123, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhang, X.; He, L.; Ma, Q.; Fang, T.; Jiang, C.; Ma, Z.; Li, Q.; Wu, C.; Tao, J. Prognostic value of ctDNA detection in patients with locally advanced rectal cancer undergoing neoadjuvant chemoradiotherapy: A systematic review and meta-analysis. Oncologist 2023, 28, e1198–e1208. [Google Scholar] [CrossRef]
- van Rees, J.M.; Wullaert, L.; Grüter, A.A.J.; Derraze, Y.; Tanis, P.J.; Verheul, H.M.W.; Martens, J.W.M.; Wilting, S.M.; Vink, G.; van Vugt, J.L.A.; et al. Circulating tumour DNA as biomarker for rectal cancer: A systematic review and meta-analyses. Front. Oncol. 2023, 13, 1083285. [Google Scholar] [CrossRef]
- Grinshpun, A.; Kustanovich, A.; Neiman, D.; Lehmann-Werman, R.; Zick, A.; Meir, K.; Vainer, E.; Granit, R.Z.; Arad, A.; Daskal, N.; et al. A universal cell-free DNA approach for response prediction to preoperative chemoradiation in rectal cancer. Int. J. Cancer 2023, 152, 1444–1451. [Google Scholar] [CrossRef]
- Shepherdson, M.; Symonds, E.L.; Byrne, S.; Gormly, K.; Karapetis, C.S.; Vatandoust, S.; Young, G.P.; Roy, A.C. Circulating tumor DNA and circumferential resection margin as key prognostic indicators for survival in rectal cancer. J. Clin. Oncol. 2021, 39 (Suppl. S15), 3579. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Li, L.; Christie, M.; Simons, K.; Elsaleh, H.; Kosmider, S.; Wong, R.; Yip, D.; et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2019, 68, 663–671. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Gonzales Ginestet, P.; Wong, R.; Shapiro, J.D.; Campell, R.; Day, F.; Hayes, T.M.; Aghmesheh, M.; et al. Circulating tumor DNA analysis informing adjuvant chemotherapy in locally advanced rectal cancer: The randomized AGITG DYNAMIC-Rectal study. J. Clin. Oncol. 2024, 42, 12. [Google Scholar] [CrossRef]
- Cervantes, A.; Adam, R.; Roselló, S.; Arnold, D.; Normanno, N.; Taïeb, J.; Seligmann, J.; De Baere, T.; Osterlund, P.; Yoshino, T.; et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 10–32. [Google Scholar] [CrossRef]
- Torresan, S.; de Scordilli, M.; Bortolot, M.; Di Nardo, P.; Foltran, L.; Fumagalli, A.; Guardascione, M.; Ongaro, E.; Puglisi, F. Liquid biopsy in colorectal cancer: Onward and upward. Crit. Rev. Oncol. Hematol. 2024, 194, 104242. [Google Scholar] [CrossRef] [PubMed]
- Schmiegel, W.; Scott, R.J.; Dooley, S.; Lewis, W.; Meldrum, C.J.; Pockney, P.; Draganic, B.; Smith, S.; Hewitt, C.; Philimore, H.; et al. Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: Concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol. Oncol. 2017, 11, 208–219. [Google Scholar] [CrossRef] [PubMed]
- van ’t Erve, I.; Greuter, M.J.E.; Bolhuis, K.; Vessies, D.C.L.; Leal, A.; Vink, G.R.; van den Broek, D.; Velculescu, V.E.; Punt, C.J.A.; Meijer, G.A.; et al. Diagnostic strategies toward clinical implementation of liquid biopsy RAS/BRAF circulating tumor DNA analyses in patients with metastatic colorectal cancer. J. Mol. Diagn. 2020, 22, 1430–1437. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, J.; Elez, E.; Caratù, G.; Matito, J.; Santos, C.; Macarulla, T.; Vidal, J.; Garcia, M.; Viéitez, J.M.; Paéz, D.; et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann. Oncol. 2017, 28, 1294–1301. [Google Scholar] [CrossRef]
- Ye, P.; Cai, P.; Xie, J.; Wei, Y. The diagnostic accuracy of digital PCR, ARMS and NGS for detecting KRAS mutation in cell-free DNA of patients with colorectal cancer: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0248775. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, R.; Yan, C.; Liu, L.; Tong, Z.; Jiang, W.; Yao, M.; Fang, W.; Chen, Z. Advantage of next-generation sequencing in dynamic monitoring of circulating tumor DNA over droplet digital PCR in cetuximab treated colorectal cancer patients. Transl. Oncol. 2019, 12, 426–431. [Google Scholar] [CrossRef]
- Thierry, A.R.; Mouliere, F.; El Messaoudi, S.; Mollevi, C.; Lopez-Crapez, E.; Rolet, F.; Gillet, B.; Gongora, C.; Dechelotte, P.; Robert, B.; et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 2014, 20, 430–435. [Google Scholar] [CrossRef]
- Procaccio, L.; Bergamo, F.; Daniel, F.; Rasola, C.; Munari, G.; Biason, P.; Crucitta, S.; Barsotti, G.; Zanella, G.; Angerilli, V.; et al. A real-world application of liquid biopsy in metastatic colorectal cancer: The Poseidon study. Cancers 2021, 13, 5128. [Google Scholar] [CrossRef]
- Vitiello, P.P.; De Falco, V.; Giunta, E.F.; Ciardiello, D.; Cardone, C.; Vitale, P.; Zanaletti, N.; Borrelli, C.; Poliero, L.; Terminiello, M.; et al. Clinical practice use of liquid biopsy to identify RAS/BRAF mutations in patients with metastatic colorectal cancer (mCRC): A single institution experience. Cancers 2019, 11, 1504. [Google Scholar] [CrossRef]
- Aoki, Y.; Nakamura, Y.; Denda, T.; Ohta, T.; Esaki, T.; Shiozawa, M.; Yamaguchi, K.; Yamazaki, K.; Sunakawa, Y.; Kato, T.; et al. Clinical validation of plasma-based genotyping for RAS and BRAF V600E mutation in metastatic colorectal cancer: SCRUM-Japan GOZILA substudy. JCO Precis. Oncol. 2023, 7, e2200688. [Google Scholar] [CrossRef]
- Bando, H.; Nakamura, Y.; Taniguchi, H.; Shiozawa, M.; Yasui, H.; Esaki, T.; Kagawa, Y.; Denda, T.; Satoh, T.; Yamazaki, K.; et al. Effects of metastatic sites on circulating tumor DNA in patients with metastatic colorectal cancer. JCO Precis. Oncol. 2022, 6, e2100535. [Google Scholar] [CrossRef] [PubMed]
- Henry, J.; Willis, J.; Parseghian, C.M.; Raghav, K.P.S.; Johnson, B.; Dasari, A.; Stone, D.; Jeyakumar, N.; Coker, O.; Raymond, V.M.; et al. NeoRAS: Incidence of RAS reversion from RAS mutated to RAS wild type. J. Clin. Oncol. 2020, 38 (Suppl. S4), 180. [Google Scholar] [CrossRef]
- Moati, E.; Blons, H.; Taly, V.; Garlan, F.; Wang-Renault, S.F.; Pietrasz, D.; Didelot, A.; Garrigou, S.; Saint, A.; Pernot, S.; et al. Plasma clearance of RAS mutation under therapeutic pressure is a rare event in metastatic colorectal cancer. Int. J. Cancer 2020, 147, 1185–1189. [Google Scholar] [CrossRef]
- Klein-Scory, S.; Wahner, I.; Maslova, M.; Al-Sewaidi, Y.; Pohl, M.; Mika, T.; Ladigan, S.; Schroers, R.; Baraniskin, A. Evolution of RAS mutational status in liquid biopsies during first-line chemotherapy for metastatic colorectal cancer. Front. Oncol. 2020, 10, 1115. [Google Scholar] [CrossRef]
- Pesola, G.; Epistolio, S.; Cefalì, M.; Trevisi, E.; De Dosso, S.; Frattini, M. Neo-RAS wild type or RAS conversion in metastatic colorectal cancer: A comprehensive narrative review. Cancers 2024, 16, 3923. [Google Scholar] [CrossRef]
- Arici, S.; Hamdard, J.; Sakin, A.; Sengiz Erhan, S.; Atci, M.M.; Cekin, R.; Saka, B.; Köse, E.; Saydam, T.; Geredeli, C.; et al. The conversion of RAS status in metastatic colorectal cancer patients after first-line biological agent treatment. Color. Dis. 2021, 23, 206–212. [Google Scholar] [CrossRef]
- Epistolio, S.; Cefalì, M.; Spina, P.; Molinari, F.; Movilia, A.; Cergnul, M.; Mazzucchelli, L.; De Dosso, S.; Frattini, M.; Saletti, P. Occurrence of RAS reversion in metastatic colorectal cancer patients treated with bevacizumab. Oncotarget 2021, 12, 1046–1056. [Google Scholar] [CrossRef]
- Vidal, J.; Muinelo, L.; Dalmases, A.; Jones, F.; Edelstein, D.; Iglesias, M.; Orrillo, M.; Abalo, A.; Rodríguez, C.; Brozos, E.; et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann. Oncol. 2017, 28, 1325–1332. [Google Scholar] [CrossRef]
- Sato, S.; Mikayama, Y.O.; Shiozawa, M.; Nukada, S.; Iguchi, K.; Okamoto, H.; Kohmura, T.; Kazama, K.; Tanaka, K.; Oshima, T.; et al. Chemotherapy-induced reversion of mutant RAS to wild-type RAS in metastatic colorectal cancer. Anticancer Res. 2022, 42, 2625–2635. [Google Scholar] [CrossRef]
- Gazzaniga, P.; Raimondi, C.; Urbano, F.; Cortesi, E. EGFR inhibitor as second-line therapy in a patient with mutant RAS metastatic colorectal cancer: Circulating tumor DNA to personalize treatment. JCO Precis. Oncol. 2018, 2, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bouchahda, M.; Saffroy, R.; Karaboué, A.; Hamelin, J.; Innominato, P.; Saliba, F.; Levi, F.; Bosselut, N.; Lemoine, A. Efficacy of an anti-EGFR after ctDNA conversion from mutated RAS status of metastatic colorectal cancer: Results of a pilot study. J. Clin. Oncol. 2021, 39 (Suppl. S15), e15574. [Google Scholar] [CrossRef]
- Nicolazzo, C.; Barault, L.; Caponnetto, S.; De Renzi, G.; Belardinilli, F.; Bottillo, I.; Bargiacchi, S.; Macagno, M.; Grammatico, P.; Giannini, G.; et al. True conversions from RAS mutant to RAS wild-type in circulating tumor DNA from metastatic colorectal cancer patients as assessed by methylation and mutational signature. Cancer Lett. 2021, 507, 89–96. [Google Scholar] [CrossRef]
- Osumi, H.; Shinozaki, E.; Nakamura, Y.; Esaki, T.; Yasui, H.; Taniguchi, H.; Satake, H.; Sunakawa, Y.; Komatsu, Y.; Kagawa, Y.; et al. Clinical features associated with NeoRAS wild-type metastatic colorectal cancer: A SCRUM-Japan GOZILA substudy. Nat. Commun. 2024, 15, 5885. [Google Scholar] [CrossRef]
- Harada, K.; Yuki, S.; Kawamoto, Y.; Nakamura, T.; Kaneko, S.; Ishida, K.; Sakamoto, N.; Komatsu, Y. Anti-epidermal growth factor receptor treatment for patients with NeoRAS wild-type metastatic colorectal cancer: A case report of two cases. Ther. Adv. Med. Oncol. 2023, 15, 17588359231216090. [Google Scholar] [CrossRef]
- Gramaça, J.; Fernandes, I.G.; Trabulo, C.; Gonçalves, J.; Dos Santos, R.G.; Baptista, A.; Pina, I. Emerging role of liquid biopsy in rat sarcoma virus mutated metastatic colorectal cancer: A case report. World J. Gastrointest. Oncol. 2024, 16, 234–243. [Google Scholar] [CrossRef]
- Hamfjord, J.; Guren, T.K.; Glimelius, B.; Sorbye, H.; Pfeiffer, P.; Dajani, O.; Lingjaerde, O.C.; Tveit, K.M.; Pallisgaard, N.; Spindler, K.G.; et al. Clinicopathological factors associated with tumour-specific mutation detection in plasma of patients with RAS-mutated or BRAF-mutated metastatic colorectal cancer. Int. J. Cancer 2021, 149, 1385–1397. [Google Scholar] [CrossRef]
- Garlan, F.; Laurent-Puig, P.; Sefrioui, D.; Siauve, N.; Didelot, A.; Sarafan-Vasseur, N.; Michel, P.; Perkins, G.; Mulot, C.; Blons, H.; et al. Early evaluation of circulating tumor DNA as marker of therapeutic efficacy in metastatic colorectal cancer patients (PLACOL study). Clin. Cancer Res. 2017, 23, 5416–5425. [Google Scholar] [CrossRef]
- Unseld, M.; Belic, J.; Pierer, K.; Zhou, Q.; Moser, T.; Bauer, R.; Piringer, G.; Gerger, A.; Siebenhüner, A.; Speicher, M.; et al. A higher ctDNA fraction decreases survival in regorafenib-treated metastatic colorectal cancer patients: Results from the regorafenib’s liquid biopsy translational biomarker phase II pilot study. Int. J. Cancer 2021, 148, 1452–1461. [Google Scholar] [CrossRef] [PubMed]
- Formica, V.; Lucchetti, J.; Doldo, E.; Riondino, S.; Morelli, C.; Argirò, R.; Renzi, N.; Nitti, D.; Nardecchia, A.; Dell’Aquila, E.; et al. Clinical utility of plasma KRAS, NRAS and BRAF mutational analysis with real time PCR in metastatic colorectal cancer patients—The importance of tissue/plasma discordant cases. J. Clin. Med. 2020, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Muendlein, A.; Geiger, K.; Heinzle, C.; Gaenger, S.; Winder, T.; Severgnini, L.; Reimann, P.; Brandtner, E.M.; Leiherer, A.; Drexel, H.; et al. Cell-free circulating RAS mutation concentrations significantly impact the survival of metastatic colorectal cancer patients. J. Cancer Res. Clin. Oncol. 2023, 149, 6435–6444. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.P.; Overman, M.J.; Dasari, A.; Kazmi, S.M.A.; Mazard, T.; Vilar, E.; Morris, V.K.; Lee, M.S.; Herron, D.; Eng, C.; et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann. Oncol. 2015, 26, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.R.; Mojtahed, A.; Schneider, J.L.; Kanter, K.; Van Seventer, E.E.; Fetter, I.J.; Thabet, A.; Fish, M.G.; Teshome, B.; Fosbenner, K.; et al. Serial ctDNA Monitoring to Predict Response to Systemic Therapy in Metastatic Gastrointestinal Cancers. Clin. Cancer Res. 2020, 26, 1877–1885. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Vidal, J.; Fernández-Rodríguez, M.C.; Casadevall, D.; García-Alfonso, P.; Páez, D.; Guix, M.; Alonso, V.; Cano, M.T.; Santos, C.; Durán, G.; et al. Liquid biopsy detects early molecular response and predicts benefit to first-line chemotherapy plus cetuximab in metastatic colorectal cancer: PLATFORM-B study. Clin. Cancer Res. 2023, 29, 379–388. [Google Scholar] [CrossRef]
- Riedesser, J.E.; Ebert, M.P.; Betge, J. Precision medicine for metastatic colorectal cancer in clinical practice. Ther. Adv. Med. Oncol. 2022, 14, 17588359211072703. [Google Scholar] [CrossRef]
- Takayama, Y.; Suzuki, K.; Muto, Y.; Ichida, K.; Fukui, T.; Kakizawa, N.; Ishikawa, H.; Watanabe, F.; Hasegawa, F.; Saito, M.; et al. Monitoring circulating tumor DNA revealed dynamic changes in KRAS status in patients with metastatic colorectal cancer. Oncotarget 2018, 9, 24398–24413. [Google Scholar] [CrossRef]
- Holm, M.; Andersson, E.; Osterlund, E.; Ovissi, A.; Soveri, L.M.; Anttonen, A.K.; Kytölä, S.; Aittomäki, K.; Osterlund, P.; Ristimäki, A. Detection of KRAS mutations in liquid biopsies from metastatic colorectal cancer patients using droplet digital PCR, Idylla, and next generation sequencing. PLoS ONE 2020, 15, e0239819. [Google Scholar] [CrossRef]
- Salvianti, F.; Gelmini, S.; Mancini, I.; Pazzagli, M.; Pillozzi, S.; Giommoni, E.; Brugia, M.; Di Costanzo, F.; Galardi, F.; De Luca, F.; et al. Circulating tumour cells and cell-free DNA as a prognostic factor in metastatic colorectal cancer: The OMITERC prospective study. Br. J. Cancer 2021, 125, 94–100. [Google Scholar] [CrossRef]
- Tan, Y.; Wu, H. The significant prognostic value of circulating tumor cells in colorectal cancer: A systematic review and meta-analysis. Curr. Probl. Cancer 2018, 42, 95–106. [Google Scholar] [CrossRef]
- Wu, F.; Zhu, J.; Mao, Y.; Li, X.; Hu, B.; Zhang, D. Associations between the epithelial-mesenchymal transition phenotypes of circulating tumor cells and the clinicopathological features of patients with colorectal cancer. Dis. Markers 2017, 2017, 9474532. [Google Scholar] [CrossRef]
- Zhong, Y.; Ma, T.; Qiao, T.; Hu, H.; Li, Z.; Luo, K.; Wang, Y.; Tang, Q.; Wang, G.; Huang, R.; et al. Role of phenotypes of circulating tumor cells in the diagnosis and treatment of colorectal cancer. Cancer Manag. Res. 2021, 13, 7077–7085. [Google Scholar] [CrossRef]
- Choi, J.; Cho, H.Y.; Jeon, J.; Kim, K.A.; Han, Y.D.; Ahn, J.B.; Wortzel, I.; Lyden, D.; Kim, H.S. Detection of circulating KRAS mutant DNA in extracellular vesicles using droplet digital PCR in patients with colon cancer. Front. Oncol. 2022, 12, 1067210. [Google Scholar] [CrossRef]
- Galbiati, S.; Damin, F.; Brambilla, D.; Ferraro, L.; Soriani, N.; Ferretti, A.M.; Burgio, V.; Ronzoni, M.; Vago, R.; Sola, L.; et al. Small EVs-associated DNA as complementary biomarker to circulating tumor DNA in plasma of metastatic colorectal cancer patients. Pharmaceuticals 2021, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Janssens, K.; Vanhoutte, G.; Lybaert, W.; Demey, W.; Decaestecker, J.; Hendrickx, K.; Rezaei Kalantari, H.; Zwaenepoel, K.; Pauwels, P.; Fransen, E.; et al. NPY methylated ctDNA is a promising biomarker for treatment response monitoring in metastatic colorectal cancer. Clin. Cancer Res. 2023, 29, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, C.B.; Hansen, T.F.; Andersen, R.F.; Lindebjerg, J.; Jensen, L.H.; Jakobsen, A. Early identification of treatment benefit by methylated circulating tumor DNA in metastatic colorectal cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835920918472. [Google Scholar] [CrossRef] [PubMed]
- Ponomaryova, A.A.; Rykova, E.Y.; Solovyova, A.I.; Tarasova, A.S.; Kostromitsky, D.N.; Dobrodeev, A.Y.; Afanasiev, S.A.; Cherdyntseva, N.V. Genomic and transcriptomic research in the discovery and application of colorectal cancer circulating markers. Int. J. Mol. Sci. 2023, 24, 12407. [Google Scholar] [CrossRef]
- Pan, Z.; Miao, L. Serum microRNA-592 serves as a novel potential biomarker for early diagnosis of colorectal cancer. Oncol. Lett. 2020, 20, 1119–1126. [Google Scholar] [CrossRef]
- Shaker, O.G.; Ayeldeen, G.; Abdelhamid, A.M. Circulating microRNA-944 and its target gene EPHA7 as a potential biomarker for colorectal cancer. Arch. Physiol. Biochem. 2022, 128, 1181–1187. [Google Scholar] [CrossRef]
- Toledano-Fonseca, M.; Gómez-España, M.A.; Élez, E.; Grávalos, C.; García-Alfonso, P.; Rodríguez, R.; Losa, F.; Alés Díaz, I.; Graña, B.; Valladares-Ayerbes, M.; et al. A signature of circulating microRNAs predicts the response to treatment with FOLFIRI plus aflibercept in metastatic colorectal cancer patients. Biomed. Pharmacother. 2023, 159, 114272. [Google Scholar] [CrossRef]
- Ren, J.; Ding, L.; Zhang, D.; Shi, G.; Xu, Q.; Shen, S.; Wang, Y.; Wang, T.; Hou, Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8, 3932–3948. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Ruan, H.; Zhang, X.; Xu, X.; Zhu, Y.; Peng, H.; Zhang, X.; Kong, F.; Guan, M. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int. J. Cancer 2020, 146, 1700–1716. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J.; et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122–PKM2 axis in colorectal cancer. Mol. Oncol. 2020, 14, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Cheng, X.; Ye, Z.; Li, Y.; Peng, W.; Wu, Y.; Xing, C. Exosome-mediated transfer of circ_0000338 enhances 5-fluorouracil resistance in colorectal cancer through regulating microRNA 217 (miR-217) and miR-485-3p. Mol. Cell. Biol. 2021, 41, e00517-20. [Google Scholar] [CrossRef]
- van Helden, E.J.; Angus, L.; Menke-van der Houven van Oordt, C.W.; Heideman, D.A.M.; Boon, E.; van Es, S.C.; Radema, S.A.; van Herpen, C.M.L.; de Groot, D.J.A.; de Vries, E.G.E.; et al. RAS and BRAF mutations in cell-free DNA are predictive for outcome of cetuximab monotherapy in patients with tissue-tested RAS wild-type advanced colorectal cancer. Mol. Oncol. 2019, 13, 2361–2374. [Google Scholar] [CrossRef]
- Rachiglio, A.M.; Sacco, A.; Lambiase, M.; Tufano, S.; Damato, A.; Raimondi, A.; Frassineti, L.; Pazzola, A.; Colombo, A.; Mosconi, S.; et al. RAS testing of circulating cell-free DNA (cfDNA) from tissue RAS/BRAF wild-type metastatic colorectal carcinoma (mCRC) patients: Preliminary data from the Liquid Biopsy Monoclonal Antibodies in mCRC (LIBImAb) study. J. Clin. Oncol. 2023, 41, 3046. [Google Scholar] [CrossRef]
- Stintzing, S.; Heinemann, V.; Fischer von Weikersthal, L.; Fuchs, M.; Kaiser, F.; Heinrich, K.; Modest, D.P.; Hofheinz, R.D.; Decker, T.; Gerger, A.; et al. Phase III FIRE-4 study (AIO KRK-0114): Influence of baseline liquid biopsy results in first-line treatment efficacy of FOLFIRI/cetuximab in patients with tissue RAS-WT mCRC. J. Clin. Oncol. 2023, 41, 3507. [Google Scholar] [CrossRef]
- Stintzing, S.; Klein-Scory, S.; Fischer von Weikersthal, L.; Fuchs, M.; Kaiser, F.; Heinrich, K.; Modest, D.P.; Hofheinz, R.D.; Decker, T.; Gerger, A.; et al. Baseline liquid biopsy in relation to tissue-based parameters in metastatic colorectal cancer: Results from the randomized FIRE-4 (AIO-KRK-0114) study. J. Clin. Oncol. 2025, 43, 1463–1473. [Google Scholar] [CrossRef]
- Morano, F.; Corallo, S.; Lonardi, S.; Raimondi, A.; Cremolini, C.; Rimassa, L.; Murialdo, R.; Zaniboni, A.; Sartore-Bianchi, A.; Tomasello, G.; et al. Negative hyperselection of patients with RAS and BRAF wild-type metastatic colorectal cancer who received panitumumab-based maintenance therapy. J. Clin. Oncol. 2019, 37, 3099–3110. [Google Scholar] [CrossRef]
- Cremolini, C.; Rossini, D.; Dell’Aquila, E.; Lonardi, S.; Conca, E.; Del Re, M.; Busico, A.; Pietrantonio, F.; Danesi, R.; Aprile, G.; et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: A phase 2 single-arm clinical trial. JAMA Oncol. 2019, 5, 343–350. [Google Scholar] [CrossRef]
- Kagawa, Y.; Kotani, D.; Bando, H.; Takahashi, N.; Hamaguchi, T.; Kanazawa, A. Plasma RAS dynamics and anti-EGFR rechallenge efficacy in patients with RAS/BRAF wild-type metastatic colorectal cancer: REMARRY and PURSUIT trials. J. Clin. Oncol. 2022, 40, 3518. [Google Scholar] [CrossRef]
- Nakajima, H.; Kotani, D.; Bando, H.; Kato, T.; Oki, E.; Shinozaki, E.; Sunakawa, Y.; Yamazaki, K.; Yuki, S.; Nakamura, Y.; et al. REMARRY and PURSUIT trials: Liquid biopsy-guided rechallenge with anti-EGFR therapy with panitumumab plus irinotecan for patients with plasma RAS wild-type metastatic colorectal cancer. BMC Cancer 2021, 21, 674. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, S.; De Falco, V.; Martini, G.; Ciardiello, D.; Martinelli, E.; Della Corte, C.M.; Esposito, L.; Famiglietti, V.; Di Liello, A.; Avallone, A.; et al. Panitumumab plus trifluridine-tipiracil as anti-EGFR rechallenge therapy for refractory RAS wild-type metastatic colorectal cancer: A phase 2 randomized clinical trial. JAMA Oncol. 2023, 9, 966–970. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, E.; Martini, G.; Famiglietti, V.; Troiani, T.; Napolitano, S.; Pietrantonio, F.; Ciardiello, D.; Terminiello, M.; Borrelli, C.; Vitiello, P.P.; et al. Cetuximab rechallenge plus avelumab in pretreated patients with RAS wild-type metastatic colorectal cancer: The phase 2 single-arm clinical CAVE trial. JAMA Oncol. 2021, 7, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Sartore-Bianchi, A.; Pietrantonio, F.; Lonardi, S.; Mussolin, B.; Rua, F.; Crisafulli, G.; Bartolini, A.; Fenocchio, E.; Amatu, A.; Manca, P.; et al. Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: The phase 2 CHRONOS trial. Nat. Med. 2022, 28, 1612–1618. [Google Scholar] [CrossRef]
- Mariani, S.; Puzzoni, M.; Giampieri, R.; Ziranu, P.; Pusceddu, V.; Donisi, C.; Persano, M.; Pinna, G.; Cimbro, E.; Parrino, A.; et al. Liquid biopsy-driven cetuximab rechallenge strategy in molecularly selected metastatic colorectal cancer patients. Front. Oncol. 2022, 12, 852583. [Google Scholar] [CrossRef]
- Napolitano, S.; Boscolo Bielo, L.; Martini, G.; Pietrantonio, F.; Pisconti, S.; Sartore-Bianchi, A.; Blasi, L.; Tortora, G.; Zaniboni, A.; Pinto, C.; et al. The use of liquid biopsy comprehensive genomic profiling to identify the molecular landscape of patients with metastatic colorectal cancer who are candidates for anti-EGFR rechallenge therapy: Findings from the CAVE-2 GOIM trial. J. Clin. Oncol. 2025, 43 (Suppl. S4), 221. [Google Scholar] [CrossRef]
- Alshammari, K.; Aung, K.L.; Zhang, T.; Razak, A.R.A.; Serra, S.; Stockley, T.; Wang, L.; Nguyen, J.; Spreafico, A.; Hansen, A.R.; et al. Phase II trial of trametinib and panitumumab in RAS/RAF wild type metastatic colorectal cancer. Clin. Color. Cancer 2021, 20, 334–341. [Google Scholar] [CrossRef]
- Bekaii-Saab, T.S.; Van Cutsem, E.; Cubillo, A.; Petorin-Lesens, C.; Rodriguez-Salas, N.; Raghav, K.P.S.; Dupuis, O.; Lòpez-Lòpez, C.; Tournigand, C.; Isambert, N.; et al. PERSPECTIVE: Tepotinib + cetuximab in patients with RAS/BRAF wild-type left-sided metastatic colorectal cancer and acquired resistance to anti-EGFR antibody therapy due to MET amplification. J. Clin. Oncol. 2021, 39 (Suppl. S15), TPS3616. [Google Scholar] [CrossRef]
- Ciracì, P.; Germani, M.M.; Pietrantonio, F.; Leonardi, S.; Businco, A.; Chiaramonte, A.; Ricagano, G.; Esposto, G.; Formica, V.; Scartozzi, M.; et al. Liquid biopsy-guided selection for anti-EGFR re-treatment in RAS/BRAF wild-type (wt) chemorefractory metastatic colorectal cancer patients (mCRC pts): Results from the phase II randomized PARERE trial. Ann. Oncol. 2025, 36, S4. [Google Scholar] [CrossRef]
- Tabernero, J.; Fernandez, E.E.; Ghiringhelli, F.; Folprecht, G.; Curigliano, G.; Siena, S.; Cremolini, C.; Sobrero, A.; Kwiatek, M.; Rosello Keranen, S.; et al. PRECISE-01 study: A phase Ib/II trial of MEN1611, a PI3K inhibitor, and cetuximab in patients with PIK3CA mutated metastatic colorectal cancer failing irinotecan, oxaliplatin, 5-FU and anti-EGFR containing regimens. Ann. Oncol. 2020, 31 (Suppl. S2), S115. [Google Scholar] [CrossRef]
- Arnold, D.; Elez Fernandez, E.; Van Cutsem, E.; Ahmad, A.R.; Lopez, C.; Rosello Keranen, S.; Velez, H.; Eng, C.; Pietrantonio, F.; Moreno Garcia, V.; et al. Rechallenge with amivantamab, an EGFR-MET bispecific antibody, after disease progression on prior EGFR inhibitor in left-sided RAS/BRAF wild-type metastatic colorectal cancer: Updated results from OrigAMI-1. Ann. Oncol. 2025, 36, S5–S6. [Google Scholar] [CrossRef]
- Sartore-Bianchi, A.; Di Nicolantonio, F.; Nichelatti, M.; Molinari, F.; De Dosso, S.; Saletti, P.; Martini, M.; Cipani, T.; Marrapese, G.; Mazzucchelli, L.; et al. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS ONE 2009, 4, e7287. [Google Scholar] [CrossRef]
- Randon, G.; Maddalena, G.; Germani, M.M.; Pircher, C.C.; Manca, P.; Bergamo, F.; Giordano, M.; Sposetti, C.; Montagna, A.; Vetere, G.; et al. Negative ultraselection of patients with RAS/BRAF wild-type, microsatellite-stable metastatic colorectal cancer receiving anti-EGFR-based therapy. JCO Precis. Oncol. 2022, 6, e2200037. [Google Scholar] [CrossRef]
- Cremolini, C.; Morano, F.; Moretto, R.; Berenato, R.; Tamborini, E.; Perrone, F.; Rossini, D.; Gloghini, A.; Busico, A.; Zucchelli, G.; et al. Negative hyper-selection of metastatic colorectal cancer patients for anti-EGFR monoclonal antibodies: The PRESSING case-control study. Ann. Oncol. 2017, 28, 3009–3014. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Peeters, M.; Thomas, A.; Gibbs, P.; Hool, K.; Zhang, J.; Ang, A.L.; Bach, B.A.; Price, T. Impact of emergent circulating tumor DNA RAS mutation in panitumumab-treated chemoresistant metastatic colorectal cancer. Clin. Cancer Res. 2018, 24, 5602–5609. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Williams, R.T.; Wu, J.; Kinde, I.; Hecht, J.R.; Berlin, J.; Allen, B.; Bozic, I.; Reiter, J.G.; Nowak, M.A.; et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012, 486, 537–540. [Google Scholar] [CrossRef]
- Raghav, K.; Ou, F.S.; Venook, A.P.; Innocenti, F.; Sun, R.; Lenz, H.J.; Kopetz, S. Acquired genomic alterations on first-line chemotherapy with cetuximab in advanced colorectal cancer: Circulating tumor DNA analysis of the CALGB/SWOG-80405 trial (Alliance). J. Clin. Oncol. 2023, 41, 472–478. [Google Scholar] [CrossRef]
- Parseghian, C.M.; Sun, R.; Woods, M.; Napolitano, S.; Lee, H.M.; Alshenaifi, J.; Willis, J.; Nunez, S.; Raghav, K.P.; Morris, V.K.; et al. Resistance mechanisms to anti-epidermal growth factor receptor therapy in RAS/RAF wild-type colorectal cancer vary by regimen and line of therapy. J. Clin. Oncol. 2023, 41, 460–471. [Google Scholar] [CrossRef]
- Bardelli, A.; Corso, S.; Bertotti, A.; Hobor, S.; Valtorta, E.; Siravegna, G.; Sartore-Bianchi, A.; Scala, E.; Cassingena, A.; Zecchin, D.; et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013, 3, 658–673. [Google Scholar] [CrossRef]
- Maurel, J.; Alonso, V.; Escudero, P.; Fernández-Martos, C.; Salud, A.; Méndez, M.; Gallego, J.; Rodriguez, J.R.; Martín-Richard, M.; Fernández-Plana, J.; et al. Clinical Impact of Circulating Tumor RAS and BRAF Mutation Dynamics in Patients with Metastatic Colorectal Cancer Treated with First-Line Chemotherapy Plus Anti-Epidermal Growth Factor Receptor Therapy. JCO Precis. Oncol. 2019, 3, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Topham, J.T.; O’Callaghan, C.J.; Feilotter, H.; Kennecke, H.F.; Lee, Y.S.; Li, W.; Banks, K.C.; Quinn, K.; Renouf, D.J.; Jonker, D.J.; et al. Circulating tumor DNA identifies diverse landscape of acquired resistance to anti-epidermal growth factor receptor therapy in metastatic colorectal cancer. J. Clin. Oncol. 2023, 41, 485–496. [Google Scholar] [CrossRef]
- Peeters, M.; Price, T.; Boedigheimer, M.; Kim, T.W.; Ruff, P.; Gibbs, P.; Thomas, A.; Demonty, G.; Hool, K.; Ang, A. Evaluation of emergent mutations in circulating cell-free DNA and clinical outcomes in patients with metastatic colorectal cancer treated with panitumumab in the ASPECCT study. Clin. Cancer Res. 2019, 25, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 827. [Google Scholar] [CrossRef] [PubMed]
- Parseghian, C.M.; Loree, J.M.; Morris, V.K.; Liu, X.; Clifton, K.K.; Napolitano, S.; Henry, J.T.; Pereira, A.A.; Vilar, E.; Johnson, B.; et al. Anti-EGFR-resistant clones decay exponentially after progression: Implications for anti-EGFR re-challenge. Ann. Oncol. 2019, 30, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Masuishi, T.; Tsuji, A.; Kotaka, M.; Nakamura, M.; Kochi, M.; Takagane, A.; Shimada, K.; Denda, T.; Segawa, Y.; Tanioka, H.; et al. Phase 2 study of irinotecan plus cetuximab rechallenge as third-line treatment in KRAS wild-type metastatic colorectal cancer: JACCRO CC-08. Br. J. Cancer 2020, 123, 1490–1495. [Google Scholar] [CrossRef]
- Santini, D.; Vincenzi, B.; Addeo, R.; Garufi, C.; Masi, G.; Scartozzi, M.; Mancuso, A.; Frezza, A.M.; Venditti, O.; Imperatori, M.; et al. Cetuximab rechallenge in metastatic colorectal cancer patients: How to come away from acquired resistance? Ann. Oncol. 2012, 23, 2313–2318. [Google Scholar] [CrossRef]
- Gazzaniga, P.; Raimondi, C.; Nicolazzo, C.; Gradilone, A.; Cortesi, E. ctDNA might expand therapeutic options for second line treatment of KRAS mutant mCRC. Ann. Oncol. 2017, 28 (Suppl. S5), v586. [Google Scholar] [CrossRef]
- Santos Vivas, C.; Vidal Barrull, J.; Fernández Rodríguez, C.; Salvà Ballabrera, F.; Alonso-Orduna, V.; García-Carbonero, R.; Losa, F.; Tarazona Llavero, N.; Safont Aguileria, M.J.; Rivera Herrero, F.; et al. Third line rechallenge with cetuximab (Cet) and irinotecan in circulating tumor DNA (ctDNA) selected metastatic colorectal cancer (mCRC) patients: The randomized phase II CITRIC trial. Ann. Oncol. 2024, 35 (Suppl. S5), S433–S434. [Google Scholar] [CrossRef]
- Loupakis, F.; Intini, R.; Cremolini, C.; Orlandi, A.; Sartore-Bianchi, A.; Pietrantonio, F.; Pella, N.; Spallanzani, A.; Dell’Aquila, E.; Scartozzi, M.; et al. A validated prognostic classifier for V600EBRAF-mutated metastatic colorectal cancer: The ‘BRAF BeCool’ study. Eur. J. Cancer 2019, 118, 121–130. [Google Scholar] [CrossRef]
- Lai, E.; Pretta, A.; Impera, V.; Mariani, S.; Giampieri, R.; Casula, L.; Pusceddu, V.; Coni, P.; Fanni, D.; Puzzoni, M.; et al. BRAF-mutant colorectal cancer, a different breed evolving. Expert Rev. Mol. Diagn. 2018, 18, 499–512. [Google Scholar] [CrossRef]
- Ueda, K.; Yamada, T.; Ohta, R.; Matsuda, A.; Sonoda, H.; Kuriyama, S.; Takahashi, G.; Iwai, T.; Takeda, K.; Miyasaka, T.; et al. BRAF V600E mutations in right-side colon cancer: Heterogeneity detected by liquid biopsy. Eur. J. Surg. Oncol. 2022, 48, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Kopetz, S.; Bekaii-Saab, T.S.; Yoshino, T.; Chung, C.-H.; Zhang, X.; Tabernero, J. SEAMARK: Randomized phase 2 study of pembrolizumab + encorafenib + cetuximab versus pembrolizumab alone for first-line treatment of BRAF V600E-mutant and microsatellite instability-high (MSI-H)/mismatch repair deficient (dMMR) metastatic colorectal cancer. J. Clin. Oncol. 2022, 40 (Suppl. S16), TPS3634. [Google Scholar] [CrossRef]
- Mas, L.; Bachet, J.B.; Taly, V.; Bouché, O.; Taieb, J.; Cohen, R.; Meurisse, A.; Normand, C.; Gornet, J.M.; Artru, P.; et al. BRAF mutation status in circulating tumor DNA from patients with metastatic colorectal cancer: Extended mutation analysis from the AGEO RASANC study. Cancers 2019, 11, 998. [Google Scholar] [CrossRef] [PubMed]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef]
- Tabernero, J.; Grothey, A.; Van Cutsem, E.; Yaeger, R.; Wasan, H.; Yoshino, T.; Desai, J.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib plus cetuximab as a new standard of care for previously treated BRAF V600E-mutant metastatic colorectal cancer: Updated survival results and subgroup analyses from the BEACON study. J. Clin. Oncol. 2021, 39, 273–284. [Google Scholar] [CrossRef]
- Grothey, A.; Fakih, M.; Tabernero, J. Management of BRAF-mutant metastatic colorectal cancer: A review of treatment options and evidence-based guidelines. Ann. Oncol. 2021, 32, 959–967. [Google Scholar] [CrossRef]
- Martinelli, E.; Arnold, D.; Cervantes, A.; Stintzing, S.; Van Cutsem, E.; Tabernero, J.; Taieb, J.; Wasan, H.; Ciardiello, F. European expert panel consensus on the clinical management of BRAFV600E-mutant metastatic colorectal cancer. Cancer Treat. Rev. 2023, 115, 102541. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Taieb, J.; Yaeger, R.; Yoshino, T.; Grothey, A.; Maiello, E.; Elez, E.; Dekervel, J.; Ross, P.; Ruiz-Casado, A.; et al. ANCHOR CRC: Results from a single-arm, phase II study of encorafenib plus binimetinib and cetuximab in previously untreated BRAFV600E-mutant metastatic colorectal cancer. J. Clin. Oncol. 2023, 41, 2628–2637. [Google Scholar] [CrossRef]
- Kopetz, S.; Yoshino, T.; Kim, T.W.; Wasan, H.S.; Van Cutsem, E.; Ciardiello, F.; Maughan, T.S.; Eng, C.; Yaeger, R.; Desai, J.; et al. BREAKWATER: An open-label, multicenter, randomized, phase 3 study, with a safety lead-in (SLI), of first-line encorafenib + cetuximab ± chemotherapy vs standard-of-care chemotherapy for BRAF V600E-mutant metastatic colorectal cancer. J. Clin. Oncol. 2023, 41 (Suppl. S16), TPS3627. [Google Scholar] [CrossRef]
- Malla, M.; Loree, J.M.; Kasi, P.M.; Parikh, A.R. Using circulating tumor DNA in colorectal cancer: Current and evolving practices. J. Clin. Oncol. 2022, 40, 2846–2857. [Google Scholar] [CrossRef]
- Ros, J.; Rodríguez-Castells, M.; Saoudi, N.; Baraibar, I.; Salva, F.; Tabernero, J.; Élez, E. Treatment of BRAF-V600E mutant metastatic colorectal cancer: New insights and biomarkers. Expert Rev. Anticancer Ther. 2023, 23, 797–806. [Google Scholar] [CrossRef]
- Xu, T.; Wang, X.; Wang, Z.; Deng, T.; Qi, C.; Liu, D.; Li, Y.; Ji, C.; Li, J.; Shen, L. Molecular mechanisms underlying the resistance of BRAF V600E-mutant metastatic colorectal cancer to EGFR/BRAF inhibitors. Ther. Adv. Med. Oncol. 2022, 14, 17588359221105022. [Google Scholar] [CrossRef]
- Ji, J.; Wang, C.; Fakih, M. Rechallenge with BRAF and anti-EGFR inhibitors in patients with metastatic colorectal cancer harboring BRAFV600E mutation who progressed on cetuximab and encorafenib with or without binimetinib: A case series. Clin. Color. Cancer 2022, 21, 267–271. [Google Scholar] [CrossRef]
- Oddo, D.; Siravegna, G.; Gloghini, A.; Vernieri, C.; Mussolin, B.; Morano, F.; Crisafulli, G.; Berenato, R.; Corti, G.; Volpi, C.C.; et al. Emergence of MET hyper-amplification at progression to MET and BRAF inhibition in colorectal cancer. Br. J. Cancer 2017, 117, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.F.; Huang, Z.Y.; Chen, X.X.; Chen, Z.G.; Wu, S.X.; Ren, C.; Hu, M.T.; Bao, H.; Jin, Y.; Wang, F.; et al. Monitoring tumour resistance to the BRAF inhibitor combination regimen in colorectal cancer patients via circulating tumour DNA. Drug Resist. Updates 2022, 65, 100883. [Google Scholar] [CrossRef] [PubMed]
- Elez, E.; Ros, J.; Fernández, J.; Villacampa, G.; Moreno-Cárdenas, A.B.; Arenillas, C.; Bernatowicz, K.; Comas, R.; Li, S.; Kodack, D.P.; et al. RNF43 mutations predict response to anti-BRAF/EGFR combinatory therapies in BRAFV600E metastatic colorectal cancer. Nat. Med. 2022, 28, 2162–2170. [Google Scholar] [CrossRef] [PubMed]
- Moretto, R.; Germani, M.M.; Ros, J.; Daniel, F.; Ghelardi, F.; Vetere, G.; Giordano, M.; Toledo, R.A.; Bergamo, F.; Randon, G.; et al. Predictive impact of RNF43 mutations in patients with proficient mismatch repair/microsatellite stable BRAFV600E-mutated metastatic colorectal cancer treated with target therapy or chemotherapy. JCO Precis. Oncol. 2023, 7, e2300255. [Google Scholar] [CrossRef]
- Wei, Q.; Zhang, Y.; Gao, J.; Li, J.; Li, J.; Li, Y.; Zhou, J.; Lu, M.; Gong, J.; Zhang, X.; et al. Clinicopathologic characteristics of HER2-positive metastatic colorectal cancer and detection of HER2 in plasma circulating tumor DNA. Clin. Color. Cancer 2019, 18, 175–182. [Google Scholar] [CrossRef]
- Siravegna, G.; Sartore-Bianchi, A.; Nagy, R.J.; Raghav, K.; Odegaard, J.I.; Lanman, R.B.; Trusolino, L.; Marsoni, S.; Siena, S.; Bardelli, A. Plasma HER2 (ERBB2) copy number predicts response to HER2-targeted therapy in metastatic colorectal cancer. Clin. Cancer Res. 2019, 25, 3046–3053. [Google Scholar] [CrossRef]
- Kopetz, S.; Murphy, D.A.; Pu, J.; Yaeger, R.; Ciardiello, F.; Desai, J.; Van Cutsem, E.; Wasan, H.S.; Yoshino, T.; Elez, E.; et al. Evaluation of baseline BRAF V600E mutation in circulating tumor DNA and efficacy response from the BEACON study. J. Clin. Oncol. 2022, 40 (Suppl. S4), 162. [Google Scholar] [CrossRef]
- Siena, S.; Raghav, K.; Masuishi, T.; Yamaguchi, K.; Nishina, T.; Elez, E.; Rodriguez, J.; Chau, I.; Di Bartolomeo, M.; Kawakami, H.; et al. Exploratory biomarker analysis of DESTINY-CRC01, a phase II, multicenter, open-label study of trastuzumab deruxtecan (T-DXd, DS-8201) in patients with HER2-expressing metastatic colorectal cancer. Ann. Oncol. 2021, 32 (Suppl. S5), S532. [Google Scholar] [CrossRef]
- Nakamura, Y.; Okamoto, W.; Kato, T.; Esaki, T.; Kato, K.; Komatsu, Y.; Yuki, S.; Masuishi, T.; Nishina, T.; Ebi, H.; et al. Circulating tumor DNA-guided treatment with pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer: A phase 2 trial. Nat. Med. 2021, 27, 1899–1903. [Google Scholar] [CrossRef] [PubMed]
- Vaghi, C.; Mauri, G.; Agostara, A.G.; Patelli, G.; Pizzutilo, E.G.; Nakamura, Y.; Yoshino, T.; Siena, S.; Sartore-Bianchi, A. The predictive role of ERBB2 point mutations in metastatic colorectal cancer: A systematic review. Cancer Treat. Rev. 2023, 112, 102488. [Google Scholar] [CrossRef]
- Siravegna, G.; Lazzari, L.; Crisafulli, G.; Sartore-Bianchi, A.; Mussolin, B.; Cassingena, A.; Martino, C.; Lanman, R.B.; Nagy, R.J.; Fairclough, S.; et al. Radiologic and genomic evolution of individual metastases during HER2 blockade in colorectal cancer. Cancer Cell 2018, 34, 148–162.e7. [Google Scholar] [CrossRef]
- Silveira, A.B.; Bidard, F.C.; Kasperek, A.; Melaabi, S.; Tanguy, M.L.; Rodrigues, M.; Bataillon, G.; Cabel, L.; Buecher, B.; Pierga, J.Y.; et al. High-accuracy determination of microsatellite instability compatible with liquid biopsies. Clin. Chem. 2020, 66, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Tieng, F.Y.F.; Abu, N.; Lee, L.H.; Ab Mutalib, N.S. Microsatellite instability in colorectal cancer liquid biopsy—Current updates on its potential in non-invasive detection, prognosis and as a predictive marker. Diagnostics 2021, 11, 544. [Google Scholar] [CrossRef]
- Willis, J.; Lefterova, M.I.; Artyomenko, A.; Kasi, P.M.; Nakamura, Y.; Mody, K.; Catenacci, D.V.T.; Fakih, M.; Barbacioru, C.; Zhao, J.; et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin. Cancer Res. 2019, 25, 7035–7045. [Google Scholar] [CrossRef]
- Ceccon, C.; Angerilli, V.; Rasola, C.; Procaccio, L.; Sabbadin, M.; Bergamo, F.; Malapelle, U.; Lonardi, S.; Fassan, M. Microsatellite instable colorectal adenocarcinoma diagnostics: The advent of liquid biopsy approaches. Front. Oncol. 2022, 12, 930108. [Google Scholar] [CrossRef]
- Joost, P.; Veurink, N.; Holck, S.; Klarskov, L.; Bojesen, A.; Harbo, M.; Baldetorp, B.; Rambech, E.; Nilbert, M. Heterogenous mismatch-repair status in colorectal cancer. Diagn. Pathol. 2014, 9, 126. [Google Scholar] [CrossRef]
- McCarthy, A.J.; Capo-Chichi, J.M.; Spence, T.; Grenier, S.; Stockley, T.; Kamel-Reid, S.; Serra, S.; Sabatini, P.; Chetty, R. Heterogenous loss of mismatch repair (MMR) protein expression: A challenge for immunohistochemical interpretation and microsatellite instability (MSI) evaluation. J. Pathol. Clin. Res. 2019, 5, 115–129. [Google Scholar] [CrossRef]
- Kumar, A.; Green, M.; Thacker, J.; Jeck, W.R.; Strickler, J.H. Circulating tumor DNA testing overcomes limitations of comprehensive genomic profiling from tumor tissue. Case Rep. Oncol. 2023, 16, 210–217. [Google Scholar] [CrossRef]
- Loupakis, F.; Maddalena, G.; Depetris, I.; Murgioni, S.; Bergamo, F.; Dei Tos, A.P.; Rugge, M.; Munari, G.; Nguyen, A.; Szeto, C.; et al. Treatment with checkpoint inhibitors in a metastatic colorectal cancer patient with molecular and immunohistochemical heterogeneity in MSI/dMMR status. J. Immunother. Cancer 2019, 7, 297. [Google Scholar] [CrossRef]
- Kasi, P.M. Mutational burden on circulating cell-free tumor-DNA testing as a surrogate marker of mismatch repair deficiency or microsatellite instability in patients with colorectal cancers. J. Gastrointest. Oncol. 2017, 8, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, A.; Durham, J.N.; Keefer, L.A.; Bartlett, B.R.; Zielonka, M.; Murphy, D.; White, J.R.; Lu, S.; Verner, E.L.; Ruan, F.; et al. Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin. Cancer Res. 2019, 25, 7024–7034. [Google Scholar] [CrossRef] [PubMed]
- André, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef] [PubMed]
- Kasi, P.M.; Budde, G.; Krainock, M.; Aushev, V.N.; Koyen Malashevich, A.; Malhotra, M.; Olshan, P.; Billings, P.R.; Aleshin, A. Circulating tumor DNA (ctDNA) serial analysis during progression on PD-1 blockade and later CTLA-4 rescue in patients with mismatch repair deficient metastatic colorectal cancer. J. Immunother. Cancer 2022, 10, e003312. [Google Scholar] [CrossRef]
- Stein, A.; Binder, M.; Goekkurt, E.; Lorenzen, S.; Riera-Knorrenschild, J.; Depenbusch, R.; Riera-Knorrenschild, J.; Lorenzen, S.; Ettrich, T.J.; Doerfel, S.; et al. Avelumab and cetuximab in combination with FOLFOX in patients with previously untreated metastatic colorectal cancer: Final results of the phase II AVETUX trial. J. Clin. Oncol. 2020, 38 (Suppl. S4), 96. [Google Scholar] [CrossRef]
- Tintelnot, J.; Ristow, I.; Sauer, M.; Simnica, D.; Schultheiß, C.; Scholz, R.; Goekkurt, E.; von Wenserski, L.; Willscher, E.; Paschold, L.; et al. Translational analysis and final efficacy of the AVETUX trial—Avelumab, cetuximab and FOLFOX in metastatic colorectal cancer. Front. Oncol. 2022, 12, 993611. [Google Scholar] [CrossRef]
- Stein, A.; Simnica, D.; Schultheiß, C.; Scholz, R.; Tintelnot, J.; Gökkurt, E.; von Wenserski, L.; Willscher, E.; Paschold, L.; Sauer, M.; et al. PD-L1 targeting and subclonal immune escape mediated by PD-L1 mutations in metastatic colorectal cancer. J. Immunother. Cancer 2021, 9, e002844. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration (FDA). Breakthrough Device Designation for Signatera MRD Test. Available online: https://www.fda.gov/medical-devices (accessed on 3 July 2025).
- National Comprehensive Cancer Network (NCCN). Clinical Practice Guidelines in Oncology—Colon Cancer. Version 2.2024. Available online: https://www.nccn.org (accessed on 3 July 2025).
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).