Loss of SVIP Results in Metabolic Reprograming and Increased Retention of Very-Low-Density Lipoproteins in Hepatocytes
Abstract
1. Introduction
2. Results
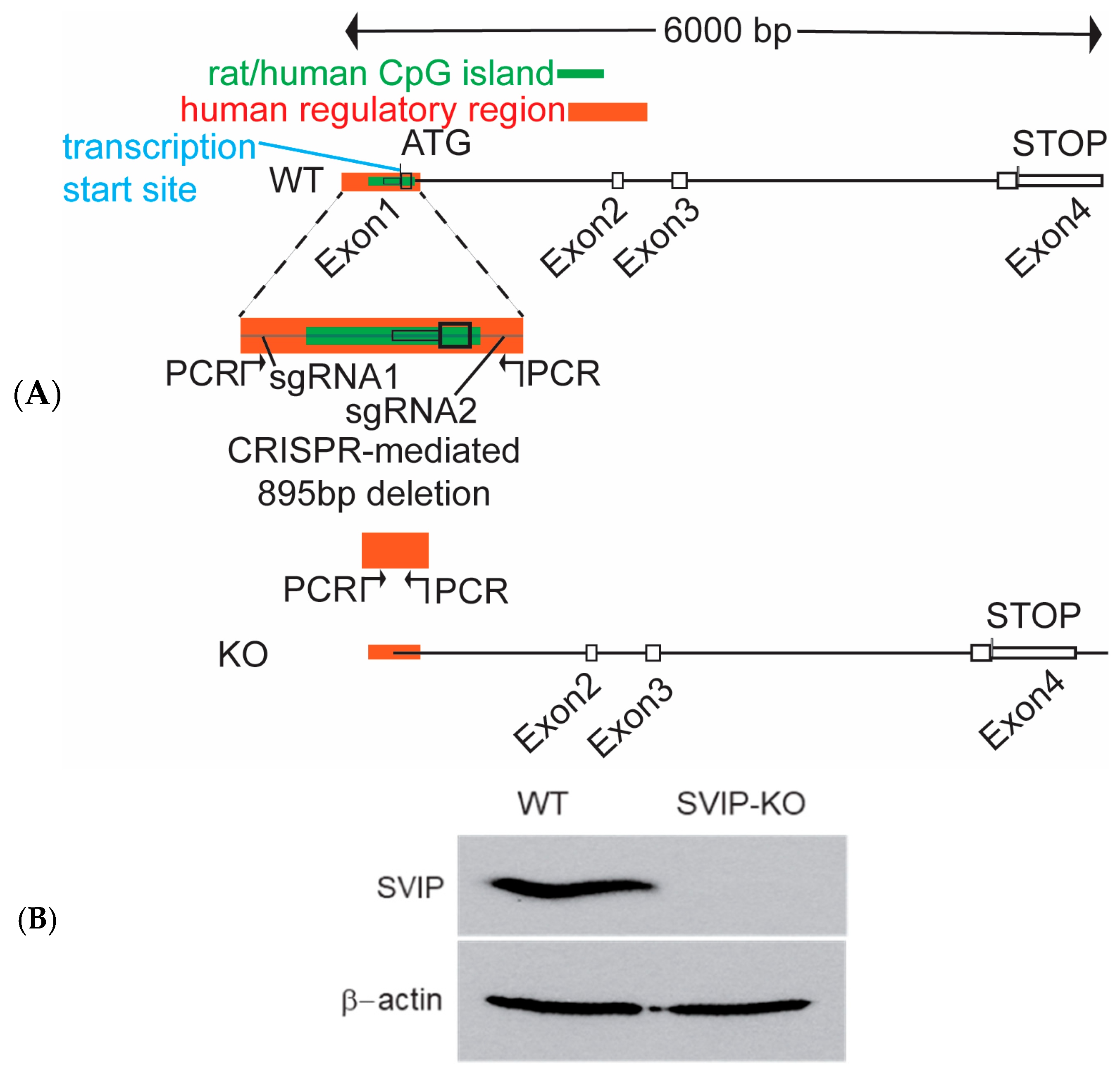
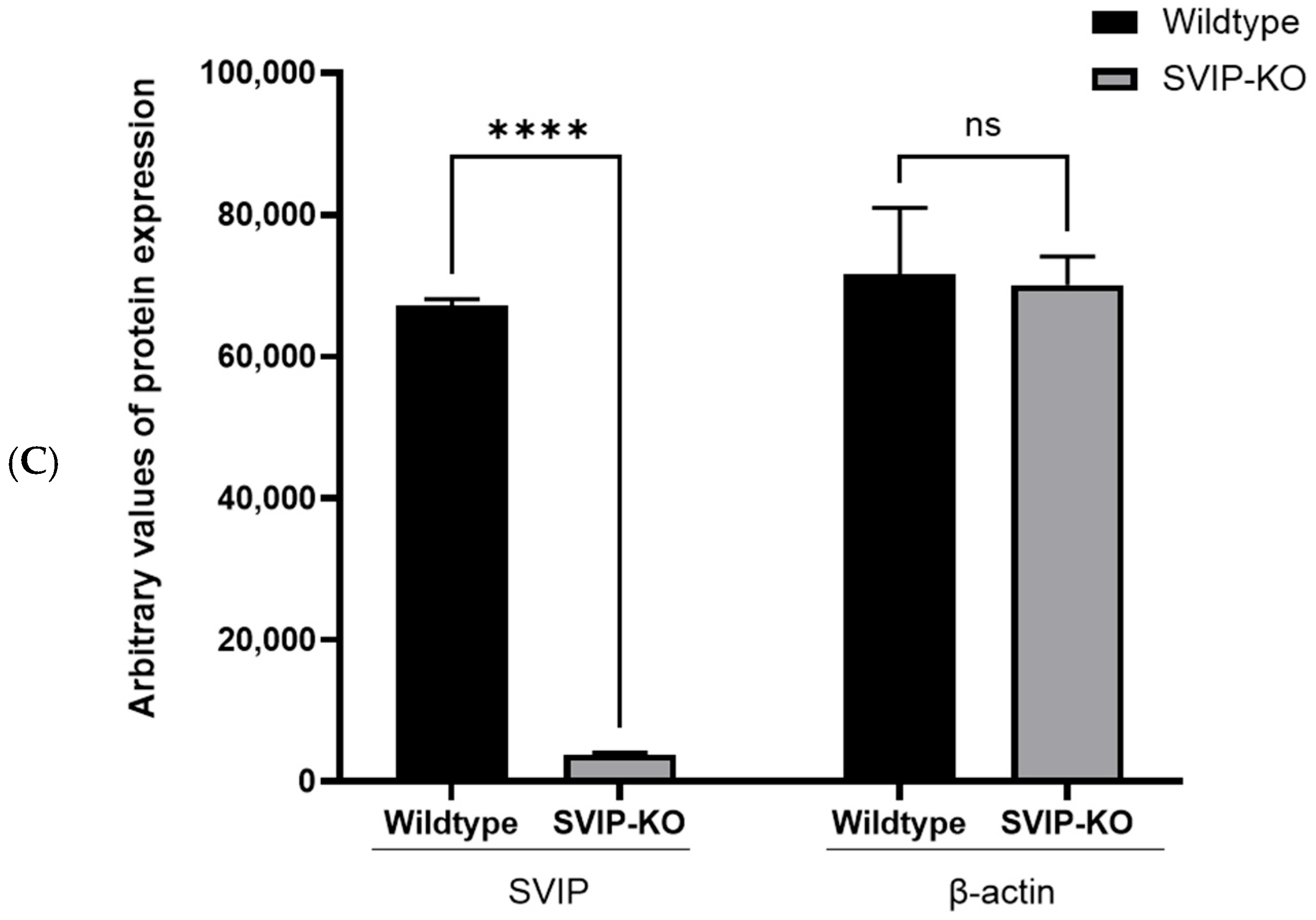
2.1. Generation of CRISPR-Cas9-Mediated SVIP-Knockout Rat Hepatoma Cells
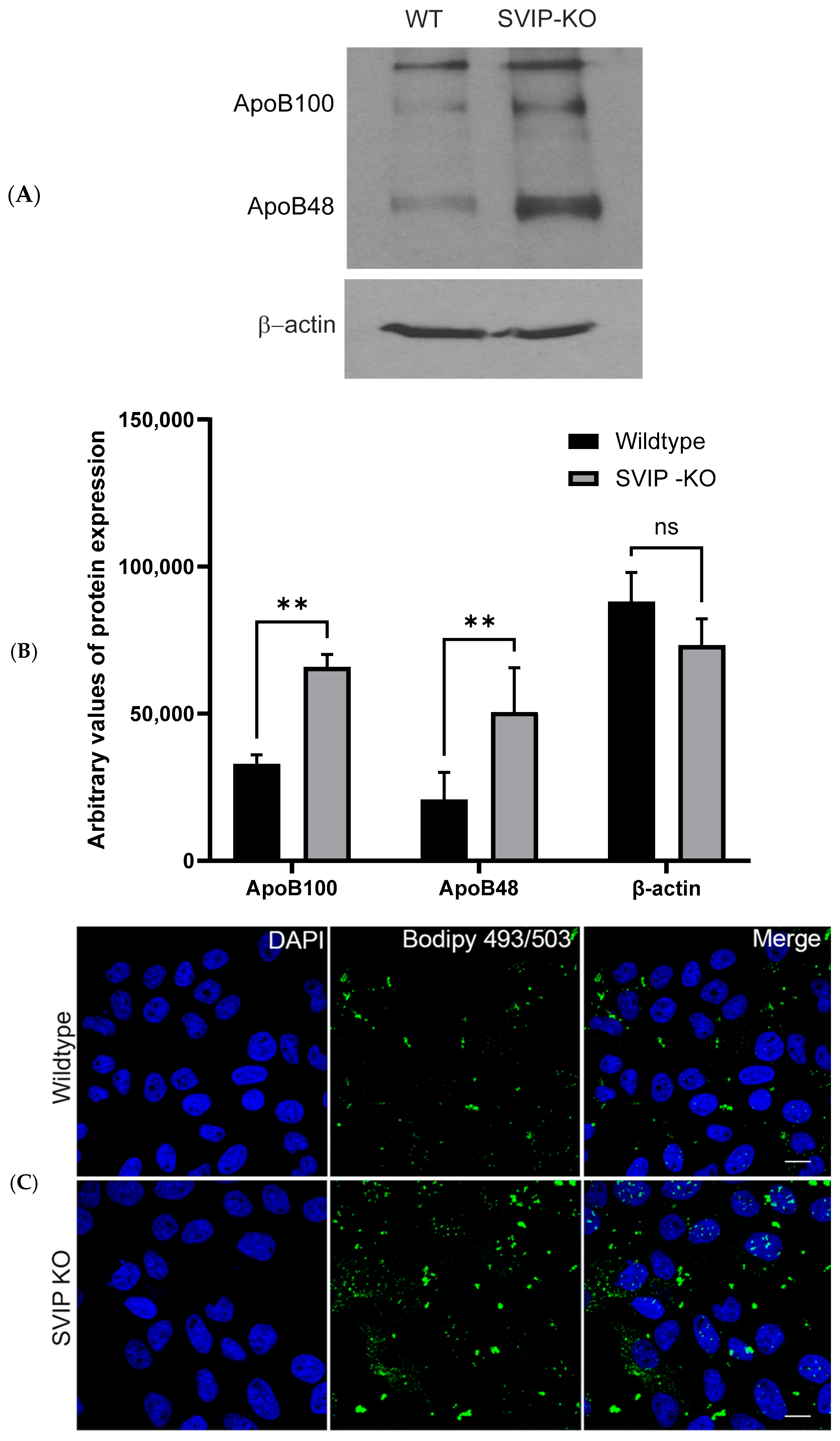
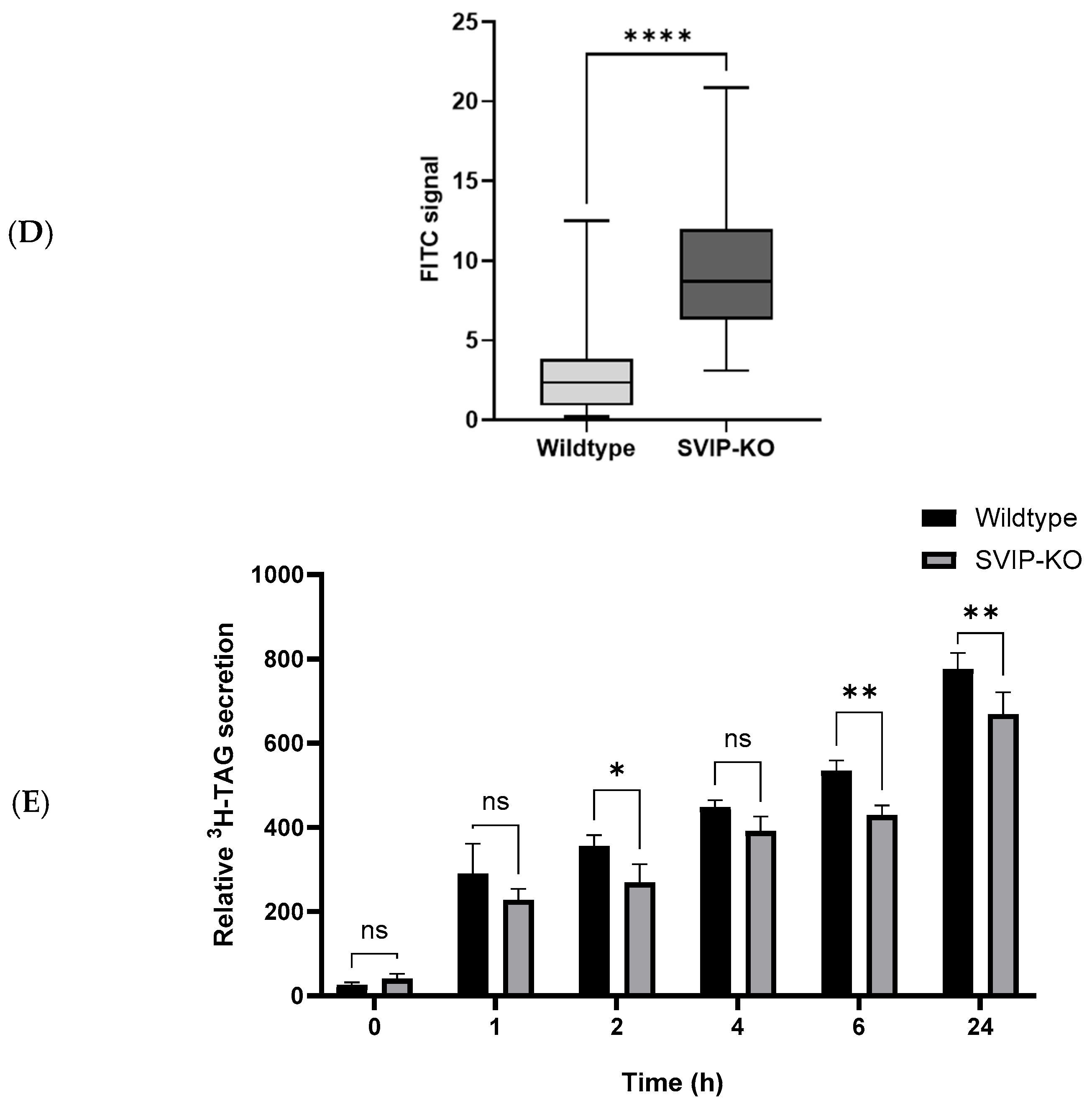
2.2. CRISPR-Cas9-Mediated SVIP Knockout Results in Increased VLDL Retention in Hepatoma Cells
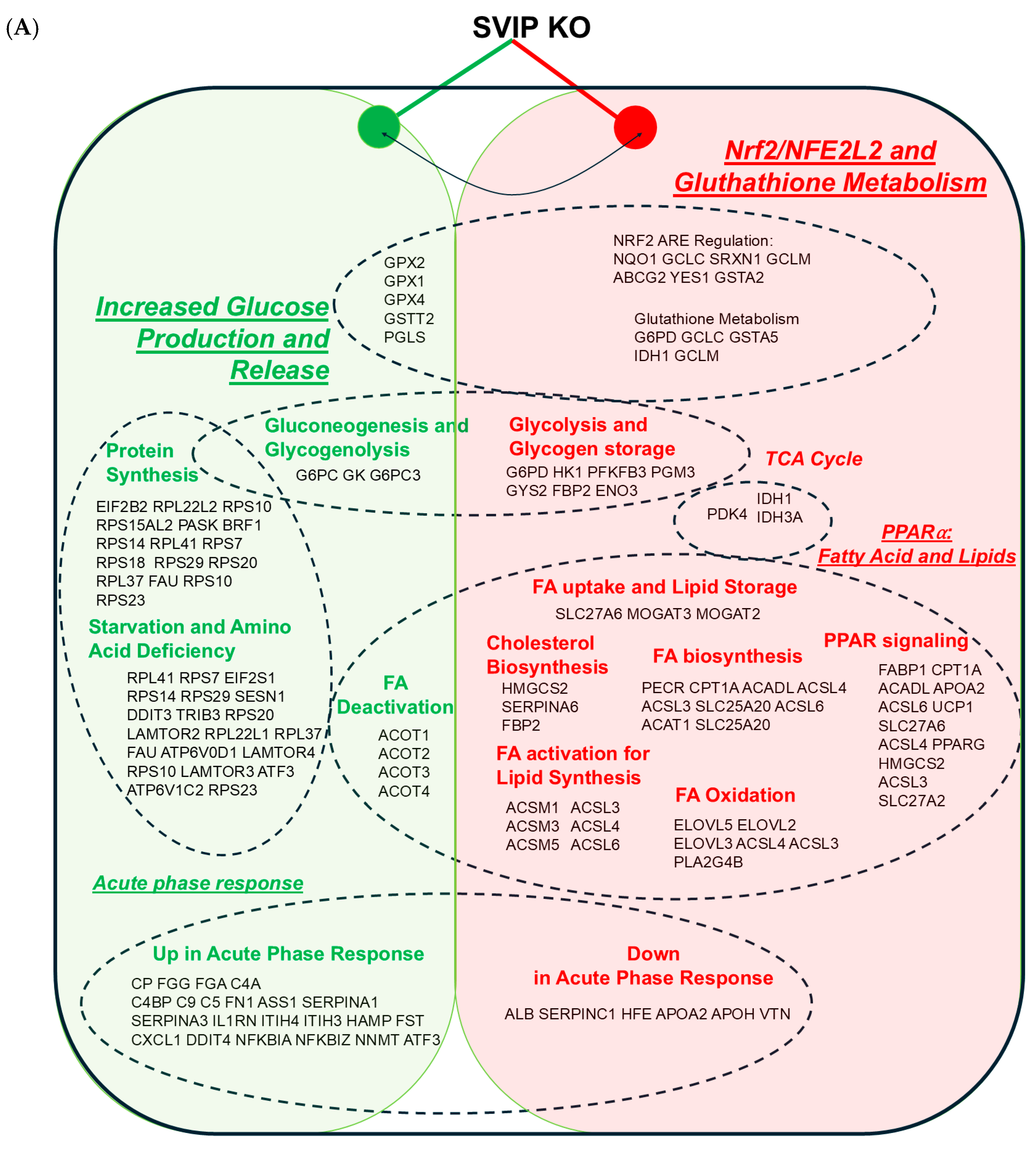
2.3. SVIP KO Cells Show a Differential Expression of Genes Involved in Fatty Acid Metabolism and the PPARα and Nrf2 Signaling Pathways
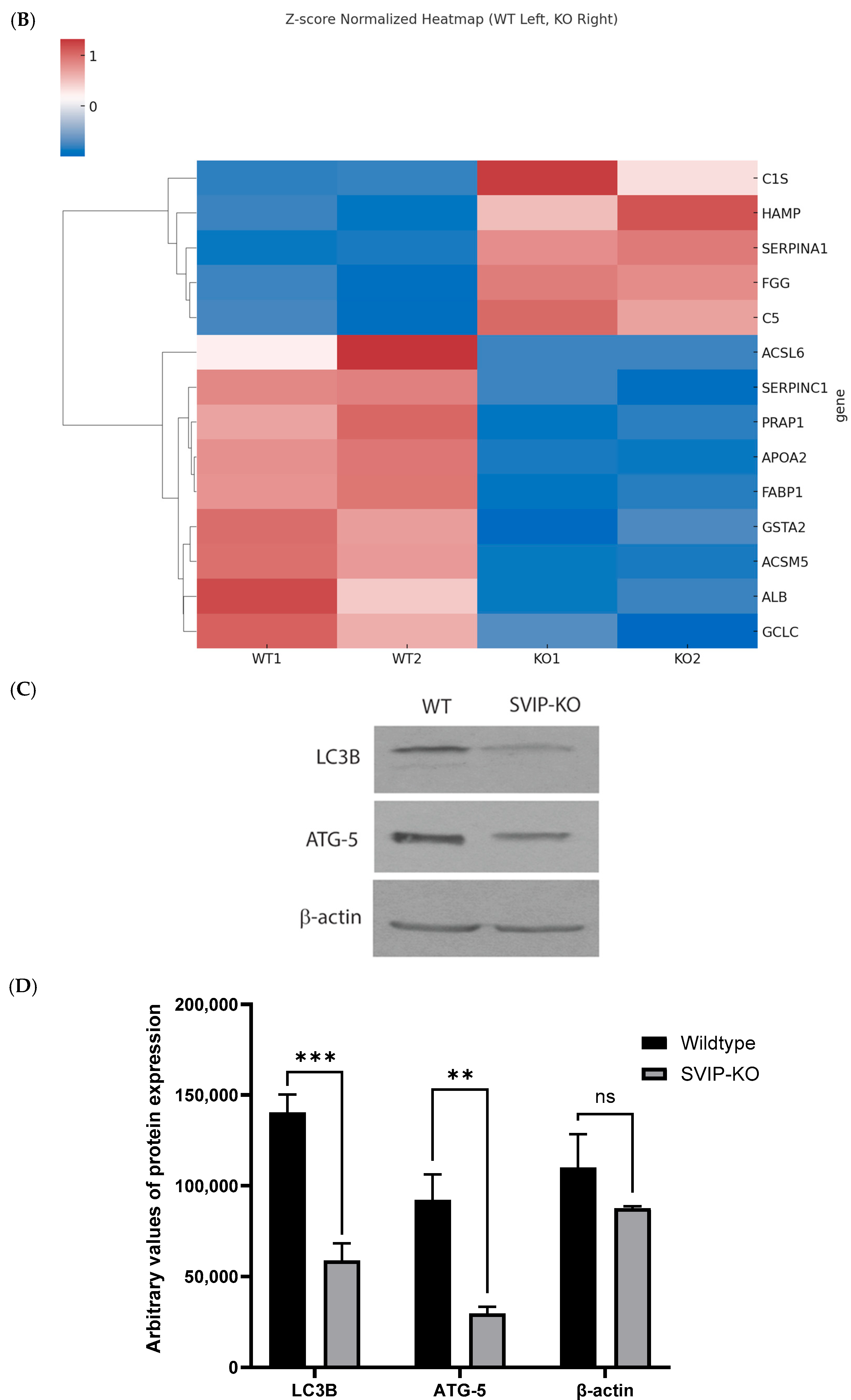
2.4. Knockout of the SVIP Gene Significantly Reduces the Intracellular Levels of the L-FABP Protein
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Antibodies
4.3. CRISPR-Cas9 Mediated SVIP Knockout
4.4. Immunoblotting
4.5. Determination of Triglycerides Secretion Using [3H] TAG Secretion Assay
4.6. RNA Preparation and Quantitative Reverse Transcription–PCR
4.7. RNA Sequencing Studies of Rat Hepatoma Cell Lines
4.8. Immunofluorescence Assay
4.9. Image Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SVIP | Small valosin-containing protein-interacting protein |
| L-FABP | Liver fatty-acid-binding protein |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
| APR | Acute phase response |
| COPII | Coat complex proteins II |
References
- Olié, V.; Gabet, A.; Grave, C.; Helft, G.; Fosse-Edorh, S.; Piffaretti, C.; Lailler, G.; Verdot, C.; Deschamps, V.; Vay-Demouy, J.; et al. Epidemiology of cardiovascular risk factors: Non-behavioural risk factors. Arch. Cardiovasc. Dis. 2024, 117, 761–769. [Google Scholar] [CrossRef] [PubMed]
- van Zwol, W.; van de Sluis, B.; Ginsberg, H.N.; Kuivenhoven, J.A. VLDL Biogenesis and Secretion: It Takes a Village. Circ. Res. 2024, 134, 226–244. [Google Scholar] [CrossRef]
- Shelness, G.S.; Ingram, M.F.; Huang, X.F.; DeLozier, J.A. Apolipoprotein B in the rough endoplasmic reticulum: Translation, translocation and the initiation of lipoprotein assembly. J. Nutr. 1999, 129 (Suppl. S2), 456S–462S. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, S.A. VLDL exits from the endoplasmic reticulum in a specialized vesicle, the VLDL transport vesicle, in rat primary hepatocytes. Biochem. J. 2008, 413, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Barlowe, C. COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 1994, 77, 895–907. [Google Scholar] [CrossRef]
- D’ARcangelo, J.G.; Stahmer, K.R.; Miller, E.A. Vesicle-mediated export from the ER: COPII coat function and regulation. Biochim. Biophys. Acta (BBA)–Mol. Cell Res. 2013, 1833, 2464–2472. [Google Scholar] [CrossRef]
- Saleem, U.; Abumrad, N.A.; Davidson, N.O.; Storch, J.; Siddiqi, S.A.; Mansbach, C.M. A novel multiprotein complex is required to generate the prechylomicron transport vesicle from intestinal ER. J. Lipid Res. 2010, 51, 1918–1928. [Google Scholar] [CrossRef]
- Rahim, A.; Nafi-valencia, E.; Siddiqi, S.; Basha, R.; Runyon, C.C.; Siddiqi, S.A. Proteomic analysis of the very low density lipoprotein (VLDL) transport vesicles. J. Proteom. 2012, 75, 2225–2235. [Google Scholar] [CrossRef]
- Tiwari, S.; Siddiqi, S.; Siddiqi, S.A. CideB protein is required for the biogenesis of very low density lipoprotein (VLDL) transport vesicle. J. Biol. Chem. 2013, 288, 5157–5165. [Google Scholar] [CrossRef]
- Ye, J.; Li, J.Z.; Liu, Y.; Li, X.; Yang, T.; Ma, X.; Li, Q.; Yao, Z.; Li, P. Cideb, an ER- and lipid droplet-associated protein, mediates VLDL lipidation and maturation by interacting with apolipoprotein B. Cell Metab. 2009, 9, 177–190. [Google Scholar] [CrossRef]
- Tiwari, S.; Siddiqi, S.; Zhelyabovska, O.; Siddiqi, S.A. Silencing of Small Valosin-containing Protein-interacting Protein (SVIP) Reduces Very Low Density Lipoprotein (VLDL) Secretion from Rat Hepatocytes by Disrupting Its Endoplasmic Reticulum (ER)-to-Golgi Trafficking. J. Biol. Chem. 2016, 291, 12514–12526. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, M.; Suzuki, M.; Hamada, Y.; Hatsuzawa, K.; Tani, K.; Yamamoto, A.; Tagaya, M.; Malhotra, V. SVIP is a novel VCP/p97-interacting protein whose expression causes cell vacuolation. Mol. Biol. Cell 2003, 14, 262–273. [Google Scholar] [CrossRef]
- Ballar, P.; Zhong, Y.; Nagahama, M.; Tagaya, M.; Shen, Y.; Fang, S. Identification of SVIP as an endogenous inhibitor of endoplasmic reticulum-associated degradation. J. Biol. Chem. 2007, 282, 33908–33914. [Google Scholar] [CrossRef]
- Kounatidis, D.; Vallianou, N.G.; Poulaki, A.; Evangelopoulos, A.; Panagopoulos, F.; Stratigou, T.; Geladari, E.; Karampela, I.; Dalamaga, M. ApoB100 and Atherosclerosis: What’s New in the 21st Century? Metabolites 2024, 14, 123. [Google Scholar] [CrossRef]
- Mandard, S.; Muller, M.; Kersten, S. Peroxisome proliferator-activated receptor alpha target genes. Cell Mol. Life Sci. 2004, 61, 393–416. [Google Scholar] [CrossRef] [PubMed]
- van der Sluis, R.; Erasmus, E. Xenobiotic/medium chain fatty acid: CoA ligase—A critical review on its role in fatty acid metabolism and the detoxification of benzoic acid and aspirin. Expert Opin. Drug Metab. Toxicol. 2016, 12, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, J.; Qiu, S.; Ni, S.; Duan, Y. ACSM5 inhibits ligamentum flavum hypertrophy by regulating lipid accumulation mediated by FABP4/PPAR signaling pathway. Biol. Direct 2023, 18, 75. [Google Scholar] [CrossRef]
- Sebastiano, M.R.; Konstantinidou, G. Targeting Long Chain Acyl-CoA Synthetases for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 3624. [Google Scholar] [CrossRef]
- Peng, H.; Chiu, T.-Y.; Liang, Y.-J.; Lee, C.-J.; Liu, C.-S.; Suen, C.-S.; Yen, J.J.-Y.; Chen, H.-T.; Hwang, M.-J.; Hussain, M.M.; et al. PRAP1 is a novel lipid-binding protein that promotes lipid absorption by facilitating MTTP-mediated lipid transport. J. Biol. Chem. 2021, 296, 100052. [Google Scholar] [CrossRef]
- Djouadi, F.; Weinheimer, C.J.; E Saffitz, J.; Pitchford, C.; Bastin, J.; Gonzalez, F.J.; Kelly, D.P. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator- activated receptor alpha- deficient mice. J. Clin. Investig. 1998, 102, 1083–1091. [Google Scholar] [CrossRef]
- Chambel, S.S.; Santos-Goncalves, A.; Duarte, T.L. The Dual Role of Nrf2 in Nonalcoholic Fatty Liver Disease: Regulation of Antioxidant Defenses and Hepatic Lipid Metabolism. BioMed Res. Int. 2015, 2015, 597134. [Google Scholar] [CrossRef]
- Qiu, S.; Liang, Z.; Wu, Q.; Wang, M.; Yang, M.; Chen, C.; Zheng, H.; Zhu, Z.; Li, L.; Yang, G. Hepatic lipid accumulation induced by a high-fat diet is regulated by Nrf2 through multiple pathways. FASEB J. 2022, 36, e22280. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Wang, Y.Y.; Wang, P.; Huang, Y.; Liang, D.Y.; Wang, D.; Cheng, C.; Zhang, C.; Guo, L.; Liang, P.; et al. SVIP alleviates CCl(4)-induced liver fibrosis via activating autophagy and protecting hepatocytes. Cell Death Dis. 2019, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Ehle, C.; Iyer-Bierhoff, A.; Wu, Y.; Xing, S.; Kiehntopf, M.; Mosig, A.S.; Godmann, M.; Heinzel, T. Downregulation of HNF4A enables transcriptomic reprogramming during the hepatic acute-phase response. Commun. Biol. 2024, 7, 589. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Lu, S.; He, Z.; Altmann, C.; Montford, J.R.; Li, A.S.; Lucia, M.S.; Orlicky, D.J.; Weiser-Evans, M.; Faubel, S. IL-6 mediates the hepatic acute phase response after prerenal azotemia in a clinically defined murine model. Am. J. Physiol. Renal Physiol. 2023, 325, F328–F344. [Google Scholar] [CrossRef]
- Gervois, P.; Kleemann, R.; Pilon, A.; Percevault, F.; Koenig, W.; Staels, B.; Kooistra, T. Global suppression of IL-6-induced acute phase response gene expression after chronic in vivo treatment with the peroxisome proliferator-activated receptor-alpha activator fenofibrate. J. Biol. Chem. 2004, 279, 16154–16160. [Google Scholar] [CrossRef]
- Ip, E.; Farrell, G.C.; Robertson, G.; Hall, P.; Kirsch, R.; Leclercq, I. Central role of PPARalpha-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology 2003, 38, 123–132. [Google Scholar] [CrossRef]
- Rutkowski, D.T.; Kaufman, R.J. That which does not kill me makes me stronger: Adapting to chronic ER stress. Trends Biochem. Sci. 2007, 32, 469–476. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Hertzel, A.V.; Bernlohr, D.A. The mammalian fatty acid-binding protein multigene family: Molecular and genetic insights into function. Trends Endocrinol. Metab. 2000, 11, 175–180. [Google Scholar] [CrossRef]
- Wang, G.; Bonkovsky, H.L.; de Lemos, A.; Burczynski, F.J. Recent insights into the biological functions of liver fatty acid binding protein 1. J. Lipid Res. 2015, 56, 2238–2247. [Google Scholar] [CrossRef]
- Huang, H.; Starodub, O.; McIntosh, A.; Atshaves, B.P.; Woldegiorgis, G.; Kier, A.B.; Schroeder, F. Liver fatty acid-binding protein colocalizes with peroxisome proliferator activated receptor alpha and enhances ligand distribution to nuclei of living cells. Biochemistry 2004, 43, 2484–2500. [Google Scholar] [CrossRef]
- Lawrence, J.W.; Kroll, D.J.; Eacho, P.I. Ligand-dependent interaction of hepatic fatty acid-binding protein with the nucleus. J. Lipid Res. 2000, 41, 1390–1401. [Google Scholar] [CrossRef]
- Nakagawa, S.; Kawashima, Y.; Hirose, A.; Kozuka, H. Regulation of hepatic level of fatty-acid-binding protein by hormones and clofibric acid in the rat. Biochem. J. 1994, 297 Pt 3, 581–584. [Google Scholar] [CrossRef] [PubMed]
- Mashek, D.G. Hepatic lipid droplets: A balancing act between energy storage and metabolic dysfunction in NAFLD. Mol. Metab. 2021, 50, 101115. [Google Scholar] [CrossRef]
- Seebacher, F.; Zeigerer, A.; Kory, N.; Krahmer, N. Hepatic lipid droplet homeostasis and fatty liver disease. Semin. Cell Dev. Biol. 2020, 108, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Régnier, M.; Polizzi, A.; Smati, S.; Lukowicz, C.; Fougerat, A.; Lippi, Y.; Fouché, E.; Lasserre, F.; Naylies, C.; Bétoulières, C.; et al. Hepatocyte-specific deletion of Pparalpha promotes NAFLD in the context of obesity. Sci. Rep. 2020, 10, 6489. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, R.M.; Bauge, E.; Staels, B.; Gervois, P. Systemic and distal repercussions of liver-specific peroxisome proliferator-activated receptor-alpha control of the acute-phase response. Endocrinology 2008, 149, 3215–3223. [Google Scholar] [CrossRef]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef]
- Coulouarn, C.; Lefebvre, G.; Derambure, C.; Lequerre, T.; Scotte, M.; Francois, A.; Cellier, D.; Daveau, M.; Salier, J.-P. Altered gene expression in acute systemic inflammation detected by complete coverage of the human liver transcriptome. Hepatology 2004, 39, 353–364. [Google Scholar] [CrossRef]

| Name | Fold Change | Average KO | Average CTRL | Function |
|---|---|---|---|---|
| (1) | ||||
| ApoA2 | 32.49 | 11.21 | 364.45 | Plasma lipoprotein assembly, PPARA target |
| Pdzk1 | 22.27 | 1.90 | 42.50 | Carnitine transport |
| Sult2a6 | 15.08 | 6.22 | 93.82 | Sulfotransferase family member involved in sulfation |
| Aadac | 11.23 | 2.87 | 32.24 | Positive regulation of triglyceride catabolic process |
| ApoH | 6.83 | 36.48 | 249.31 | Component of circulating plasma lipoproteins |
| Fabp1 | 6.77 | 72.08 | 488.10 | Regulation of lipid metabolism by PPARα, fatty acid transporters |
| Serpina6 | 6.56 | 27.82 | 182.54 | Metabolism of lipids involved in glucocorticoid metabolic process |
| Cldn2 | 4.88 | 5.18 | 25.33 | Vitamin D receptor pathway, cell–cell adhesion |
| Cryl1 | 4.44 | 6.03 | 26.80 | Carbohydrate metabolism, D-glucuronate catabolic process to D-xylulose 5-phosphate |
| Prap1 | 4.43 | 16.31 | 72.27 | Triglyceride binding, DNA damage response, endoplasmic reticulum |
| Dpp4 | 4.35 | 16.20 | 70.44 | Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1), ferroptosis |
| Gys2 | 4.34 | 9.05 | 39.28 | Glycogen synthesis |
| Serpind1 | 3.83 | 5.90 | 22.64 | Formation of fibrin clot (clotting cascade), complement and coagulation cascades |
| Atp10a | 3.29 | 8.54 | 28.07 | ATPase-coupled intramembrane lipid transporter activity, phosphatidylcholine flippase activity |
| SVIP | Inf. | 0 | 11.72 | Negative regulation of ERAD pathway, positive regulation of autophagy |
| (2) | ||||
| Paics | Inf. | 55.29 | 0.00 | Nucleotide biosynthesis |
| S100g | 10.73 | 68.41 | 6.38 | Vitamin D receptor pathway |
| Lcn2 | 7.33 | 65.58 | 8.95 | Iron uptake and transport |
| Fuca2 | 5.69 | 20.62 | 3.63 | Plasma fucosidase |
| Cp | 4.33 | 26.62 | 6.14 | Iron uptake and transport, ferroptosis, metal ion SLC transporters, copper ion binding; up in APR |
| Wfdc21 | 4.16 | 58.83 | 14.13 | Antibacterial humoral response |
| Fst | 4.00 | 89.02 | 22.23 | Hepatokine (other hepatokines such as Selenop, Shbg, Smoc1, and Clu are also upregulated), pro skeletal muscle growth, antagonism of activin by Follistatin, up in APR |
| Hamp | 3.98 | 328.06 | 82.44 | Iron metabolism, defense response to bacterium, up in APR |
| C4bpb | 3.85 | 65.24 | 16.97 | Regulation of complement cascade, up in APR |
| Cyp2c6v1 | 3.70 | 27.35 | 7.40 | Arachidonate metabolic process, xenobiotic catabolic process, long-chain fatty acid omega-1 hydroxylase activity |
| Itih4 | 3.43 | 21.32 | 6.21 | Up in APR |
| Ifi27l2b | 3.33 | 438.58 | 131.81 | Innate immune response |
| Arhgef2 | 3.32 | 27.31 | 8.23 | Regulation of RhoA and RAC1 activity, “Innate Immune Sensor” |
| Adhfe1 | 3.27 | 20.87 | 6.37 | Oxidation of 4-hydroxybutyrate, aerobic respiration and respiratory electron transport, pyruvate metabolism and citric acid (TCA) cycle, tyrosine metabolism, glutamate catabolic process via 2-hydroxyglutarate |
| Ass1 | 3.24 | 28.39 | 8.75 | Citrulline–nitric oxide cycle, amino acid and derivative metabolism, urea cycle, arginine and proline metabolism, up in APR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sekhar, V.; Andl, T.; Siddiqi, S.A. Loss of SVIP Results in Metabolic Reprograming and Increased Retention of Very-Low-Density Lipoproteins in Hepatocytes. Int. J. Mol. Sci. 2025, 26, 7465. https://doi.org/10.3390/ijms26157465
Sekhar V, Andl T, Siddiqi SA. Loss of SVIP Results in Metabolic Reprograming and Increased Retention of Very-Low-Density Lipoproteins in Hepatocytes. International Journal of Molecular Sciences. 2025; 26(15):7465. https://doi.org/10.3390/ijms26157465
Chicago/Turabian StyleSekhar, Vandana, Thomas Andl, and Shadab A. Siddiqi. 2025. "Loss of SVIP Results in Metabolic Reprograming and Increased Retention of Very-Low-Density Lipoproteins in Hepatocytes" International Journal of Molecular Sciences 26, no. 15: 7465. https://doi.org/10.3390/ijms26157465
APA StyleSekhar, V., Andl, T., & Siddiqi, S. A. (2025). Loss of SVIP Results in Metabolic Reprograming and Increased Retention of Very-Low-Density Lipoproteins in Hepatocytes. International Journal of Molecular Sciences, 26(15), 7465. https://doi.org/10.3390/ijms26157465






