Hormonal Balance in Relation to Expression of Selected Genes Connected with Hormone Biosynthesis and Signalling—The Effect of Deacclimation Process in Oilseed Rape
Abstract
1. Introduction
2. Results
2.1. Phytohormones in Leaves
2.2. Phytohormones in Cell Sap
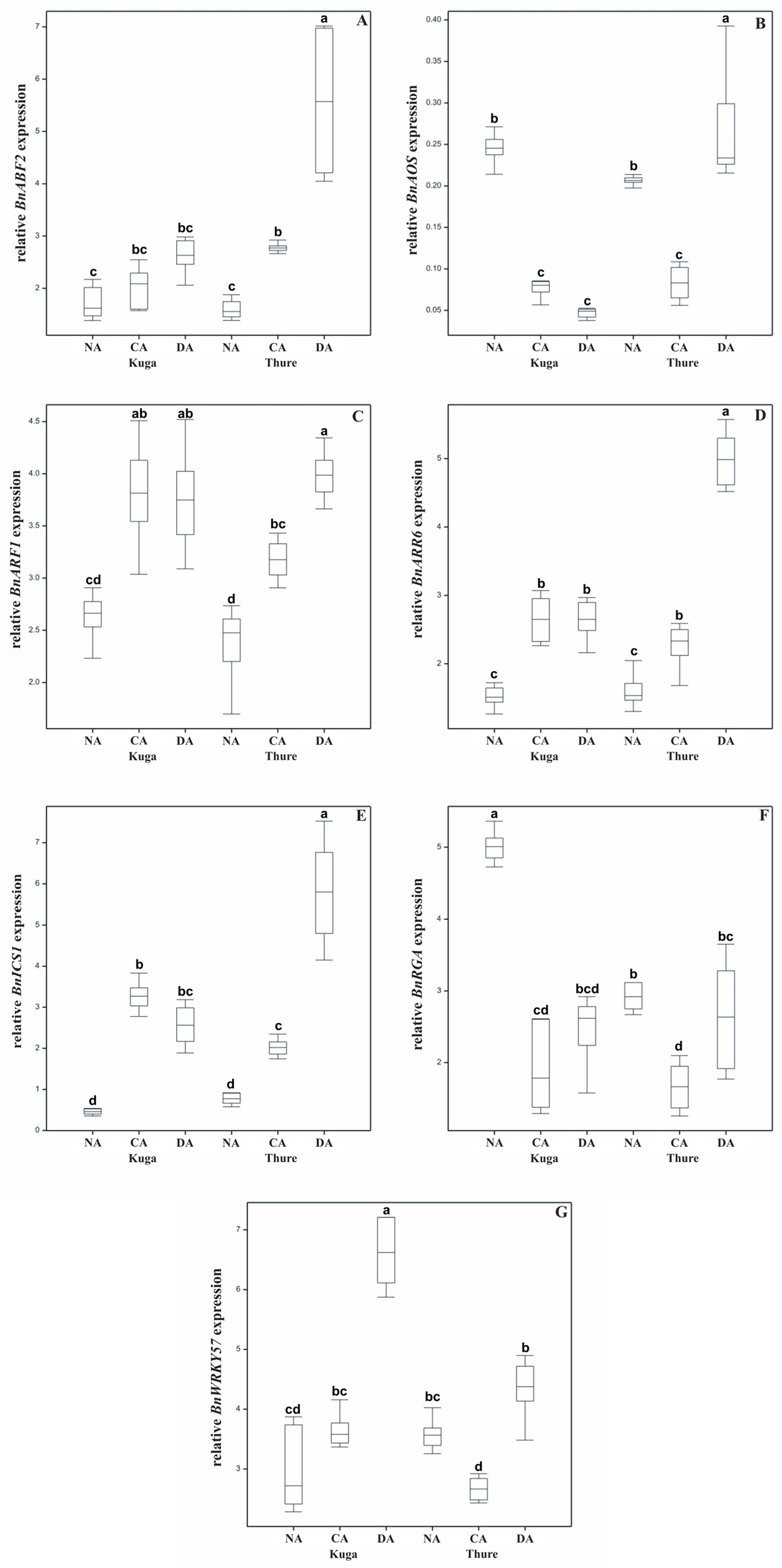
2.3. Expression of the BnRGA, BnARF1, BnARR6, BnABF2, BnISC1, BnAOS, and BnWRKY57
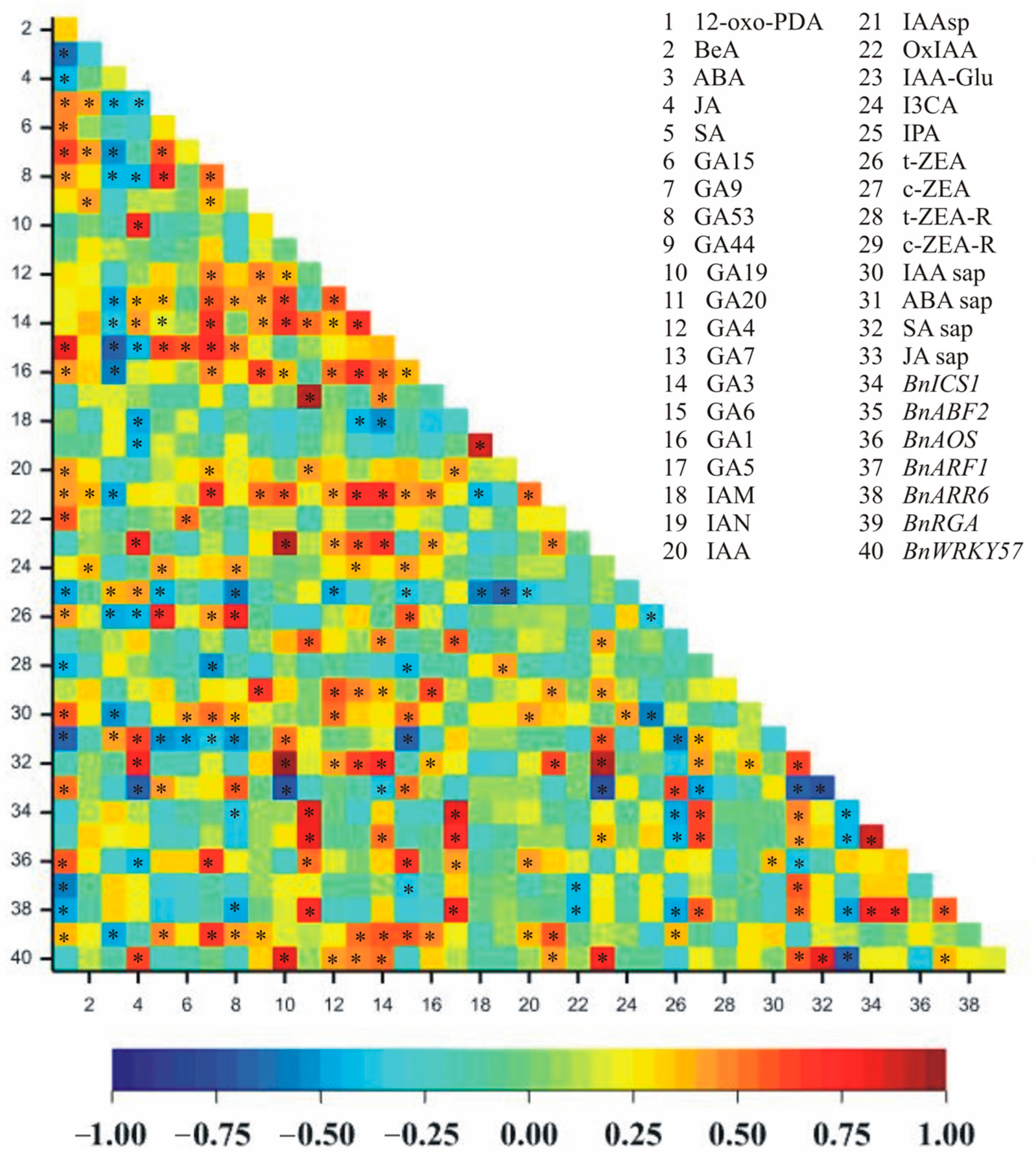
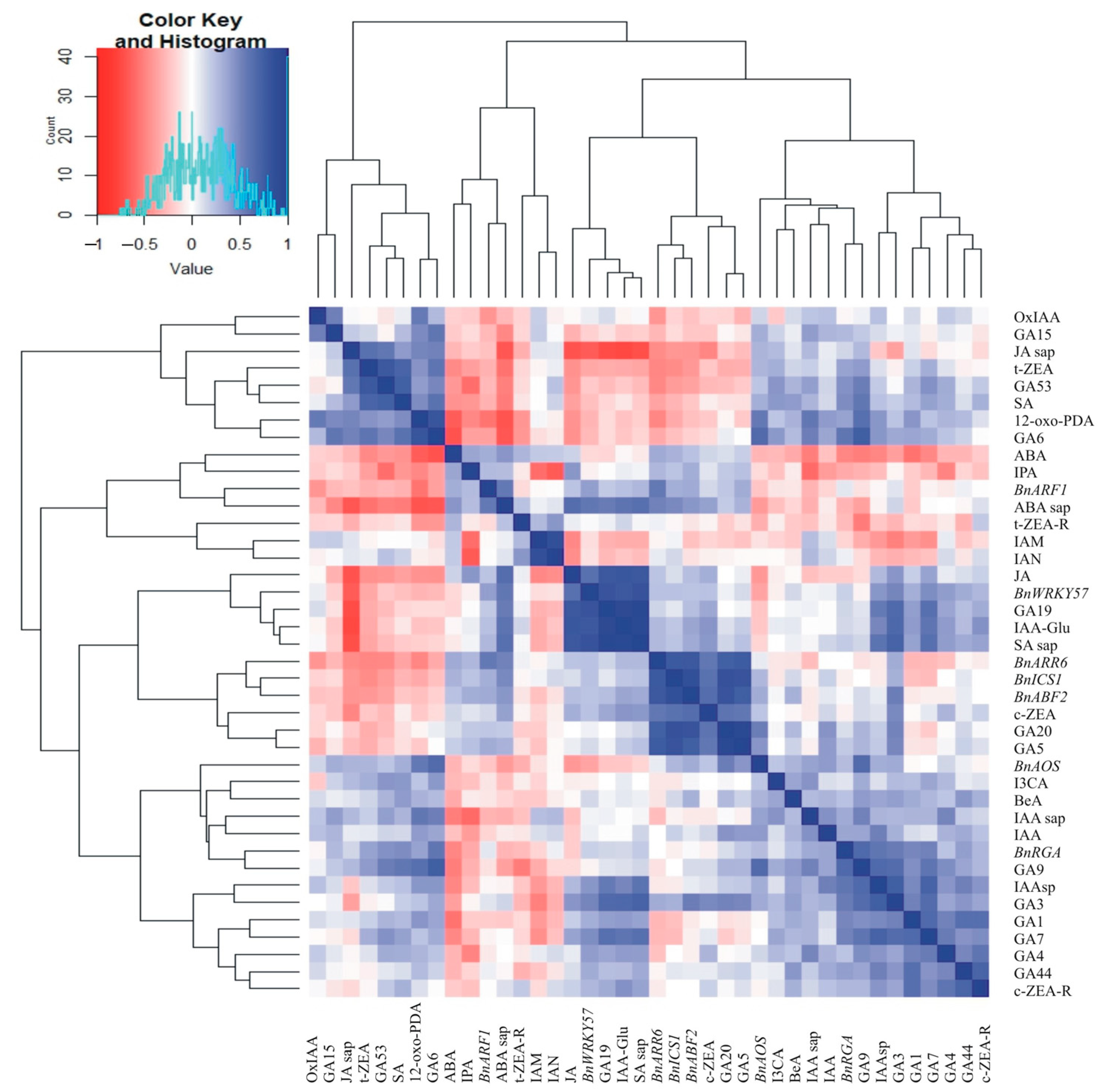
2.4. Correlations Between Hormone Concentrations in the Leaves, in the Cell Sap, and the Relative Expression of Hormone-Related Transcript Genes
3. Discussion
4. Materials and Methods
4.1. Plant Naterial
4.2. Experimental Design and Sampling
4.3. Analysis of the Plant Hormones and Related Metabolites in Leaves and in Cell Sap
4.4. Accumulation of the BnRGA, BnARF1, BnARR6, BnABF2, BnISC1, BnAOS, and BnWRKY57 Transcripts: RNA Isolation, cDNA Synthesis, and qRT-PCR Reaction
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yong, K.J.; Wu, T.Y. Second-Generation Bioenergy from Oilseed Crop Residues: Recent Technologies, Techno-Economic Assessments and Policies. Energy Convers. Manag. 2022, 267, 115869. [Google Scholar] [CrossRef]
- Fang, Y.; Ren, T.; Zhang, S.; Liu, Y.; Liao, S.; Li, X.; Cong, R.; Lu, J. Rotation with Oilseed Rape as the Winter Crop Enhances Rice Yield and Improves Soil Indigenous Nutrient Supply. Soil Tillage Res. 2021, 212, 105065. [Google Scholar] [CrossRef]
- Rudolphi-Szydło, E.; Dyba, B.; Janeczko, A.; Latowski, D.; Sadura, I.; Filek, M. Brassinosteroid-Lipid Membrane Interaction under Low and High Temperature Stress in Model Systems. BMC Plant Biol. 2022, 22, 246. [Google Scholar] [CrossRef] [PubMed]
- Rys, M.; Stachurska, J.; Rudolphi-Szydło, E.; Dziurka, M.; Waligórski, P.; Filek, M.; Janeczko, A. Does Deacclimation Reverse the Changes in Structural/Physicochemical Properties of the Chloroplast Membranes That Are Induced by Cold Acclimation in Oilseed Rape? Plant Physiol. Biochem. 2024, 214, 108961. [Google Scholar] [CrossRef]
- Sasaki, H.; Ichimura, K.; Oda, M. Changes in Sugar Content during Cold Acclimation and Deacclimation of Cabbage Seedlings. Ann. Bot. 1996, 78, 365–369. [Google Scholar] [CrossRef]
- Pagter, M.; Hausman, J.F.; Arora, R. Deacclimation Kinetics and Carbohydrate Changes in Stem Tissues of Hydrangea in Response to an Experimental Warm Spell. Plant Sci. 2011, 180, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Rys, M.; Pociecha, E.; Oliwa, J.; Ostrowska, A.; Jurczyk, B.; Saja, D.; Janeczko, A. Deacclimation of Winter Oilseed Rape-Insight into Physiological Changes. Agronomy 2020, 10, 1565. [Google Scholar] [CrossRef]
- Kosová, K.; Prášil, I.T.; Vítámvás, P.; Dobrev, P.; Motyka, V.; Floková, K.; Novák, O.; Turečková, V.; Rolčik, J.; Pešek, B.; et al. Complex Phytohormone Responses during the Cold Acclimation of Two Wheat Cultivars Differing in Cold Tolerance, Winter Samanta and Spring Sandra. J. Plant Physiol. 2012, 169, 567–576. [Google Scholar] [CrossRef]
- Sadura, I.; Libik-Konieczny, M.; Jurczyk, B.; Gruszka, D.; Janeczko, A. HSP Transcript and Protein Accumulation in Brassinosteroid Barley Mutants Acclimated to Low and High Temperatures. Int. J. Mol. Sci. 2020, 21, 1889. [Google Scholar] [CrossRef]
- Stachurska, J.; Sadura, I.; Rys, M.; Dziurka, M.; Janeczko, A. Insight into Hormonal Homeostasis and the Accumulation of Selected Heat Shock Proteins in Cold Acclimated and Deacclimated Winter Oilseed Rape. Agriculture 2023, 13, 641. [Google Scholar] [CrossRef]
- Pociecha, E.; Dziurka, M.; Waligórski, P.; Krępski, T.; Janeczko, A. 24-Epibrassinolide Pre-Treatment Modifies Cold-Induced Photosynthetic Acclimation Mechanisms and Phytohormone Response of Perennial Ryegrass in Cultivar-Dependent Manner. J. Plant Growth Regul. 2017, 36, 618–628. [Google Scholar] [CrossRef]
- Janeczko, A.; Dziurka, M.; Pociecha, E. Increased Leaf Tocopherol and β-Carotene Content Is Associated with the Tolerance of Winter Wheat Cultivars to Frost. J. Agron. Crop Sci. 2018, 204, 594–602. [Google Scholar] [CrossRef]
- Vyse, K.; Pagter, M.; Zuther, E.; Hincha, D.K. Deacclimation after Cold Acclimation- A Crucial, but Widely Neglected Part of Plant Winter Survival. J. Exp. Bot. 2019, 70, 4595–4604. [Google Scholar] [CrossRef]
- Stachurska, J.; Rys, M.; Pociecha, E.; Kalaji, H.M.; Dąbrowski, P.; Oklestkova, J.; Jurczyk, B.; Janeczko, A. Deacclimation-Induced Changes of Photosynthetic Efficiency, Brassinosteroid Homeostasis and BRI1 Expression in Winter Oilseed Rape (Brassica napus L.)—Relation to Frost Tolerance. Int. J. Mol. Sci. 2022, 23, 5224. [Google Scholar] [CrossRef]
- Stachurska, J.; Sadura, I.; Jurczyk, B.; Rudolphi-Szydło, E.; Dyba, B.; Pociecha, E.; Ostrowska, A.; Rys, M.; Kvasnica, M.; Oklestkova, J.; et al. Cold Acclimation and Deacclimation of Winter Oilseed Rape, with Special Attention Being Paid to the Role of Brassinosteroids. Int. J. Mol. Sci. 2024, 25, 6010. [Google Scholar] [CrossRef] [PubMed]
- Vaitkevičiūtė, G.; Aleliūnas, A.; Gibon, Y.; Armonienė, R. The Effect of Cold Acclimation, Deacclimation and Reacclimation on Metabolite Profiles and Freezing Tolerance in Winter Wheat. Front. Plant Sci. 2022, 13, 959118. [Google Scholar] [CrossRef] [PubMed]
- Byun, Y.J.; Koo, M.Y.; Joo, H.J.; Ha-Lee, Y.M.; Lee, D.H. Comparative Analysis of Gene Expression under Cold Acclimation, Deacclimation and Reacclimation in Arabidopsis. Physiol. Plant 2014, 152, 256–274. [Google Scholar] [CrossRef]
- Vítámvás, P.; Prášil, I.T. WCS120 Protein Family and Frost Tolerance during Cold Acclimation, Deacclimation and Reacclimation of Winter Wheat. Plant Physiol. Biochem. 2008, 46, 970–976. [Google Scholar] [CrossRef]
- Vaitkevičiūtė, G.; Aleliūnas, A.; Gibon, Y.; Armonienė, R. Comparative Analysis of Antioxidant Accumulation under Cold Acclimation, Deacclimation and Reacclimation in Winter Wheat. Plants 2022, 11, 2818. [Google Scholar] [CrossRef]
- Zuther, E.; Juszczak, I.; Ping Lee, Y.; Baier, M.; Hincha, D.K. Time-Dependent Deacclimation after Cold Acclimation in Arabidopsis Thaliana Accessions. Sci. Rep. 2015, 5, 12199. [Google Scholar] [CrossRef]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin Action in Response to Abiotic and Biotic Stresses in Plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Rapacz, M.; Waligórski, P.; Janowiak, F. ABA and Gibberellin-like Substances during Prehardening, Cold Acclimation, de- and Reacclimation of Oilseed Rape. Acta Physiol. Plant 2003, 25, 151–161. [Google Scholar] [CrossRef]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, Action, and Interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef]
- Majláth, I.; Szalai, G.; Soós, V.; Sebestyén, E.; Balázs, E.; Vanková, R.; Dobrev, P.I.; Tari, I.; Tandori, J.; Janda, T. Effect of Light on the Gene Expression and Hormonal Status of Winter and Spring Wheat Plants during Cold Hardening. Physiol. Plant 2012, 145, 296–314. [Google Scholar] [CrossRef]
- Gavelienė, V.; Novickienė, L.; Pakalniškytė, L. Effect of Auxin Physiological Analogues on Rapeseed (Brassica napus) Cold Hardening, Seed Yield and Quality. J. Plant Res. 2013, 126, 283–292. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated Analysis of the Effects of Cold and Dehydration on Rice Metabolites, Phytohormones, and Gene Transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yu, M.; Xia, J.; Ren, Z.; Xing, J.; Li, C.; Xu, Q.; Cang, J.; Zhang, D. Cold Stress Triggers Freezing Tolerance in Wheat (Triticum Aestivum L.) via Hormone Regulation and Transcription of Related Genes. Plant Biol. 2023, 25, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin Response Factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Wang, S.; Bai, Y.; Shen, C.; Wu, Y.; Zhang, S.; Jiang, D.; Guilfoyle, T.J.; Chen, M.; Qi, Y. Auxin-Related Gene Families in Abiotic Stress Response in Sorghum bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) Are Potential Mediators of Auxin Action in Tomato Response to Biotic and Abiotic Stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Yarra, R.; Zhou, L.; Cao, H. The Auxin Response Factor (ARF) Gene Family in Oil Palm (Elaeis guineensis Jacq.): Genome-Wide Identification and Their Expression Profiling under Abiotic Stresses. Protoplasma 2022, 259, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmülling, T. Cytokinin Action in Plant Development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef]
- Mughal, N.; Shoaib, N.; Chen, J.; Li, Y.; He, Y.; Fu, M.; Li, X.; He, Y.; Guo, J.; Deng, J.; et al. Adaptive Roles of Cytokinins in Enhancing Plant Resilience and Yield against Environmental Stressors. Chemosphere 2024, 364, 143189. [Google Scholar] [CrossRef]
- Paul, S.; Wildhagen, H.; Janz, D.; Polle, A. Drought Effects on the Tissue- and Cell-Specific Cytokinin Activity in Poplar. AoB Plants 2018, 10, plx067. [Google Scholar] [CrossRef]
- Reguera, M.; Peleg, Z.; Abdel-Tawab, Y.M.; Tumimbang, E.B.; Delatorre, C.A.; Blumwald, E. Stress-Induced Cytokinin Synthesis Increases Drought Tolerance through the Coordinated Regulation of Carbon and Nitrogen Assimilation in Rice. Plant Physiol. 2013, 163, 1609–1622. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, N.Y.; Kim, S.; Kang, N.Y.; Novák, O.; Ku, S.J.; Cho, C.; Lee, D.J.; Lee, E.J.; Strnad, M.; et al. A Subset of Cytokinin Two-Component Signaling System Plays a Role in Cold Temperature Stress Response in Arabidopsis. J. Biol. Chem. 2010, 285, 23371–23386. [Google Scholar] [CrossRef]
- Karunadasa, S.S.; Kurepa, J.; Shull, T.E.; Smalle, J.A. Cytokinin-Induced Protein Synthesis Suppresses Growth and Osmotic Stress Tolerance. New Phytol. 2020, 227, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Liu, J.; Yuan, Y.; Chen, L.; Ma, J.; Li, X.; Li, J. The Mechanism of Abscisic Acid Regulation of Wild Fragaria Species in Response to Cold Stress. BMC Genom. 2022, 23, 670. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kidokoro, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulatory Networks in Plant Responses to Drought and Cold Stress. Plant Physiol. 2024, 195, 170–189. [Google Scholar] [CrossRef]
- Pál, M.; Janda, T.; Majláth, I.; Szalai, G. Involvement of Salicylic Acid and Other Phenolic Compounds in Light-Dependent Cold Acclimation in Maize. Int. J. Mol. Sci. 2020, 21, 1942. [Google Scholar] [CrossRef]
- Song, W.; Shao, H.; Zheng, A.; Zhao, L.; Xu, Y. Advances in Roles of Salicylic Acid in Plant Tolerance Responses to Biotic and Abiotic Stresses. Plants 2023, 12, 3475. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Charagh, S.; Najafi-Kakavand, S.; Abbas, S.; Shoaib, Y.; Anwar, S.; Sharifi, S.; Lu, G.; Siddique, K.H.M. Role of Phytohormones in Regulating Cold Stress Tolerance: Physiological and Molecular Approaches for Developing Cold-Smart Crop Plants. Plant Stress 2023, 8, 100152. [Google Scholar] [CrossRef]
- Gutierrez-Larruscain, D.; Krüger, M.; Abeyawardana, O.A.J.; Belz, C.; Dobrev, P.I.; Vaňková, R.; Eliášová, K.; Vondráková, Z.; Juříček, M.; Štorchová, H. The High Concentrations of Abscisic, Jasmonic, and Salicylic Acids Produced under Long Days Do Not Accelerate Flowering in Chenopodium ficifolium 459. Plant Sci. 2022, 320, 111279. [Google Scholar] [CrossRef]
- Arif, Y.; Sami, F.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salicylic Acid in Relation to Other Phytohormones in Plant: A Study towards Physiology and Signal Transduction under Challenging Environment. Environ. Exp. Bot. 2020, 175, 104040. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of Salicylic Acid Synthesis and Systemic Acquired Resistance by Two Members of a Plant-Specific Family of Transcription Factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse Roles of Jasmonates and Ethylene in Abiotic Stress Tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Sharma, M.; Laxmi, A. Jasmonates: Emerging Players in Controlling Temperature Stress Tolerance. Front. Plant Sci. 2016, 6, 1129. [Google Scholar] [CrossRef]
- Du, H.; Liu, H.; Xiong, L. Endogenous Auxin and Jasmonic Acid Levels Are Differentially Modulated by Abiotic Stresses in Rice. Front. Plant Sci. 2013, 4, 297. [Google Scholar] [CrossRef]
- Wang, F.; Guo, Z.; Li, H.; Wang, M.; Onac, E.; Zhou, J.; Xia, X.; Shi, K.; Yu, J.; Zhou, Y. Phytochrome a and b Function Antagonistically to Regulate Cold Tolerance via Abscisic Acid-Dependent Jasmonate Signaling. Plant Physiol. 2016, 170, 459–471. [Google Scholar] [CrossRef]
- Smoleńska-Sym, G.; Gawrońska, H.; Kacperska, A. Modifications of Abscisic Acid Level in Winter Oilseed Rape Leaves during Acclimation of Plants to Freezing Temperatures. Plant Growth Regul. 1995, 17, 61–65. [Google Scholar] [CrossRef]
- Lalk, I.; Dörffling, K. Hardening, Abscisic Acid, Proline and Freezing Resistance in Two Winter Wheat Varieties. Physiol. Plant 1985, 63, 287–292. [Google Scholar] [CrossRef]
- Tuteja, N. Abscisic Acid and Abiotic Stress Signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef]
- Qian, Z.; He, L.; Li, F. Understanding Cold Stress Response Mechanisms in Plants: An Overview. Front. Plant Sci. 2024, 15, 1443317. [Google Scholar] [CrossRef]
- Kalberer, S.R.; Wisniewski, M.; Arora, R. Deacclimation and Reacclimation of Cold-Hardy Plants: Current Understanding and Emerging Concepts. Plant Sci. 2006, 171, 3–16. [Google Scholar] [CrossRef]
- Sauter, A.; Davies, W.J.; Hartung, W. The Long-Distance Abscisic Acid Signal in the Droughted Plant: The Fate of the Hormone on Its Way from Root to Shoot. J. Exp. Bot. 2001, 52, 1991–1997. [Google Scholar] [CrossRef]
- Knight, H.; Zarka, D.G.; Okamoto, H.; Thomashow, M.F.; Knight, M.R. Abscisic Acid Induces CBF Gene Transcription and Subsequent Induction of Cold-Regulated Genes via the CRT Promoter Element. Plant Physiol. 2004, 135, 1710–1717. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Lastdrager, J.; Hanson, J.; Smeekens, S. Sugar Signals and the Control of Plant Growth and Development. J. Exp. Bot. 2014, 65, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate Regulates the Inducer of Cbf Expression-C-Repeat Binding Factor/Dre Binding Factor1 Cascade and Freezing Tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef]
- Ding, F.; Wang, X.; Li, Z.; Wang, M. Jasmonate Positively Regulates Cold Tolerance by Promoting ABA Biosynthesis in Tomato. Plants 2023, 12, 60. [Google Scholar] [CrossRef]
- Yan, Y.; Stolz, S.; Chételat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A Downstream Mediator in the Growth Repression Limb of the Jasmonate Pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef]
- Liu, X.; Ma, D.; Zhang, Z.; Wang, S.; Du, S.; Deng, X.; Yin, L. Plant Lipid Remodeling in Response to Abiotic Stresses. Environ. Exp. Bot. 2019, 165, 174–184. [Google Scholar] [CrossRef]
- Wang, X.; Miao, J.; Kang, W.; Shi, S. Exogenous Application of Salicylic Acid Improves Freezing Stress Tolerance in Alfalfa. Front. Plant Sci. 2023, 14, 1091077. [Google Scholar] [CrossRef] [PubMed]
- Janda, T.; Gondor, O.K.; Yordanova, R.; Szalai, G.; Pál, M. Salicylic Acid and Photosynthesis: Signalling and Effects. Acta Physiol. Plant 2014, 36, 2537–2546. [Google Scholar] [CrossRef]
- Miura, K.; Tada, Y. Regulation of Water, Salinity, and Cold Stress Responses by Salicylic Acid. Front. Plant Sci. 2014, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Eremina, M.; Rozhon, W.; Poppenberger, B. Hormonal Control of Cold Stress Responses in Plants. Cell. Mol. Life Sci. 2016, 73, 797–810. [Google Scholar] [CrossRef]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschika, P. The Cold-Inducible CBF1 Factor-Dependent Signaling Pathway Modulates the Accumulation of the Growth-Repressing DELLA Proteins via Its Effect on Gibberellin Metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef]
- Sun, T.-P. Gibberellin-GID1-DELLA: A Pivotal Regulatory Module for Plant Growth and Development. Plant Physiol. 2010, 154, 567–570. [Google Scholar] [CrossRef]
- Rahman, A. Auxin: A Regulator of Cold Stress Response. Physiol. Plant 2013, 147, 28–35. [Google Scholar] [CrossRef]
- Shibasaki, K.; Uemura, M.; Tsurumi, S.; Rahman, A. Auxin Response in Arabidopsis under Cold Stress: Underlying Molecular Mechanisms. Plant Cell 2009, 21, 3823–3838. [Google Scholar] [CrossRef]
- Yu, M.; Luobu, Z.; Zhuoga, D.; Wei, X.; Tang, Y. Advances in Plant Response to Low-Temperature Stress. Plant Growth Regul. 2024, 105, 167–185. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, J.; Ha, X.; Ma, H. Genome-Wide Identification and Expression Analysis of the Auxin-Response Factor (ARF) Gene Family in Medicago Sativa under Abiotic Stress. BMC Genom. 2023, 24, 498. [Google Scholar] [CrossRef]
- Zwack, P.J.; Rashotte, A.M. Interactions between Cytokinin Signalling and Abiotic Stress Responses. J. Exp. Bot. 2015, 66, 4863–4871. [Google Scholar] [CrossRef]
- EL Sabagh, A.; Islam, M.S.; Hossain, A.; Iqbal, M.A.; Mubeen, M.; Waleed, M.; Reginato, M.; Battaglia, M.; Ahmed, S.; Rehman, A.; et al. Phytohormones as Growth Regulators During Abiotic Stress Tolerance in Plants. Front. Agron. 2022, 4, 765068. [Google Scholar] [CrossRef]
- Prerostova, S.; Černý, M.; Dobrev, P.I.; Motyka, V.; Hluskova, L.; Zupkova, B.; Gaudinova, A.; Knirsch, V.; Janda, T.; Brzobohatý, B.; et al. Light Regulates the Cytokinin-Dependent Cold Stress Responses in Arabidopsis. Front. Plant Sci. 2021, 11, 608711. [Google Scholar] [CrossRef] [PubMed]
- Dziurka, M.; Janeczko, A.; Juhász, C.; Gullner, G.; Oklestková, J.; Novák, O.; Saja, D.; Skoczowski, A.; Tóbiás, I.; Barna, B. Local and Systemic Hormonal Responses in Pepper Leaves during Compatible and Incompatible Pepper-Tobamovirus Interactions. Plant Physiol. Biochem. 2016, 109, 355–364. [Google Scholar] [CrossRef]
- Ivanov Dobrev, P.; Kamínek, M. Fast and Efficient Separation of Cytokinins from Auxin and Abscisic Acid and Their Purification Using Mixed-Mode Solid-Phase Extraction. J. Chromatogr. A 2002, 950, 21–29. [Google Scholar] [CrossRef]
- Żur, I.; Dubas, E.; Krzewska, M.; Waligórski, P.; Dziurka, M.; Janowiak, F. Hormonal Requirements for Effective Induction of Microspore Embryogenesis in Triticale (×Triticosecale wittm.) Anther Cultures. Plant Cell Rep. 2015, 34, 47–62. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, J.; Zhao, Y.; Li, T.; Wang, M. Molecular Cloning and Expression Analysis of a RGA-like Gene Responsive to Plant Hormones in Brassica napus. Mol. Biol. Rep. 2012, 39, 1957–1962. [Google Scholar] [CrossRef]
- Jiang, J.J.; Li, N.; Chen, W.J.; Wang, Y.; Rong, H.; Xie, T.; Wang, Y.P. Genome-Wide Analysis of the Type-B Authentic Response Regulator Gene Family in Brassica Napus. Genes 2022, 13, 1449. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-H.; Lee, B.-R.; La, V.H.; Mamun, M.A.; Bae, D.-W.; Kim, T.-H. Drought Intensity-Responsive Salicylic Acid and Abscisic Acid Crosstalk with the Sugar Signaling and Metabolic Pathway in Brassica napus. Plants 2021, 10, 610. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Guo, P.; Ke, Y.; Liu, M.; Li, P.; Wu, Y.; Ran, F.; Wang, M.; Li, J.; Du, H. The Auxin Response Factor Gene Family in Allopolyploid Brassica Napus. PLoS ONE 2019, 14, e0214885. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591. [Google Scholar] [CrossRef]
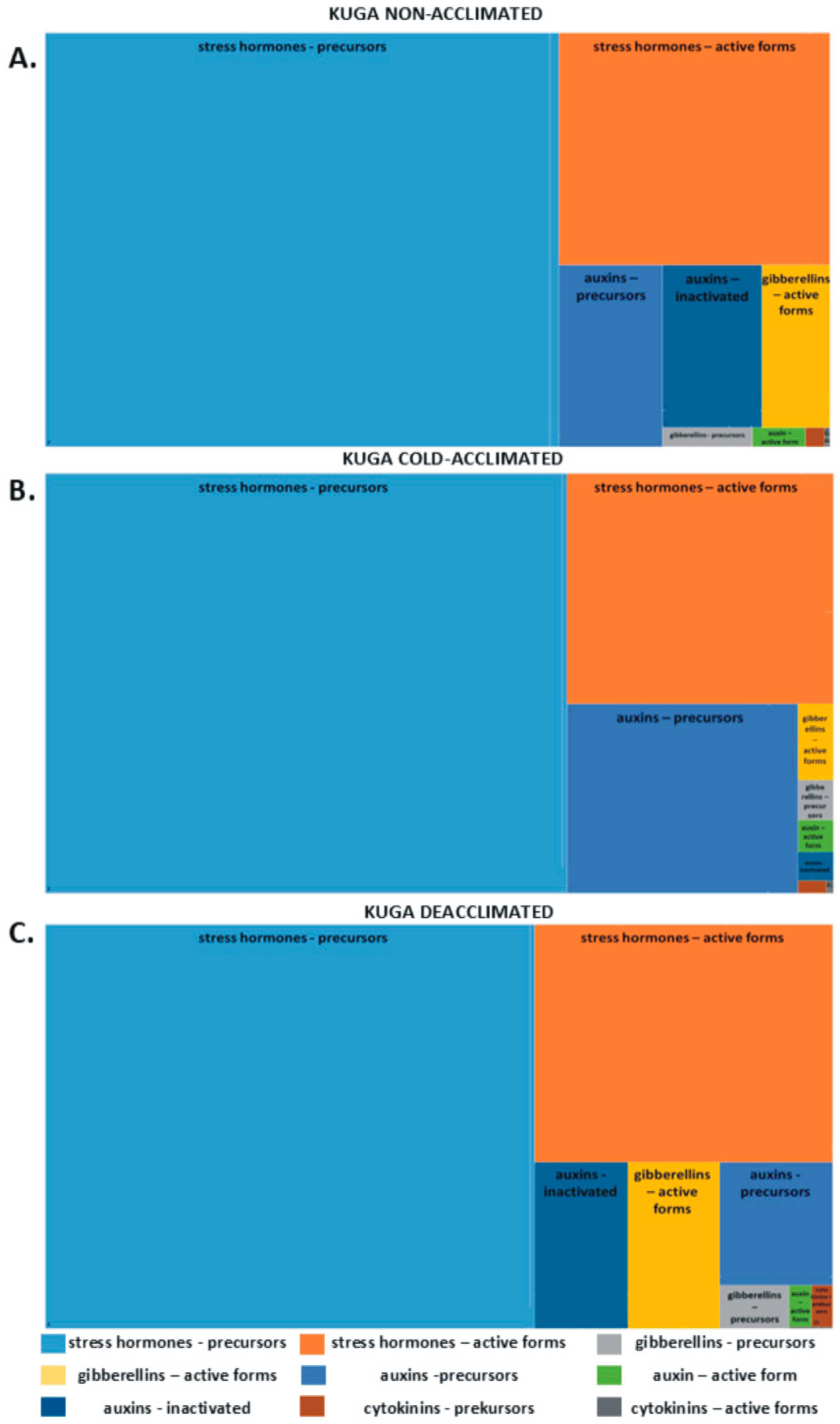
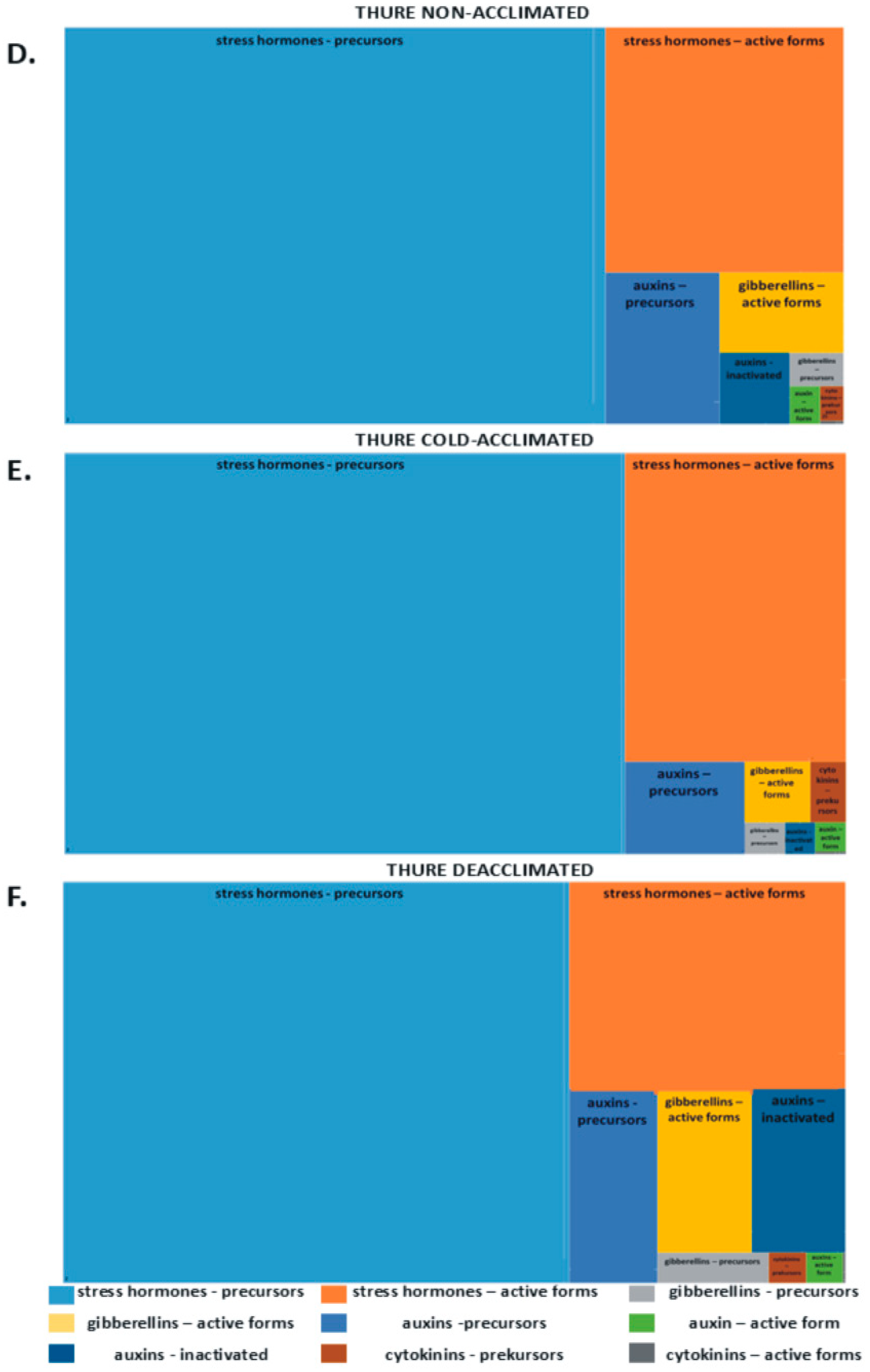


| Kuga | Thure | Cultivar Mean | Treatment Mean | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NA | CA | DA | NA | CA | DA | LSD0.05 | F | Kuga | Thure | LSD0.05 | F | NA | CA | DA | LSD0.05 | F | |||
| stress hormones—precursors | 12-oxo-PDA | Mean | 721 | 306 | 336 | 820 | 210 | 409 | nsd | 0.12 | 454 | 480 | nsd | 0.54 | 770X | 258Z | 372Y | 101 | <0.01 |
| s.d. | 55 | 57 | 73 | 115 | 50 | 211 | 204 | 294 | 100 | 72 | 153 | ||||||||
| BeA | Mean | 17040 | 14571 | 15684 | 15407 | 15276 | 16390 | nsd | 0.09 | 15765 | 15691 | nsd | 0.88 | 16223 | 14923 | 16037 | nsd | 0.08 | |
| s.d. | 1060 | 1113 | 1532 | 1185 | 397 | 1997 | 1561 | 1360 | 1365 | 871 | 1718 | ||||||||
| sum | Mean | 17761 | 14878 | 16019 | 16227 | 15486 | 16798 | nsd | 0.12 | 16219 | 16170 | nsd | 0.92 | 16994X | 15182Y | 16409XY | 1233 | 0.02 | |
| s.d. | 1104 | 1087 | 1505 | 1197 | 426 | 2103 | 1685 | 1426 | 1354 | 842 | 1772 | ||||||||
| stress hormones—active forms | ABA | Mean | 183 | 432 | 294 | 191 | 446 | 362 | nsd | 0.73 | 303 | 333 | nsd | 0.38 | 187Z | 439X | 328Y | 85 | <0.01 |
| s.d. | 24 | 54 | 129 | 38 | 172 | 2 | 130 | 145 | 31 | 120 | 93 | ||||||||
| JA | Mean | 507c | 519c | 3259a | 685c | 1763b | 1421b | 490 | <0.01 | 1428 | 1290 | nsd | 0.32 | 596Z | 1141Y | 2340X | 347 | <0.01 | |
| s.d. | 10 | 47 | 772 | 102 | 447 | 195 | 1402 | 537 | 116 | 721 | 1105 | ||||||||
| SA | Mean | 5365a | 4090b | 4110b | 4299b | 3889b | 4130b | 483 | 0.01 | 4522A | 4106B | 279 | 0.01 | 4832X | 3990Y | 4120Y | 342 | <0.01 | |
| s.d. | 446 | 344 | 261 | 550 | 252 | 268 | 701 | 394 | 734 | 303 | 250 | ||||||||
| sum | Mean | 6056b | 5041c | 7663a | 5175c | 6098b | 5914b | 679 | <0.01 | 6253A | 5729B | 392 | 0.01 | 5615Y | 5570Y | 6788X | 480 | <0.01 | |
| s.d. | 466 | 339 | 910 | 493 | 341 | 322 | 1257 | 550 | 648 | 643 | 1124 | ||||||||
| gibberelins—precursors | GA15 | Mean | 5.14 | 4.46 | 3.92 | 8.30 | 3.86 | 4.70 | nsd | 0.09 | 4.51 | 5.62 | nsd | 0.11 | 6.72X | 4.16Y | 4.31Y | 1.71 | 0.01 |
| s.d. | 2.32 | 1.29 | 0.34 | 2.74 | 0.98 | 2.21 | 1.52 | 2.79 | 2.92 | 1.12 | 1.54 | ||||||||
| GA9 | Mean | 87a | 60d | 72b | 75c | 60bc | 79d | 6.14 | <0.01 | 73 | 72 | nsd | 0.35 | 81X | 60Z | 76Y | 4.34 | <0.01 | |
| s.d. | 5.59 | 4.83 | 4.62 | 2.52 | 5.61 | 4.36 | 12 | 9.37 | 7.54 | 4.94 | 5.73 | ||||||||
| GA53 | Mean | 111a | 42b | 27c | 21cd | 16d | 20cd | 8.34 | <0.01 | 60A | 19B | 4.81 | <0.01 | 66X | 29Y | 23Y | 5.90 | <0.01 | |
| s.d. | 6.59 | 8.31 | 8.67 | 2.29 | 6.36 | 3.38 | 39 | 4.66 | 47 | 15 | 7.06 | ||||||||
| GA44 | Mean | 62 | 58 | 63 | 62 | 58 | 62 | nsd | 0.99 | 61 | 60 | nsd | 0.75 | 62X | 58Y | 62X | 3.43 | 0.04 | |
| s.d. | 3.19 | 3.86 | 2.20 | 5.31 | 4.64 | 1.75 | 3.54 | 4.32 | 4.13 | 4.03 | 1.92 | ||||||||
| GA19 | Mean | 86c | 73c | 504a | 155b | 71c | 178b | 36.87 | <0.01 | 221A | 135B | 21.29 | <0.01 | 121Y | 72Z | 341X | 26.07 | <0.01 | |
| s.d. | 14 | 11 | 36 | 29 | 11 | 47 | 208 | 56 | 42 | 10 | 176 | ||||||||
| GA20 | Mean | 39b | 30b | 30b | 50b | 33b | 415a | 27.35 | <0.01 | 33B | 166A | 15.79 | <0.01 | 44Y | 31Y | 222X | 19.34 | <0.01 | |
| s.d. | 5.59 | 0.29 | 5.57 | 8 | 6 | 50 | 6 | 184 | 8 | 4 | 206 | ||||||||
| sum | Mean | 390 | 268 | 699 | 371 | 242 | 759 | nsd | 0.06 | 452 | 457 | nsd | 0.74 | 380Y | 255Y | 729X | 38 | <0.01 | |
| s.d. | 19 | 13 | 30 | 24 | 25 | 89 | 189 | 233 | 23 | 23 | 70 | ||||||||
| gibberelins—active forms | GA4 | Mean | 74 | 66 | 78 | 67 | 48 | 66 | nsd | 0.54 | 73A | 60B | 8.08 | 0.004 | 71X | 57Y | 72X | 10 | 0.01 |
| s.d. | 6.36 | 5.97 | 12 | 12 | 15 | 9 | 9 | 15 | 10 | 14 | 12 | ||||||||
| GA7 | Mean | 114a | 36 d | 129a | 65b | 41cd | 59.07bc | 18.27 | <0.01 | 93A | 55B | 10.55 | <0.01 | 90X | 39Y | 94X | 13 | <0.01 | |
| s.d. | 13 | 4.69 | 11 | 20 | 14 | 16 | 43 | 19 | 31 | 10 | 39 | ||||||||
| GA3 | Mean | 2118 | 274 | 3086 | 1727 | 546 | 3098 | nsd | 0.10 | 1826 | 1790 | nsd | 0.77 | 1923Y | 410Z | 3092X | 305 | <0.01 | |
| s.d. | 223 | 47 | 542 | 193 | 80 | 516 | 1248 | 1120 | 284 | 156 | 499 | ||||||||
| GA6 | Mean | 152 | 56 | 69 | 140 | 51 | 98 | nsd | 0.07 | 92 | 96 | nsd | 0.58 | 146X | 53Z | 83Y | 18 | <0.01 | |
| s.d. | 23 | 9 | 10 | 37 | 13 | 10 | 46 | 43 | 30 | 11 | 18 | ||||||||
| GA1 | Mean | 41 | 36 | 41 | 40 | 37 | 38 | nsd | 0.16 | 40 | 38 | nsd | 0.14 | 41X | 37Y | 40X | 2 | 0.001 | |
| s.d. | 2.86 | 1.76 | 1.98 | 3.17 | 2.05 | 0.38 | 3.20 | 2.39 | 2.91 | 1.83 | 2 | ||||||||
| GA5 | Mean | 38bc | 36bc | 43b | 26c | 36bc | 168 a | 16.27 | <0.01 | 39B | 77A | 9.4 | <0.01 | 32Y | 36Y | 106X | 12 | <0.01 | |
| s.d. | 13 | 7 | 8 | 2.63 | 11 | 23 | 9 | 69 | 11 | 8 | 68 | ||||||||
| sum | Mean | 2538 | 504 | 3446 | 2065 | 759 | 3527 | nsd | 0.05 | 2163 | 2117 | nsd | 0.71 | 2302Y | 632Z | 3487X | 306 | <0.01 | |
| s.d. | 244 | 45 | 531 | 212 | 100 | 510 | 1311 | 1208 | 330 | 153 | 493 | ||||||||
| auxins—precursors | IAM | Mean | 142c | 540a | 115c | 263b | 81c | 139c | 94 | <0.01 | 266A | 161B | 55 | <0.01 | 202.3Y | 310.2X | 127.2Z | 66.8 | <0.01 |
| s.d. | 43 | 144 | 29 | 66 | 37 | 49 | 217 | 92 | 82.9 | 261.5 | 40 | ||||||||
| IAN | Mean | 1838b | 3268a | 1323cd | 1390bc | 894d | 1649bc | 451 | <0.01 | 2143A | 1311B | 261 | <0.01 | 1614Y | 2081X | 1486Y | 319.06 | 0.002 | |
| s.d. | 134 | 744 | 223 | 59 | 157 | 261 | 950 | 364 | 256 | 1350 | 286 | ||||||||
| sum | Mean | 1980b | 3808a | 1439cd | 1653bc | 974d | 1788bc | 511 | <0.01 | 2409A | 1472B | 295 | <0.01 | 1816Y | 2391X | 1613Y | 361.54 | <0.01 | |
| s.d. | 150 | 865 | 216 | 124 | 144 | 259 | 1155 | 407 | 215.7 | 1603.9 | 290.5 | ||||||||
| auxin—active form | IAA | Mean | 119ab | 109bc | 108bc | 121ab | 89c | 130a | 20.91 | 0.02 | 112 | 113 | nsd | 0.78 | 120X | 99Y | 119X | 15 | 0.01 |
| s.d. | 9 | 22 | 9 | 17 | 3.32 | 24 | 15 | 24 | 13 | 18 | 21 | ||||||||
| auxins—inactivated | IAAsp | Mean | 225 | 63 | 262 | 213 | 61 | 207 | nsd | 0.44 | 183 | 160 | nsd | 0.20 | 219X | 62Y | 234X | 44 | <0.01 |
| s.d. | 66 | 15 | 53 | 42 | 17 | 63 | 101 | 84 | 52 | 15 | 62 | ||||||||
| OxIAA | Mean | 23b | 24b | 25b | 32a | 22b | 22b | 5.39 | 0.01 | 24 | 25 | nsd | 0.50 | 27X | 23Y | 24XY | 3.81 | 0.04 | |
| s.d. | 5.05 | 2.41 | 4.60 | 2.91 | 2.59 | 5.87 | 3.97 | 6.01 | 5.82 | 2.54 | 5.28 | ||||||||
| IAA-Glu | Mean | 78c | 67c | 321a | 65c | 64c | 152b | 37.08 | <0.01 | 155A | 94B | 21.41 | <0.01 | 72Y | 65Y | 237X | 26 | <0.01 | |
| s.d. | 22 | 10 | 60 | 5.14 | 8.88 | 23 | 126 | 45 | 17 | 9.09 | 99 | ||||||||
| I3CA | Mean | 4347 | 0 | 1307 | 304 | 0 | 1428 | nsd | 0.14 | 1884 | 577 | nsd | 0.17 | 2325 | 0 | 1367 | nsd | 0.15 | |
| s.d. | 6253 | 0 | 189 | 95 | 0 | 170 | 3839 | 644 | 4682 | 0 | 181 | ||||||||
| sum | Mean | 4674 | 153 | 1914 | 613 | 146 | 1810 | nsd | 0.15 | 2247 | 856 | nsd | 0.15 | 2643 | 149 | 1862 | nsd | 0.10 | |
| s.d. | 6214 | 12 | 149 | 88 | 15 | 133 | 3840 | 730 | 4663 | 14 | 144 | ||||||||
| cytokinins—precursor | IPA | Mean | 83d | 67d | 192c | 165c | 407a | 252b | 41.73 | <0.01 | 114B | 275A | 24.09 | <0.01 | 124Y | 237X | 222X | 30 | <0.01 |
| s.d. | 15 | 4.75 | 30 | 25 | 20 | 63 | 60 | 110 | 47 | 180 | 56 | ||||||||
| cytokinins—active forms | t-ZEA | Mean | 4.73a | 0.94b | 0.32b | 1.18b | 0.96b | 0b | 1.35 | 0.001 | 2A | 0.71B | 0.78 | 0.002 | 2.96X | 0.95Y | 0.16Y | 0.96 | <0.01 |
| s.d. | 0.79 | 1.32 | 0.50 | 1.32 | 1.45 | 0 | 2.20 | 1.18 | 2.13 | 1.31 | 0.38 | ||||||||
| c-ZEA | Mean | 1.11 | 1.41 | 2.16 | 1.04 | 1.24 | 2.88 | nsd | 0.38 | 1.56 | 1.72 | nsd | 0.57 | 1.07Y | 1.32Y | 2.52X | 0.71 | <0.01 | |
| s.d. | 0.24 | 0.59 | 0.49 | 1.04 | 1.10 | 0.77 | 0.63 | 1.25 | 0.71 | 0.83 | 0.72 | ||||||||
| t-ZEA-R | Mean | 1.67 | 2.35 | 2.02 | 1.64 | 1.90 | 1.71 | nsd | 0.50 | 2.01 | 1.75 | nsd | 0.09 | 1.66 | 2.12 | 1.86 | nsd | 0.05 | |
| s.d. | 0.28 | 0.74 | 0.40 | 0.18 | 0.34 | 0.23 | 0.56 | 0.26 | 0.22 | 0.59 | 0.35 | ||||||||
| c-ZEA-R | Mean | 11 | 11 | 12 | 11 | 11 | 11 | nsd | 0.99 | 11 | 11 | nsd | 0.07 | 11 | 11 | 11 | nsd | 0.11 | |
| s.d. | 0.72 | 0.62 | 0.42 | 0.62 | 0.81 | 0.24 | 0.61 | 0.61 | 0.67 | 0.71 | 0.40 | ||||||||
| sum | Mean | 19 | 16 | 16 | 15 | 15 | 16 | nsd | 0.04 | 17A | 15B | 1.22 | 0.01 | 17 | 15 | 16 | nsd | 0.09 | |
| s.d. | 1.24 | 1.85 | 1 | 2.12 | 2.23 | 0.55 | 1.97 | 1.74 | 2.7 | 2 | 0.79 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rys, M.; Bocianowski, J.; Dziurka, M.; Jurczyk, B.; Stachurska, J.; Waligórski, P.; Janeczko, A. Hormonal Balance in Relation to Expression of Selected Genes Connected with Hormone Biosynthesis and Signalling—The Effect of Deacclimation Process in Oilseed Rape. Int. J. Mol. Sci. 2025, 26, 7408. https://doi.org/10.3390/ijms26157408
Rys M, Bocianowski J, Dziurka M, Jurczyk B, Stachurska J, Waligórski P, Janeczko A. Hormonal Balance in Relation to Expression of Selected Genes Connected with Hormone Biosynthesis and Signalling—The Effect of Deacclimation Process in Oilseed Rape. International Journal of Molecular Sciences. 2025; 26(15):7408. https://doi.org/10.3390/ijms26157408
Chicago/Turabian StyleRys, Magdalena, Jan Bocianowski, Michał Dziurka, Barbara Jurczyk, Julia Stachurska, Piotr Waligórski, and Anna Janeczko. 2025. "Hormonal Balance in Relation to Expression of Selected Genes Connected with Hormone Biosynthesis and Signalling—The Effect of Deacclimation Process in Oilseed Rape" International Journal of Molecular Sciences 26, no. 15: 7408. https://doi.org/10.3390/ijms26157408
APA StyleRys, M., Bocianowski, J., Dziurka, M., Jurczyk, B., Stachurska, J., Waligórski, P., & Janeczko, A. (2025). Hormonal Balance in Relation to Expression of Selected Genes Connected with Hormone Biosynthesis and Signalling—The Effect of Deacclimation Process in Oilseed Rape. International Journal of Molecular Sciences, 26(15), 7408. https://doi.org/10.3390/ijms26157408









