Investigating the Cellular Responses to Combined Nisin and Urolithin B Treatment (7:3) in HKB-11 Lymphoma Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. The Antiproliferative Activity of the Postbiotics (N and UB), and Their Synergistic Combination (7:3) Against the HKB-11 (BL) Human Cell Line
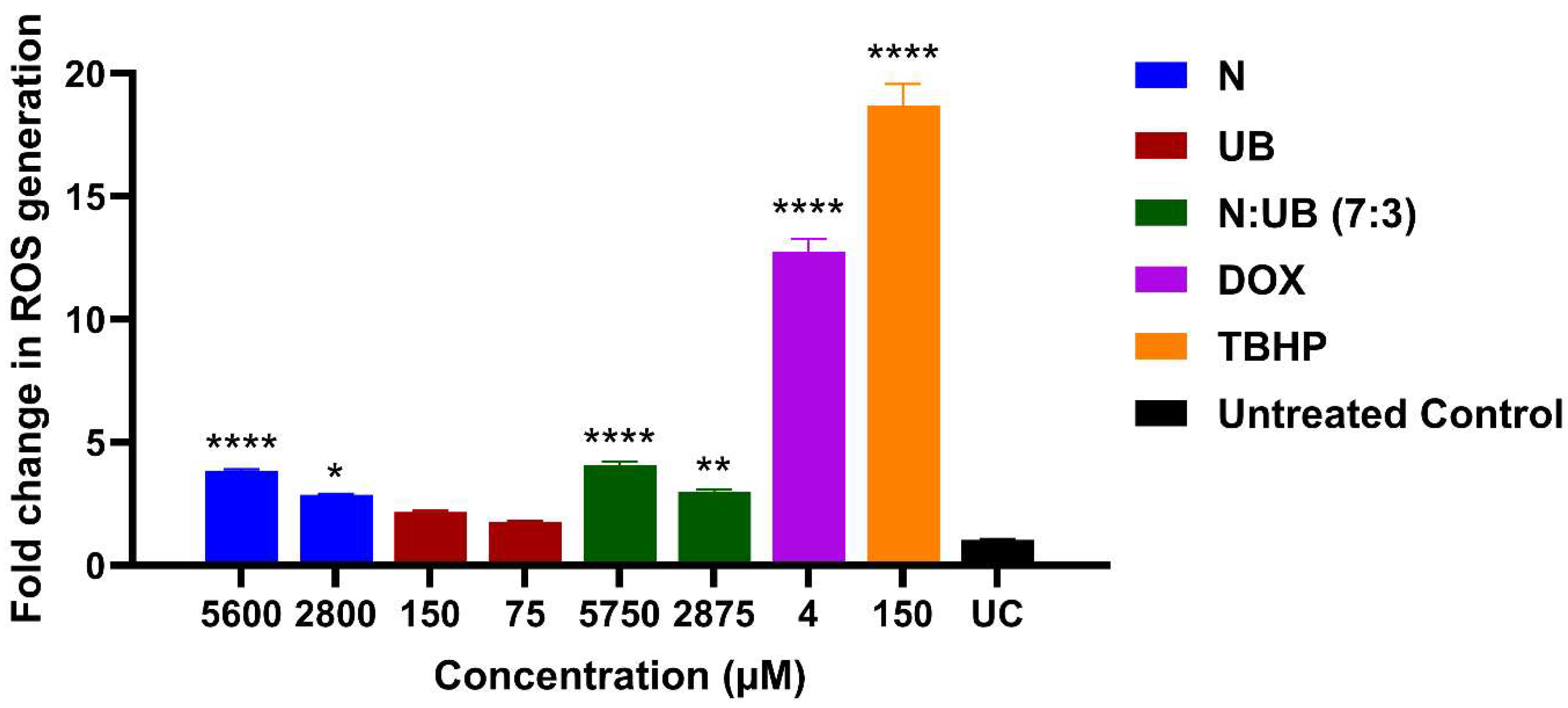
2.2. ROS Production in the HKB-11 Lymphoma Cells After Treatment with Different Concentrations of N, UB, and N: UB (7:3)
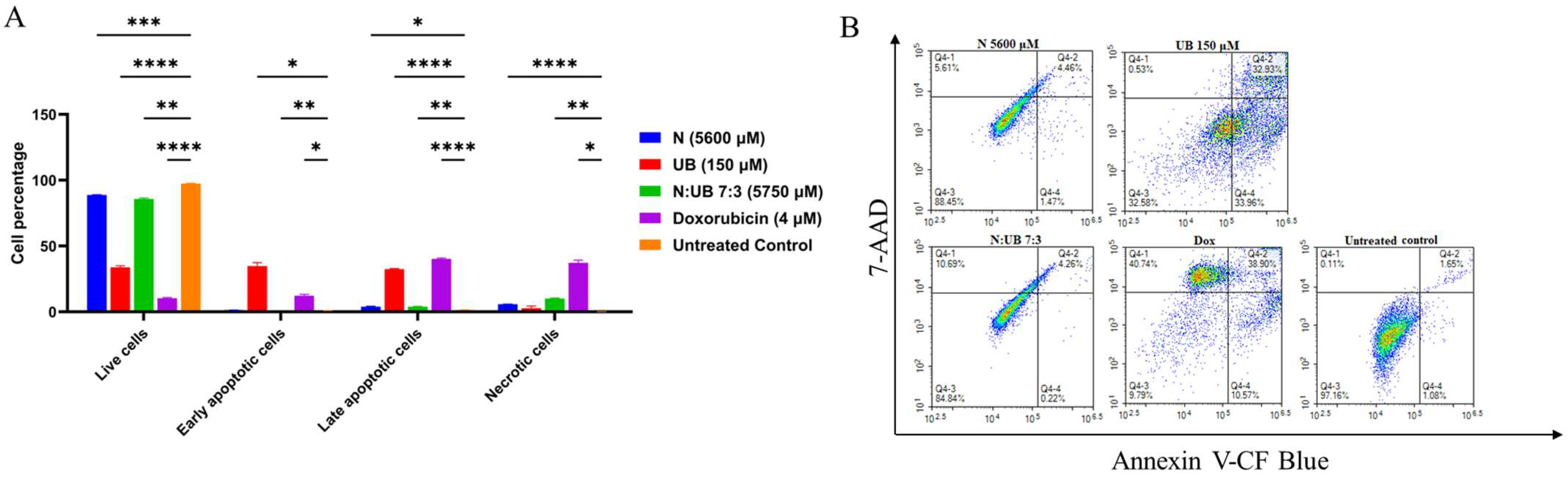
2.3. Flow Cytometric Analyses of Apoptotic Profiles of Mono and Combination Therapies
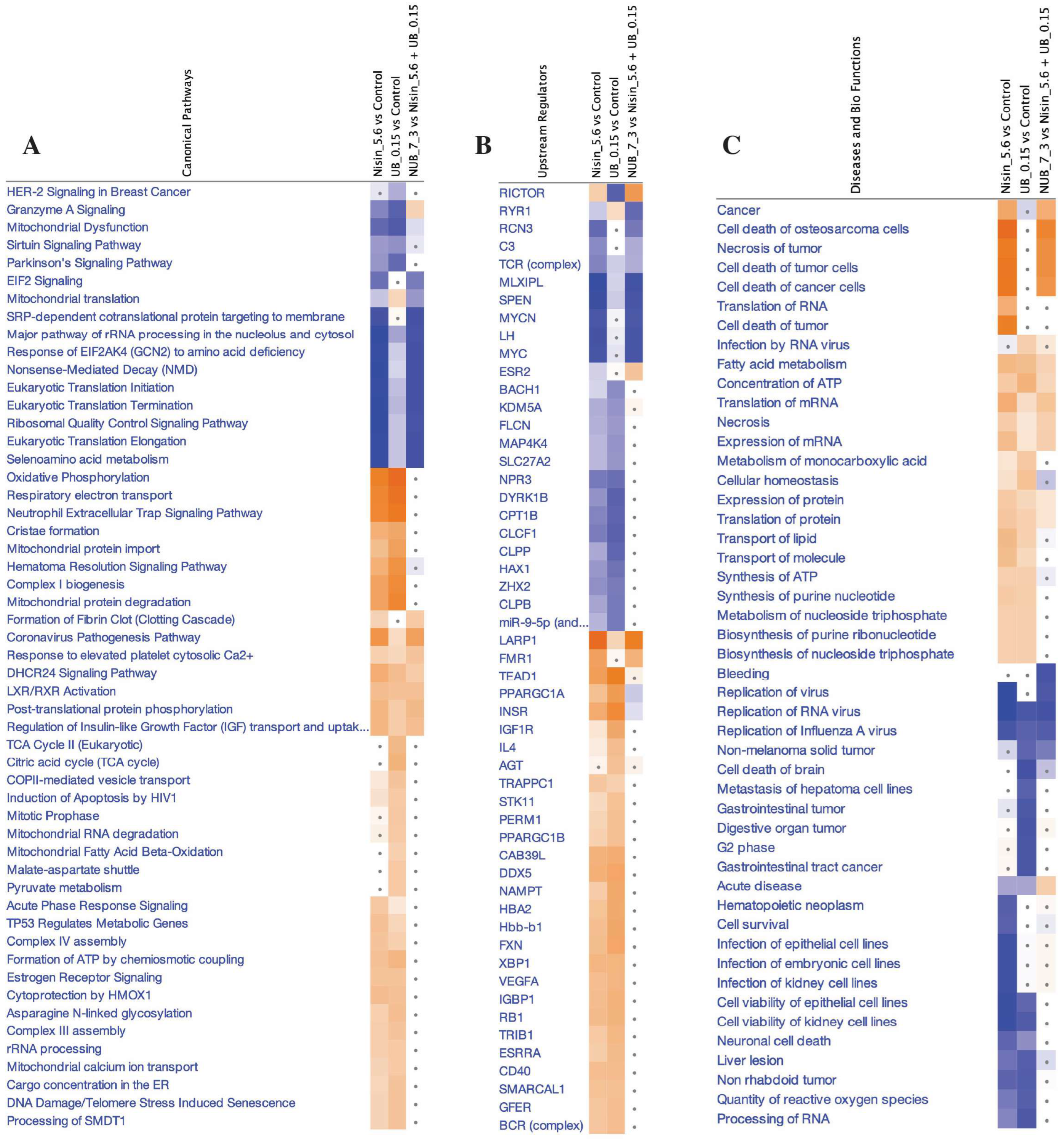
2.4. Proteomics Study of the HKB-11 Lymphoma Cells Treated with the Synergistic Combination vs. Mono Treatments
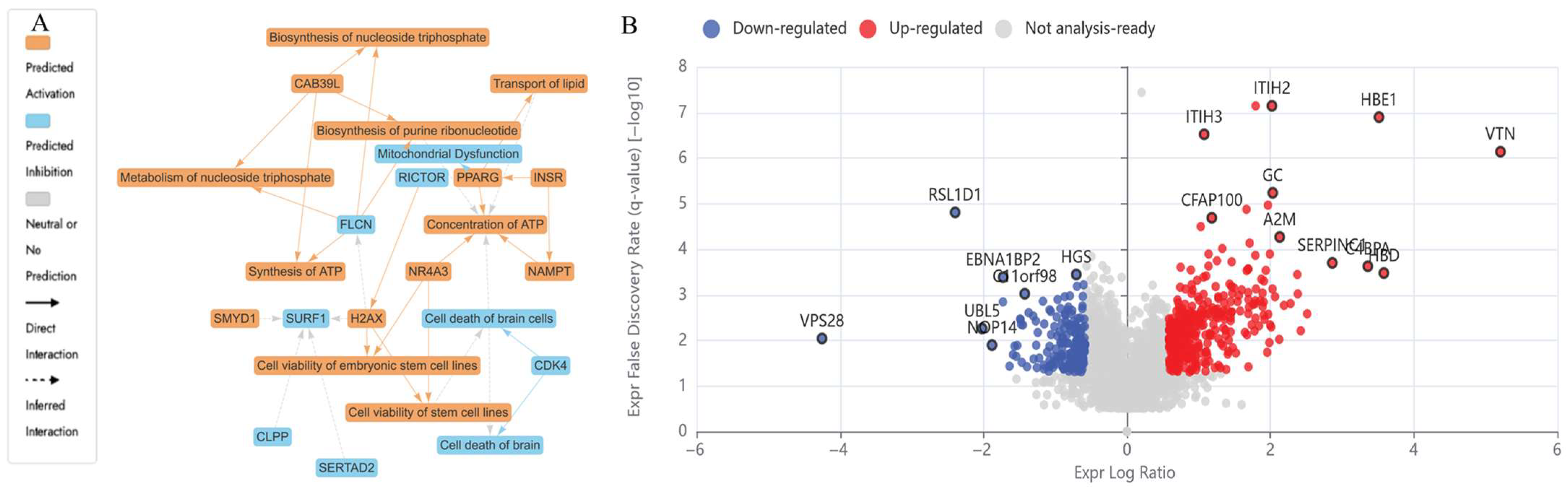
2.4.1. Differentially Expressed Proteins (DEPs) in N (5600 µM) Treated HKB-11 Lymphoma Cells Compared to Untreated Control (abs log2FC ≥ 0.58 and Q ≤ 0.05)
| Treatment | Log2FC | Gene ID | Protein Descriptions | Molecular Pathway | Mechanism of Action | Reference |
|---|---|---|---|---|---|---|
| N 5600 μM | 8.62 | HBA1 | Hemoglobin subunit alpha | Oxidative stress response, heme metabolism, hypoxia-inducible factor-1 (HIF-1) signalling | Upregulation is associated with reduced tumour progression and enhanced oxidative stress, which promotes apoptosis in tumour cells. Upregulation reduces cancer growth via redox disruption and ferroptosis. | [36,44] |
| 1.98 | FN1 | Fibronectin | Extracellular Matrix (ECM) and integrin signalling | Enhances cell adhesion, migration, epithelial–mesenchymal transition (EMT), and invasion. | [45] | |
| 0.66 | NRAS | GTPase NRas | RAS/MAPK and PI3K-AKT signalling | NRAS encodes a small GTPase regulating growth signals via MAPK and AKT cascades, inducing apoptosis at high expression. | [46] | |
| 0.70 | CYC1 | Cytochrome c1, heme protein, mitochondrial | Mitochondrial Electron Transport Chain (Complex III) | Involved in oxidative phosphorylation and ROS generation, intrinsic apoptosis. | [47] | |
| −1.66 | DDX21 | Nucleolar RNA helicase 2 | RNA processing | Involved in ribosomal RNA regulation; downregulation supports the inhibition of tumour survival. | [48] | |
| −1.91 | GNL3 | Guanine nucleotide-binding protein-like 3 | Nucleolar stress | Promotes proliferation; downregulation linked to low-grade lymphoma. | [49] | |
| −1.04 | PDCD4 | Programmed cell death protein 4 | PI3K pathway inhibition | Tumour suppressor downregulation facilitates proliferation and chemoresistance. | [50] | |
| −2.18 | SERBP1 | SERPINE1 mRNA-binding protein 1 | mRNA stability | Suppresses tumour suppressor mRNA degradation; downregulation is tumour-permissive. | [51] | |
| −1.35 | RPSA | Small ribosomal subunit protein uS2 | Cell adhesion | Loss reduces laminin receptor activity and decreases metastatic potential. | [56] | |
| −0.79 | CDK1 | Cyclin-dependent kinase 1 | Cell cycle | Catalyses mitotic onset via phosphorylation of downstream mitotic regulators | [52] | |
| −1.15 | CKS1B | Cyclin-dependent kinases regulatory subunit 1 | Cell cycle regulation | Binds CDK1/CDK2; regulates p27 degradation | [53] | |
| −0.80 | FBXL5 | F-box/LRR-repeat protein 5 | Iron homeostasis and DNA repair | Degrades iron regulatory proteins; supports replication. | [53] | |
| −1.06 | SKP1 | S-phase kinase-associated protein 1 | Ubiquitin-proteasome pathway | Core component of SCF E3 ligase for cell cycle proteins. | [55] | |
| −0.60 | TOP2A | DNA topoisomerase 2-alpha | DNA replication and mitosis | Resolves DNA supercoiling during replication. | [57] | |
| −1.02 | UBE2C | Ubiquitin-conjugating enzyme E2 C | Ubiquitination/mitosis exit | Catalyses the degradation of mitotic cyclins. | [54] | |
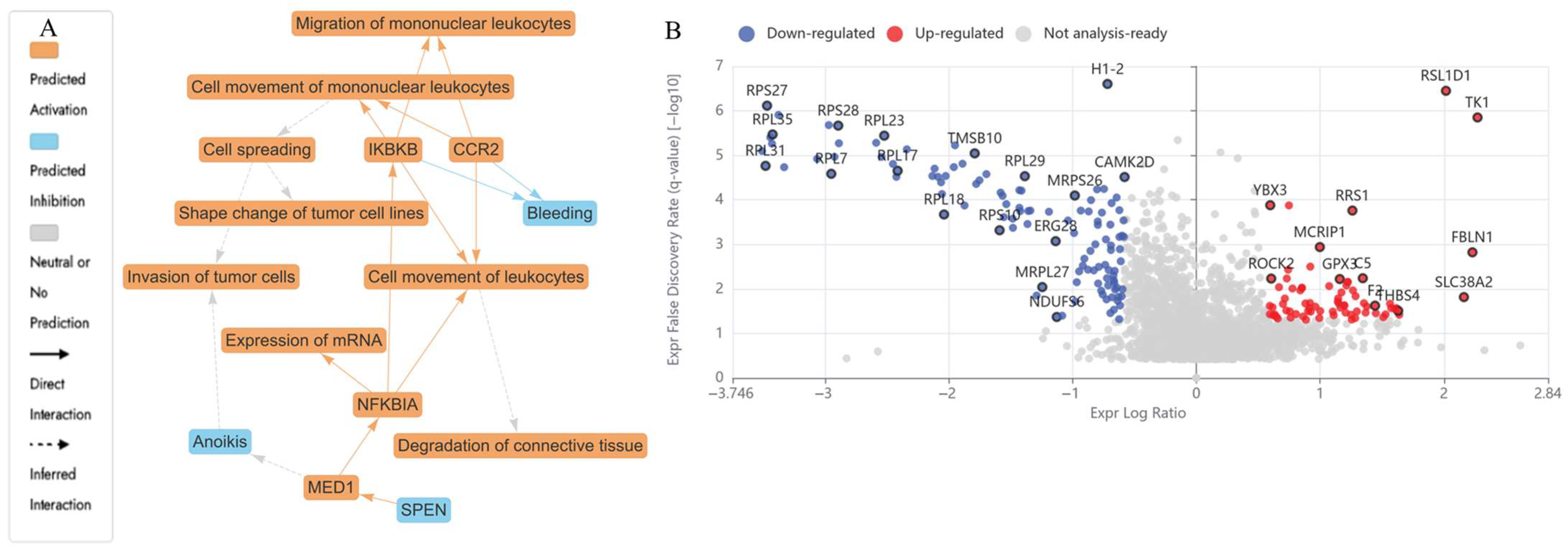
| UB 150 μM | 2.28 | MT-ND2 | NADH-ubiquinone oxidoreductase chain 2 | Oxidative phosphorylation | Mitochondrial respiratory chain dysfunction in cancer cells. | [58] |
| 2.86 | SERPINC1 | Antithrombin-III | Coagulation cascade | Associated with thrombosis in cancer, a potential biomarker for metastasis risk. | [59] | |
| 5.21 | VTN | Vitronectin | ECM-receptor interaction, FAK/AKT pathway | Promotes metastasis via EMT and integrin signalling. | [42] | |
| 0.64 | CCNB1 | G2/mitotic-specific cyclin-B1 | G2/M checkpoint of cell cycle | Forms a complex with CDK1 to drive mitosis. | [60] | |
| 1.26 | CYCS | Cytochrome c | Intrinsic apoptotic pathway (mitochondria) | Released from mitochondria to activate caspases (Apaf1 → caspase-9 → caspase-3). | [61] | |
| −0.73 | DNMT1 | DNA (cytosine-5)-methyltransferase 1 | Epigenetic regulation | Silencing of tumour suppressors via DNA methylation. | [62] | |
| −0.68 | G3BP1 | Ras GTPase-activating protein-binding protein 1 | Stress response and RNA metabolism | Modulates stress granule formation, inhibits apoptosis under stress. | [63] | |
| −2.40 | RSL1D1 | Ribosomal L1 domain-containing protein 1 | Ribosome biogenesis | Potential suppressor of uncontrolled protein synthesis in cancer. | [64,65] | |
| −0.93 | CDK4 | Cyclin-dependent kinase 4 | Cell cycle (G1-S transition) | Downregulating CDK4 maintains pRB in its active, hypophosphorylated form, which blocks E2F, causing cell cycle arrest. | [66] | |
| −0.59 | UBA2 | SUMO-activating enzyme subunit 2 | SUMOylation pathway | Regulates oncogenic transcription factors and genome stability. | [67] | |
| −0.68 | CUL1 | Cullin-1 | SCF complex (E3 ubiquitin ligase) | Scaffold for E3 ligase that degrades cell cycle regulators. | [68] | |
| −0.92 | HDAC1 | Histone deacetylase 1 | Epigenetic silencing | Deacetylates histones to repress gene expression. | [69] | |
| −0.87 | RAC1 | Ras-related C3 botulinum toxin substrate 1 | Rho GTPase signalling | Controls cytoskeleton, proliferation, and survival. | [70] | |
| −0.67 | RAC3 | Ras-related C3 botulinum toxin substrate 3 | Cell motility and metastasis (GTPase signalling) | Similar to RAC1, with roles in aggressive tumours. | [71] | |
| −4.23 | RPL27A | Large ribosomal subunit protein uL15 | Ribosome biogenesis, mTOR, p53-MDM2 | Protein synthesis, p53 stabilisation via ribosomal stress. | [72] | |
| −1.04 | RPS14 | Small ribosomal subunit protein uS11 | Regulates erythropoiesis and apoptosis | Loss activates p53, which is linked to 5q-syndrome. Mutated in 5q-syndrome (MDS). | [73] | |
| N: UB 7:3 (5600: 150 μM) | −0.87 | RPL5 | Large ribosomal subunit protein uL18 | p53-mediated cell cycle pathway | Ribosomal dysfunction, tumorigenesis. | [74] |
| −1.48 | RPL11 | Large ribosomal subunit protein uL5 | p53-mediated cell cycle pathway | Ribosomal dysfunction, tumorigenesis. | [75] | |
| −2.03 | RPS19 | Small ribosomal subunit protein eS19 | Ribosome biogenesis, possibly p53 pathway | Ribosomal dysfunction. | [76] | |
| −3.47 | RPS27 | Small ribosomal subunit protein eS27 | Ribosome biogenesis/p53 regulation | Suppression leads to reduced ribosomal activity, possibly reducing protein synthesis, but may also destabilise p53, counteracting anticancer effects. | [72] | |
| −0.60 | UBE2N | Ubiquitin-conjugating enzyme E2 N | DNA damage response, NF-κB signalling | Downregulation inhibits DNA repair and pro-survival signalling, leading to sensitisation to chemotherapy and apoptosis. | [77] | |
| 0.59 | PSMD10 | 26S proteasome non-ATPase regulatory subunit 10 | 26S proteasome, cell cycle | shRNA targeting PSMD10 significantly reduces the viability of cancer cells. miR-214 and miR-137 inhibit proliferation by suppressing PSMD10. | [78] |
2.4.2. DEPs in UB (150 µM) Treated HKB-11 Lymphoma Cells Compared to Untreated Control (abs log2FC > 0.58 and Q < 0.05)
2.4.3. DEPs in Combination 7:3 (5750 µM), Which Are N (5600 µM) and (150 µM), Respectively, Treated HKB-11 Lymphoma Cells Compared to Untreated Control (abs log2FC > 0.58 and Q < 0.05)
3. Materials and Methods
3.1. Chemicals and Drug Preparation
3.2. Cell Culture
3.3. Cell Viability Assays
3.4. Synergy Analysis
3.5. Analysis of ROS Production
3.6. Flow Cytometry Analyses of the Apoptotic Profiles
3.7. Liquid Chromatography–Mass Spectrometry (LC–MS)–Driven Bottom-Up Proteomics Analysis
3.7.1. Cell Culture, Treatment, and Protein Extraction
3.7.2. Protein Quantification
3.7.3. Peptides Preparation and Clean-Up
3.7.4. Label-Free Quantitative Proteomics Using Micro-High-Performance Liquid Chromatography Coupled with Quadruple Time-of-Flight Mass Spectrometry (Micro-HPLC-QTOF-MS)
Liquid Chromatography and Mass Spectrometry Setup
Mass Spectrometry Acquisition Parameters
Mass Calibration and Library Generation
Data Processing and Statistical Analysis
Data Availability
3.8. Statistical Analysis
4. Conclusions, Limitations, and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood J. Am. Soc. Hematol. 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Cancer Tomorrow, 2021. [Google Scholar]
- Al-Khazaleh, A.K.; Chang, D.; Münch, G.W.; Bhuyan, D.J. The gut connection: Exploring the possibility of implementing gut microbial metabolites in lymphoma treatment. Cancers 2024, 16, 1464. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Cheng, L.; Cao, X.; Liu, C. Gut microbiota in colorectal cancer: A review of its influence on tumor immune surveillance and therapeutic response. Front. Oncol. 2025, 15, 1557959. [Google Scholar] [CrossRef]
- Sadrekarimi, H.; Gardanova, Z.R.; Bakhshesh, M.; Ebrahimzadeh, F.; Yaseri, A.F.; Thangavelu, L.; Hasanpoor, Z.; Zadeh, F.A.; Kahrizi, M.S. Emerging role of human microbiome in cancer development and response to therapy: Special focus on intestinal microflora. J. Transl. Med. 2022, 20, 301. [Google Scholar] [CrossRef]
- O’keefe, S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef]
- Zitvogel, L.; Daillère, R.; Roberti, M.P.; Routy, B.; Kroemer, G. Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 2017, 15, 465–478. [Google Scholar] [CrossRef]
- Peluzio, M.d.C.G.; Martinez, J.A.; Milagro, F.I. Postbiotics: Metabolites and mechanisms involved in microbiota-host interactions. Trends Food Sci. Technol. 2021, 108, 11–26. [Google Scholar] [CrossRef]
- Jaye, K.; Alsherbiny, M.A.; Chang, D.; Li, C.-G.; Bhuyan, D.J. Mechanistic insights into the anti-proliferative action of gut microbial metabolites against breast adenocarcinoma cells. Int. J. Mol. Sci. 2023, 24, 15053. [Google Scholar] [CrossRef] [PubMed]
- Lewies, A.; Wentzel, J.F.; Miller, H.C.; Du Plessis, L.H. The antimicrobial peptide nisin Z induces selective toxicity and apoptotic cell death in cultured melanoma cells. Biochimie 2018, 144, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Sadri, H.; Aghaei, M.; Akbari, V. Nisin induces apoptosis in cervical cancer cells via reactive oxygen species generation and mitochondrial membrane potential changes. Biochem. Cell Biol. 2022, 100, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Hasheminezhad, S.H.; Boozari, M.; Iranshahi, M.; Yazarlu, O.; Sahebkar, A.; Hasanpour, M.; Iranshahy, M. A mechanistic insight into the biological activities of urolithins as gut microbial metabolites of ellagitannins. Phytother. Res. 2022, 36, 112–146. [Google Scholar] [CrossRef]
- Gandhi, G.R.; Antony, P.J.; Ceasar, S.A.; Vasconcelos, A.B.S.; Montalvão, M.M.; Farias de Franca, M.N.; Resende, A.d.S.; Sharanya, C.S.; Liu, Y.; Hariharan, G. Health functions and related molecular mechanisms of ellagitannin-derived urolithins. Crit. Rev. Food Sci. Nutr. 2024, 64, 280–310. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zheng, J.; Lu, Y.; Mai, Z.; Lin, Y.; Lin, P.; Zheng, Y.; Chen, X.; Xu, R.; Zhao, X. Optimizing CD8+ T cell-based immunotherapy via metabolic interventions: A comprehensive review of intrinsic and extrinsic modulators. Exp. Hematol. Oncol. 2024, 13, 103. [Google Scholar] [CrossRef] [PubMed]
- Kuru-Yaşar, R.; Üstün-Aytekin, Ö. The crucial roles of diet, microbiota, and postbiotics in colorectal cancer. Curr. Nutr. Rep. 2024, 13, 126–151. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Park, J.-S.; Lee, E.-J.; Ahn, J.-H.; Kim, H.-S. Anti-inflammatory and antioxidant mechanisms of urolithin B in activated microglia. Phytomedicine 2019, 55, 50–57. [Google Scholar] [CrossRef]
- Alzahrani, A.M.; Shait Mohammed, M.R.; Alghamdi, R.A.; Ahmad, A.; Zamzami, M.A.; Choudhry, H.; Khan, M.I. Urolithin a and b alter cellular metabolism and induce metabolites associated with apoptosis in leukemic cells. Int. J. Mol. Sci. 2021, 22, 5465. [Google Scholar] [CrossRef]
- Al-Khazaleh, A.K.; Alsherbiny, M.A.; Münch, G.; Chang, D.; Bhuyan, D.J. Gut Microbial Postbiotics as Potential Therapeutics for Lymphoma: Proteomics Insights of the Synergistic Effects of Nisin and Urolithin B Against Human Lymphoma Cells. Int. J. Mol. Sci. 2025, 26, 6829. [Google Scholar] [CrossRef]
- Tremblay, A.R.; Delbes, G. In vitro study of doxorubicin-induced oxidative stress in spermatogonia and immature Sertoli cells. Toxicol. Appl. Pharmacol. 2018, 348, 32–42. [Google Scholar] [CrossRef]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef]
- Jafari, M.; Sriram, V.; Premnauth, G.; Merino, E.; Lee, J.-Y. Modified peroxamide-based reactive oxygen species (ROS)-responsive doxorubicin prodrugs. Bioorg. Chem. 2022, 127, 105990. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, D.; Asaduzzaman, M.; Young, F. Real time monitoring and quantification of reactive oxygen species in breast cancer cell line MCF-7 by 2′, 7′–dichlorofluorescin diacetate (DCFDA) assay. J. Pharmacol. Toxicol. Methods 2018, 94, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Dong, W.; Sun, D.; Zhao, X.; Huang, Z.; Liu, C.; Sheng, Y. Bacterial metabolites: Effects on the development of breast cancer and therapeutic efficacy. Oncol. Lett. 2025, 29, 210. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yan, L.; Xu, X.; Jiang, C.; Shi, J.; Zhang, Y.; Liu, L.; Lei, S.; Shao, D.; Huang, Q. Potential of Bacillus subtilis lipopeptides in anti-cancer I: Induction of apoptosis and paraptosis and inhibition of autophagy in K562 cells. Amb Express 2018, 8, 78. [Google Scholar] [CrossRef]
- Joo, N.E.; Ritchie, K.; Kamarajan, P.; Miao, D.; Kapila, Y.L. Nisin, an apoptogenic bacteriocin and food preservative, attenuates HNSCC tumorigenesis via CHAC 1. Cancer Med. 2012, 1, 295–305. [Google Scholar] [CrossRef]
- Thanjavur, N.; Sangubotla, R.; Lakshmi, B.A.; Rayi, R.; Mekala, C.D.; Reddy, A.S.; Viswanath, B. Evaluating the antimicrobial and apoptogenic properties of bacteriocin (nisin) produced by Lactococcus lactis. Process Biochem. 2022, 122, 76–86. [Google Scholar] [CrossRef]
- Mohan Latha Kumari, A.; Biju Latha, C.; Padmanabhan, R.A.; Vijayakumar, N.; Ramachandran, R. Antibacterial Peptide Nisin Activates p53 Gene Expression and Triggers Apoptosis in Human Colon Cancer Cells. Pept. Sci. 2025, 117, e24379. [Google Scholar] [CrossRef]
- Huang, F.; Teng, K.; Liu, Y.; Wang, T.; Xia, T.; Yun, F.; Zhong, J. Nisin Z attenuates lipopolysaccharide-induced mastitis by inhibiting the ERK1/2 and p38 mitogen-activated protein kinase signaling pathways. J. Dairy Sci. 2022, 105, 3530–3543. [Google Scholar] [CrossRef]
- Stanisławska, I.; Piwowarski, J.; Granica, S.; Kiss, A. Urolithins, gut microbiota metabolites of ellagitannins, in prostate cancer chemoprevention. Planta Medica Int. Open 2017, 4, Mo-PO-16. [Google Scholar]
- Kujawska, M.; Jodynis-Liebert, J. Potential of the ellagic acid-derived gut microbiota metabolite–Urolithin A in gastrointestinal protection. World J. Gastroenterol. 2020, 26, 3170. [Google Scholar] [CrossRef]
- Liang, Y.; Zhao, Y.; Qi, Z.; Li, X.; Zhao, Y. Ferroptosis: CD8+ T cells’ blade to destroy tumor cells or poison for self-destruction. Cell Death Discov. 2025, 11, 128. [Google Scholar] [CrossRef]
- Maxwell, S.A.; Mousavi-Fard, S. Non-Hodgkin’s B-cell lymphoma: Advances in molecular strategies targeting drug resistance. Exp. Biol. Med. 2013, 238, 971–990. [Google Scholar] [CrossRef] [PubMed]
- Radaic, A.; Malone, E.; Kamarajan, P.; Kapila, Y.L. Solid lipid nanoparticles loaded with nisin (sln-nisin) are more effective than free nisin as antimicrobial, antibiofilm, and anticancer agents. J. Biomed. Nanotechnol. 2022, 18, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Mazzara, S.; Salemi, D.; Zanetti, S.; Sapienza, M.R.; Orecchioni, S.; Talarico, G.; Falvo, P.; Davini, A.; Ceccarelli, C. Downregulation of rRNA synthesis by BCL-2 induces chemoresistance in Diffuse Large B-cell Lymphoma. iScience 2025, 28, 112333. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Chen, X.; Li, J.; Comish, P.; Kang, R.; Tang, D. Transcription factors in ferroptotic cell death. Cancer Gene Ther. 2020, 27, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Eladwy, R.A.; Alsherbiny, M.A.; Chang, D.; Fares, M.; Li, C.-G.; Bhuyan, D.J. The postbiotic sodium butyrate synergizes the antiproliferative effects of dexamethasone against the AGS gastric adenocarcinoma cells. Front. Nutr. 2024, 11, 1372982. [Google Scholar] [CrossRef]
- Alsherbiny, M.A.; Bhuyan, D.J.; Low, M.N.; Chang, D.; Li, C.G. Synergistic interactions of cannabidiol with chemotherapeutic drugs in mcf7 cells: Mode of interaction and proteomics analysis of mechanisms. Int. J. Mol. Sci. 2021, 22, 10103. [Google Scholar] [CrossRef]
- Prindle, V.; Richardson, A.E.; Sher, K.R.; Kongpachith, S.; Kentala, K.; Petiwala, S.; Cheng, D.; Widomski, D.; Le, P.; Torrent, M. Synthetic lethality of mRNA quality control complexes in cancer. Nature 2025, 638, 1095–1103. [Google Scholar] [CrossRef]
- Rong, F.; Liu, L.; Zou, C.; Zeng, J.; Xu, Y. MALAT1 promotes cell tumorigenicity through regulating miR-515-5p/EEF2 axis in non-small cell lung cancer. Cancer Manag. Res. 2020, 12, 7691–7701. [Google Scholar] [CrossRef]
- Yip, K.-P.D.; Au-Yeung, C.-L.; Zhang, L.; Mok, S.C.; Sham, J.S. Upregulation of ryanodine receptor-1 dependent calcium signaling contributes to the malignance of high-grade serous ovarian cancer (HGSOC). Biophys. J. 2023, 122, 163a. [Google Scholar] [CrossRef]
- Lin, Y.; Bian, L.; Zhu, G.; Zhang, B. Vitronectin promotes proliferation and metastasis of cervical cancer cells via the epithelial-mesenchymal transition. Front. Oncol. 2024, 14, 1466264. [Google Scholar] [CrossRef]
- Kim, K.; Choi, E.-Y.; Ahn, H.-M.; Kim, D.-G.; Kim, Y.-J. Hemoglobin Subunit Theta 1 promotes proliferation by reducing reactive Oxygen species in Lung Adenocarcinoma. Cancers 2023, 15, 5504. [Google Scholar] [CrossRef]
- Li, X.; Wu, Z.; Wang, Y.; Mei, Q.; Fu, X.; Han, W. Characterization of adult α-and β-globin elevated by hydrogen peroxide in cervical cancer cells that play a cytoprotective role against oxidative insults. PLoS ONE 2013, 8, e54342. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Shen, W.; Peng, H.; Li, Y.; Chen, F.; Zheng, L.; Xu, J.; Jia, L. Fibronectin 1 promotes melanoma proliferation and metastasis by inhibiting apoptosis and regulating EMT. OncoTargets Ther. 2019, 12, 3207–3221. [Google Scholar] [CrossRef] [PubMed]
- Ghetti, M.; Vannini, I.; Storlazzi, C.T.; Martinelli, G.; Simonetti, G. Linear and circular PVT1 in hematological malignancies and immune response: Two faces of the same coin. Mol. Cancer 2020, 19, 69. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-S.; Wang, C.-Y.; Chen, P.-S.; Hung, J.-H.; Yen, J.-H.; Wu, M.-J. 8-Hydroxydaidzein Downregulates JAK/STAT, MMP, Oxidative Phosphorylation, and PI3K/AKT Pathways in K562 Cells. Biomedicines 2021, 9, 1907. [Google Scholar] [CrossRef]
- Wang, S.; Yang, R.; Song, M.; Li, J.; Zhou, Y.; Dai, C.; Song, T. Current understanding of the role of DDX21 in orchestrating gene expression in health and diseases. Life Sci. 2024, 349, 122716. [Google Scholar] [CrossRef]
- Kumar, S.; Shuaib, M.; AlAsmari, A.F.; Alqahtani, F.; Gupta, S. GNL3 and PA2G4 as prognostic biomarkers in prostate cancer. Cancers 2023, 15, 2723. [Google Scholar] [CrossRef]
- Wang, W.-Q.; Zhang, H.; Wang, H.-B.; Sun, Y.-G.; Peng, Z.-H.; Zhou, G.; Yang, S.-M.; Wang, R.-Q.; Fang, D.-C. Programmed cell death 4 (PDCD4) enhances the sensitivity of gastric cancer cells to TRAIL-induced apoptosis by inhibiting the PI3K/Akt signaling pathway. Mol. Diagn. Ther. 2010, 14, 155–161. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhou, D.; Chen, F.; Yang, Z.; Gu, W.; Zhang, K. SIX5-activated LINC01468 promotes lung adenocarcinoma progression by recruiting SERBP1 to regulate SERPINE1 mRNA stability and recruiting USP5 to facilitate PAI1 protein deubiquitylation. Cell Death Dis. 2022, 13, 312. [Google Scholar] [CrossRef]
- Anguelov, R.; Goddard, M.; Hlophe, Y.; Letsoalo, K.; Serem, J. A Mathematical Model of the Cell Cycle: Exploring the Impact of Zingerone on Cancer Cell Proliferation. arXiv 2025, arXiv:2502.18589. [Google Scholar]
- Ha, L.; Qian, Y.; Zhang, S.; Ju, X.; Sun, S.; Guo, H.; Wang, Q.; Li, K.; Fan, Q.; Zheng, Y.; et al. Synthesis and Biological Evaluation of Scutellaria Flavone Cyclaneaminol Mannich Base Derivatives as Novel CDK1 Inhibitors. Anticancer Agents Med Chem 2016, 16, 914–924. [Google Scholar] [CrossRef]
- Sakane, T.; Okuda, K.; Hattori, H.; Watanabe, T.; Oda, R.; Tatematsu, T.; Yokota, K.; Haneda, H.; Inagaki, H.; Nakanishi, R. Blastomatoid pulmonary carcinosarcoma: A rare case report and review of the literature. Thorac Cancer 2018, 9, 1323–1326. [Google Scholar] [CrossRef]
- Spring, K.; Chabot, C.; Langlois, S.; Lapointe, L.; Trinh, N.T.; Caron, C.; Hebda, J.K.; Gavard, J.; Elchebly, M.; Royal, I. Tyrosine phosphorylation of DEP-1/CD148 as a mechanism controlling Src kinase activation, endothelial cell permeability, invasion, and capillary formation. Blood 2012, 120, 2745–2756. [Google Scholar] [CrossRef] [PubMed]
- Limone, A.; Maggisano, V.; Sarnataro, D.; Bulotta, S. Emerging roles of the cellular prion protein (PrPC) and 37/67 kDa laminin receptor (RPSA) interaction in cancer biology. Cell. Mol. Life Sci. 2023, 80, 207. [Google Scholar] [CrossRef] [PubMed]
- Merenbakh-Lamin, K.; Ben-Baruch, N.; Yeheskel, A.; Dvir, A.; Soussan-Gutman, L.; Jeselsohn, R.; Yelensky, R.; Brown, M.; Miller, V.A.; Sarid, D.; et al. D538G mutation in estrogen receptor-α: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res 2013, 73, 6856–6864. [Google Scholar] [CrossRef] [PubMed]
- Abril, J.; de Heredia, M.L.; González, L.; Cleries, R.; Nadal, M.; Condom, E.; Aguiló, F.; Gómez-Zaera, M.; Nunes, V. Altered expression of 12S/MT-RNR1, MT-CO2/COX2, and MT-ATP6 mitochondrial genes in prostate cancer. Prostate 2008, 68, 1086–1096. [Google Scholar] [CrossRef]
- Conti, A.; Fredolini, C.; Tamburro, D.; Magagnoli, G.; Zhou, W.; Liotta, L.A.; Picci, P.; Luchini, A.; Benassi, M.S. Identification of novel candidate circulating biomarkers for malignant soft tissue sarcomas: Correlation with metastatic progression. Proteomics 2016, 16, 689–697. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, H.; Woo, J.; Yue, W.; Kim, K.; Choi, S.; Jang, J.-J.; Kim, Y.; Park, I.A.; Han, D.; et al. Reconstruction of pathway modification induced by nicotinamide using multi-omic network analyses in triple negative breast cancer. Sci. Rep. 2017, 7, 3466. [Google Scholar] [CrossRef]
- Recasens, A.; Humphrey, S.J.; Ellis, M.; Hoque, M.; Abbassi, R.H.; Chen, B.; Longworth, M.; Needham, E.J.; James, D.E.; Johns, T.G.; et al. Global phosphoproteomics reveals DYRK1A regulates CDK1 activity in glioblastoma cells. Cell Death Discov. 2021, 7, 81. [Google Scholar] [CrossRef]
- Robert, M.-F.; Morin, S.; Beaulieu, N.; Gauthier, F.; Chute, I.C.; Barsalou, A.; MacLeod, A.R. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 2003, 33, 61–65. [Google Scholar] [CrossRef]
- Bittencourt, L.; Negreiros-Lima, G.; Sousa, L.; Silva, A.; Souza, I.; Ribeiro, R.; Dutra, M.; Silva, R.; Dias, A.; Soriani, F. G3BP1 knockdown sensitizes U87 glioblastoma cell line to Bortezomib by inhibiting stress granules assembly and potentializing apoptosis. J. Neuro-Oncol. 2019, 144, 463–473. [Google Scholar] [CrossRef]
- Ding, L.; Zhang, Z.; Zhao, C.; Chen, L.; Chen, Z.; Zhang, J.; Liu, Y.; Nie, Y.; He, Y.; Liao, K. Ribosomal L1 domain-containing protein 1 coordinates with HDM2 to negatively regulate p53 in human colorectal Cancer cells. J. Exp. Clin. Cancer Res. 2021, 40, 245. [Google Scholar] [CrossRef]
- Jin, Y.; Zhao, L.; Wang, S.; Zhang, X.; Quan, J.; Lin, Z.; Piao, J. RSL1D1 knockdown induces ferroptosis and mediates ferrous iron accumulation in senescent cells by inhibiting FTH1 mRNA stability. Carcinogenesis 2023, 44, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Don, M.-J.; Chang, Y.-H.; Chen, K.-K.; Ho, L.-K.; Chau, Y.-P. Induction of CDK inhibitors (p21WAF1 and p27KIP1) and Bak in the β-lapachone-induced apoptosis of human prostate cancer cells. Mol. Pharmacol. 2001, 59, 784–794. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Lin, N.; Ye, Y.; Zhuang, H.; Zou, S.; Song, Y.; Chen, X.; Wang, Q. The prognosis, chemotherapy and immunotherapy efficacy of the SUMOylation pathway signature and the role of UBA2 in lung adenocarcinoma. Aging 2024, 16, 4378. [Google Scholar] [CrossRef] [PubMed]
- Gieffers, C.; Schleiffer, A.; Peters, J.-M. Cullins and cell cycle control. Protoplasma 2000, 211, 20–28. [Google Scholar] [CrossRef]
- Dannenberg, L.O.; Edenberg, H.J. Epigenetics of gene expression in human hepatoma cells: Expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genom. 2006, 7, 181. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, W.; Wang, G.; Wang, A.; Du, L.; Lou, W. Silencing ras-related C3 botulinum toxin substrate 1 inhibits growth and migration of hypopharyngeal squamous cell carcinoma via the P38 mitogen-activated protein kinase signaling pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 768. [Google Scholar] [CrossRef]
- De, P.; Rozeboom, B.J.; Aske, J.C.; Dey, N. Active RAC1 promotes tumorigenic phenotypes and therapy resistance in solid tumors. Cancers 2020, 12, 1541. [Google Scholar] [CrossRef]
- Xiong, X.; Zhao, Y.; Tang, F.; Wei, D.; Thomas, D.; Wang, X.; Liu, Y.; Zheng, P.; Sun, Y. Ribosomal protein S27-like is a physiological regulator of p53 that suppresses genomic instability and tumorigenesis. eLife 2014, 3, e02236. [Google Scholar] [CrossRef]
- Ebert, B. Deletion 5q in myelodysplastic syndrome: A paradigm for the study of hemizygous deletions in cancer. Leukemia 2009, 23, 1252–1256. [Google Scholar] [CrossRef]
- Oršolić, I.; Bursać, S.; Jurada, D.; Drmić Hofman, I.; Dembić, Z.; Bartek, J.; Mihalek, I.; Volarević, S. Cancer-associated mutations in the ribosomal protein L5 gene dysregulate the HDM2/p53-mediated ribosome biogenesis checkpoint. Oncogene 2020, 39, 3443–3457. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, K.M.; Lindström, M.S. Role of ribosomal protein mutations in tumor development. Int. J. Oncol. 2016, 48, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, N.; Kessel, R.; Bhagat, T.D.; Bhattacharyya, S.; Yu, Y.; Mcmahon, C.; Verma, A. Alterations in the ribosomal machinery in cancer and hematologic disorders. J. Hematol. Oncol. 2012, 5, 32. [Google Scholar] [CrossRef]
- Koo, S.-Y.; Park, E.-J.; Noh, H.-J.; Jo, S.-M.; Ko, B.-K.; Shin, H.-J.; Lee, C.-W. Ubiquitination links DNA damage and repair signaling to cancer metabolism. Int. J. Mol. Sci. 2023, 24, 8441. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Mohan, C.D.; Rangappa, S.; Zarrabi, A.; Hushmandi, K.; Kumar, A.P.; Sethi, G.; Rangappa, K.S. Noncoding RNAs as regulators of STAT3 pathway in gastrointestinal cancers: Roles in cancer progression and therapeutic response. Med. Res. Rev. 2023, 43, 1263–1321. [Google Scholar] [CrossRef]
- McAndrew, C.W.; Gastwirt, R.F.; Meyer, A.N.; Porter, L.A.; Donoghue, D.J. Spy1 enhances phosphorylation and degradation of the cell cycle inhibitor p27. Cell Cycle 2007, 6, 1937–1945. [Google Scholar] [CrossRef][Green Version]
- Xie, C.-M.; Wei, W.; Sun, Y. Role of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases in skin cancer. J. Genet. Genom. 2013, 40, 97–106. [Google Scholar] [CrossRef]
- Dissanayake, I.H.; Alsherbiny, M.A.; Chang, D.; Li, C.G.; Bhuyan, D.J. Antiproliferative effects of Australian native plums against the MCF7 breast adenocarcinoma cells and UPLC-qTOF-IM-MS-driven identification of key metabolites. Food Biosci. 2023, 54, 102864. [Google Scholar] [CrossRef]
- Pino, L.K.; Just, S.C.; MacCoss, M.J.; Searle, B.C. Acquiring and analyzing data independent acquisition proteomics experiments without spectrum libraries. Mol. Cell. Proteom. 2020, 19, 1088–1103. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, gkw937. [Google Scholar] [CrossRef]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C. Pathway enrichment analysis and visualization of omics data using g: Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B. The reactome pathway knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Kamburov, A.; Cavill, R.; Ebbels, T.M.; Herwig, R.; Keun, H.C. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics 2011, 27, 2917–2918. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bandla, C.; Kundu, D.J.; Kamatchinathan, S.; Bai, J.; Hewapathirana, S.; John, N.S.; Prakash, A.; Walzer, M.; Wang, S. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 2025, 53, D543–D553. [Google Scholar] [CrossRef] [PubMed]

| Concentration (μM) N:UB 7:3 | Cell Growth Inhibition (%) | Cell Viability (%) | |
|---|---|---|---|
| HKB-11 | Hs 313.T | HS-5 | |
| 5750 (5600:150) | 100.16 ± 0.02 a | 100.45 ± 0.11 a | 1.08 ± 1.43 |
| 2875 (2800:75) | 100.11 ± 0.03 a | 99.05 ± 1.16 a | 10.71 ± 1.96 |
| 1437.5 (1400:37.5) | 80.27 ± 3.41 a | 89.25 ± 0.71 b | 28.11 ± 5.36 |
| 718.75 (700:18.75) | 49.15 ± 7.03 a | 82.71 ± 0.88 b | 72.96 ± 9.29 |
| 359.38 (350:9.375) | 14.40 ± 5.52 a | 56.49 ± 3.83 b | 90.69 ± 7.06 |
| 179.69 (175:4.6875) | 8.53 ± 3.03 a | 21.35 ± 1.53 b | 93.63 ± 6.49 |
| IC50 | 820 μM | 332.2 μM | 1091 μM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Khazaleh, A.K.; Alsherbiny, M.A.; Chang, D.; Münch, G.; Bhuyan, D.J. Investigating the Cellular Responses to Combined Nisin and Urolithin B Treatment (7:3) in HKB-11 Lymphoma Cells. Int. J. Mol. Sci. 2025, 26, 7369. https://doi.org/10.3390/ijms26157369
Al-Khazaleh AK, Alsherbiny MA, Chang D, Münch G, Bhuyan DJ. Investigating the Cellular Responses to Combined Nisin and Urolithin B Treatment (7:3) in HKB-11 Lymphoma Cells. International Journal of Molecular Sciences. 2025; 26(15):7369. https://doi.org/10.3390/ijms26157369
Chicago/Turabian StyleAl-Khazaleh, Ahmad K., Muhammad A. Alsherbiny, Dennis Chang, Gerald Münch, and Deep Jyoti Bhuyan. 2025. "Investigating the Cellular Responses to Combined Nisin and Urolithin B Treatment (7:3) in HKB-11 Lymphoma Cells" International Journal of Molecular Sciences 26, no. 15: 7369. https://doi.org/10.3390/ijms26157369
APA StyleAl-Khazaleh, A. K., Alsherbiny, M. A., Chang, D., Münch, G., & Bhuyan, D. J. (2025). Investigating the Cellular Responses to Combined Nisin and Urolithin B Treatment (7:3) in HKB-11 Lymphoma Cells. International Journal of Molecular Sciences, 26(15), 7369. https://doi.org/10.3390/ijms26157369








