Transcriptomic Reprogramming and Key Molecular Pathways Underlying Huanglongbing Tolerance and Susceptibility in Six Citrus Cultivars
Abstract
1. Introduction
2. Results
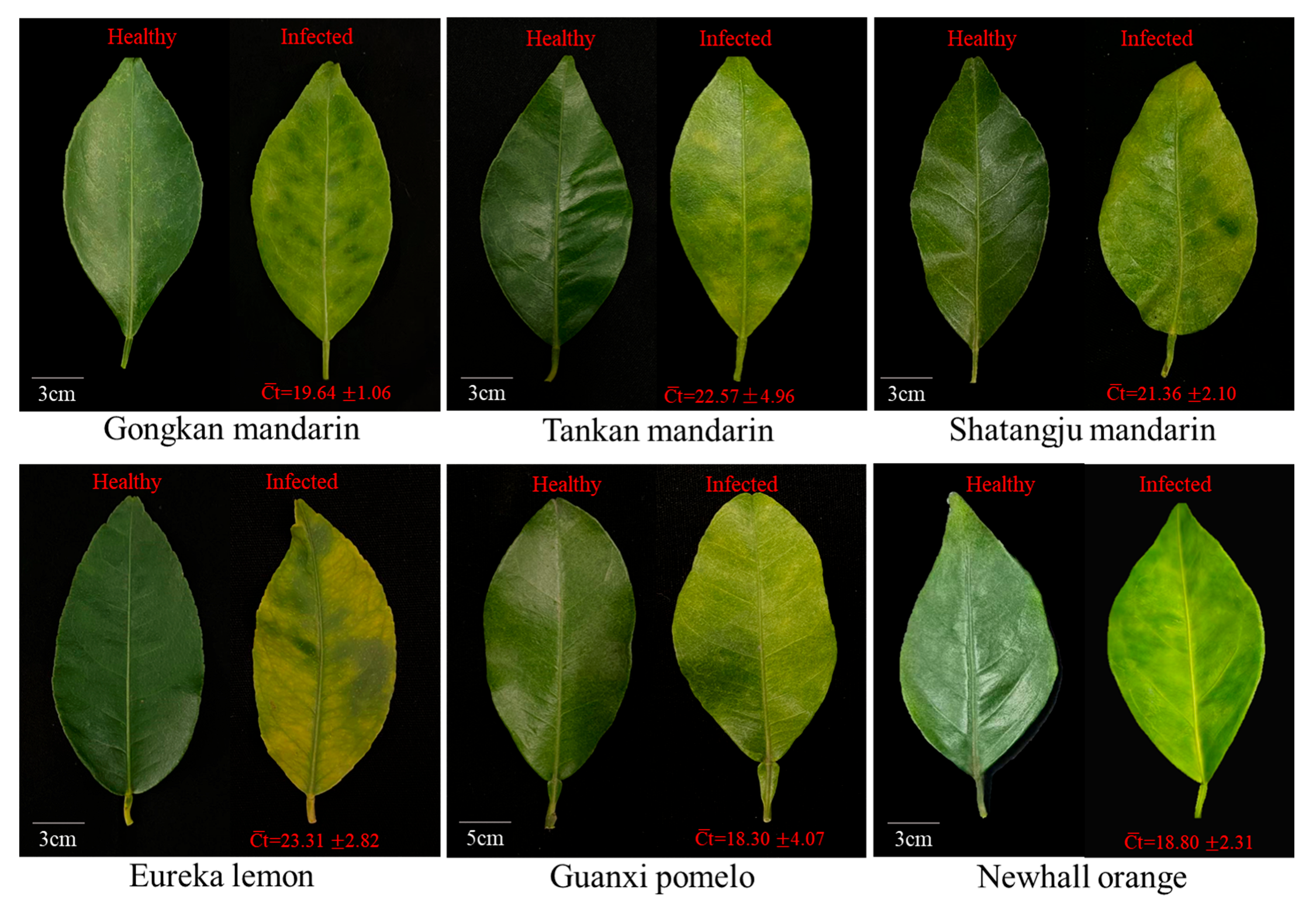
2.1. Symptoms and CLas Quantification of Leaf Samples from Six HLB-Affected Citrus Cultivars
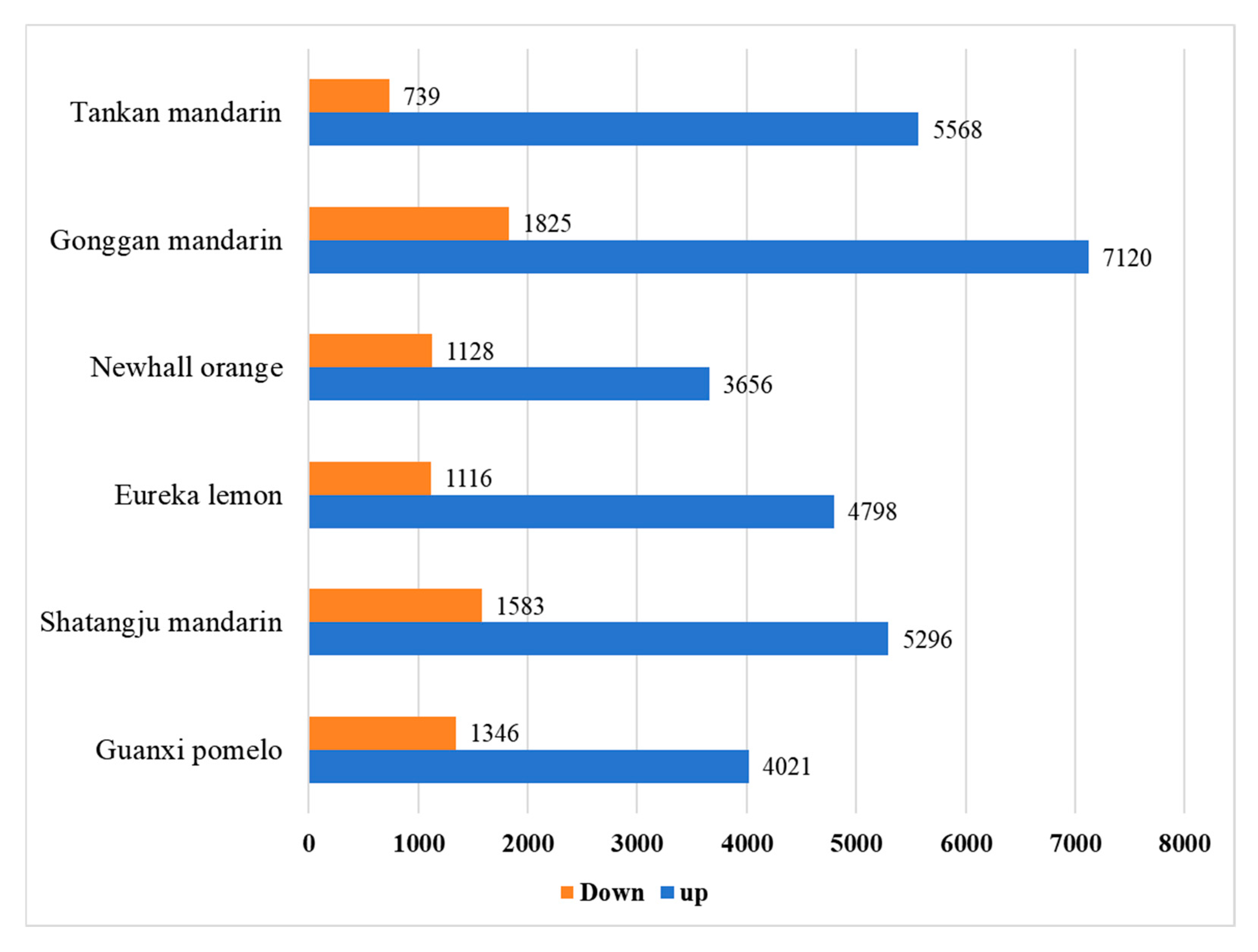
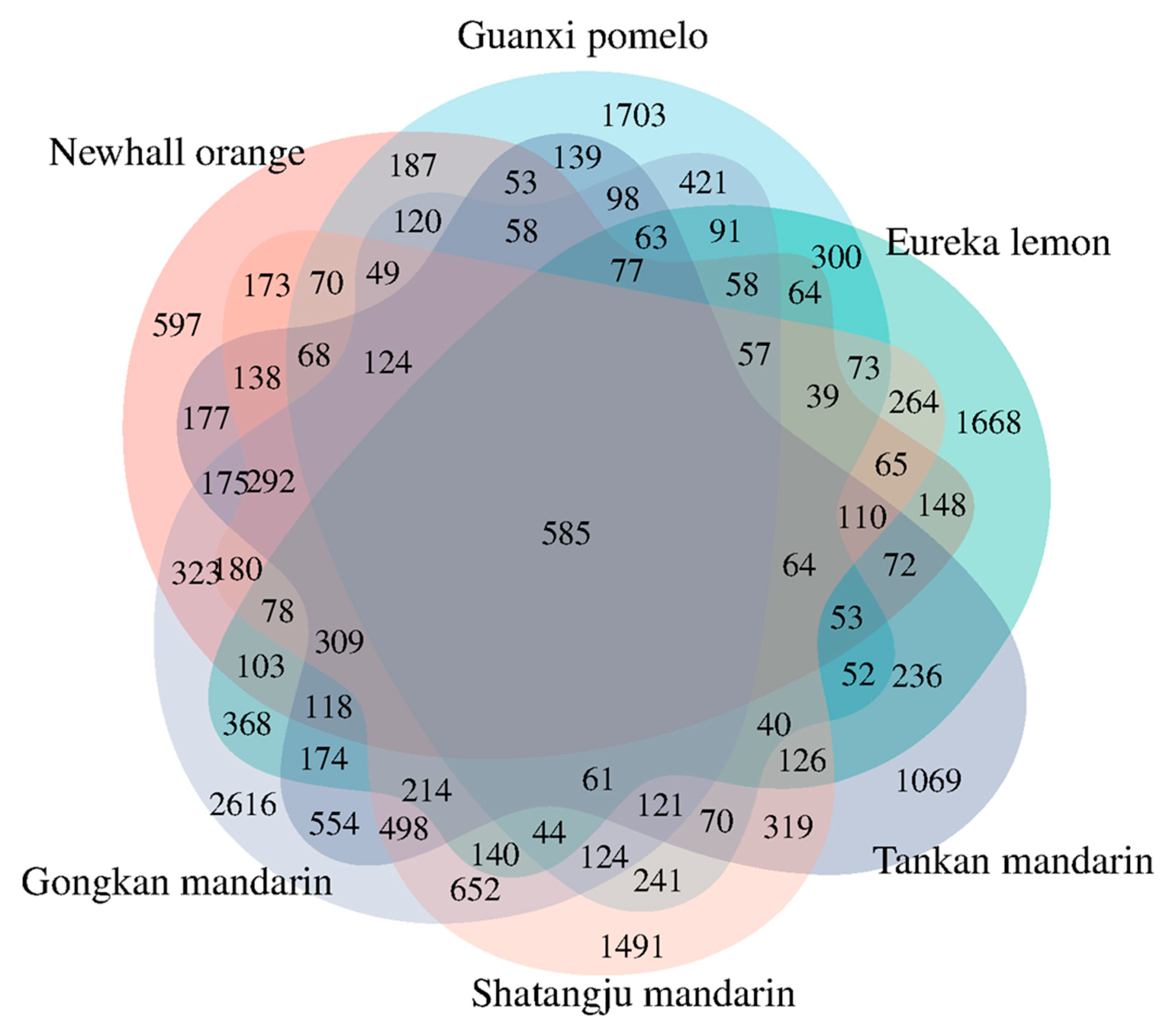
2.2. Identification of DEGs
2.3. Gene Ontology (GO) and KEGG Enrichment Analysis
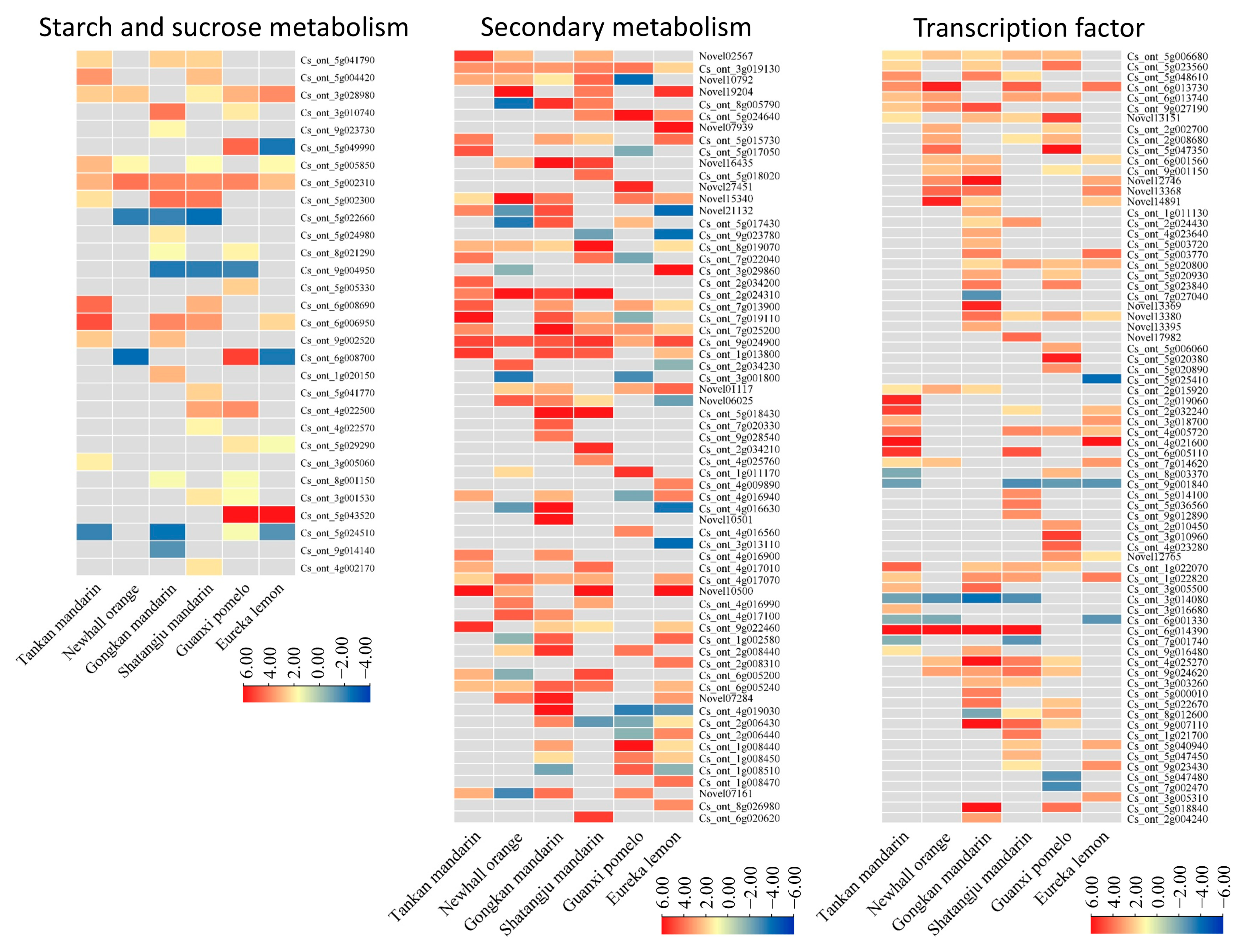
2.4. Starch and Sucrose Metabolism
2.5. Secondary Metabolism
2.6. Transcription Factors
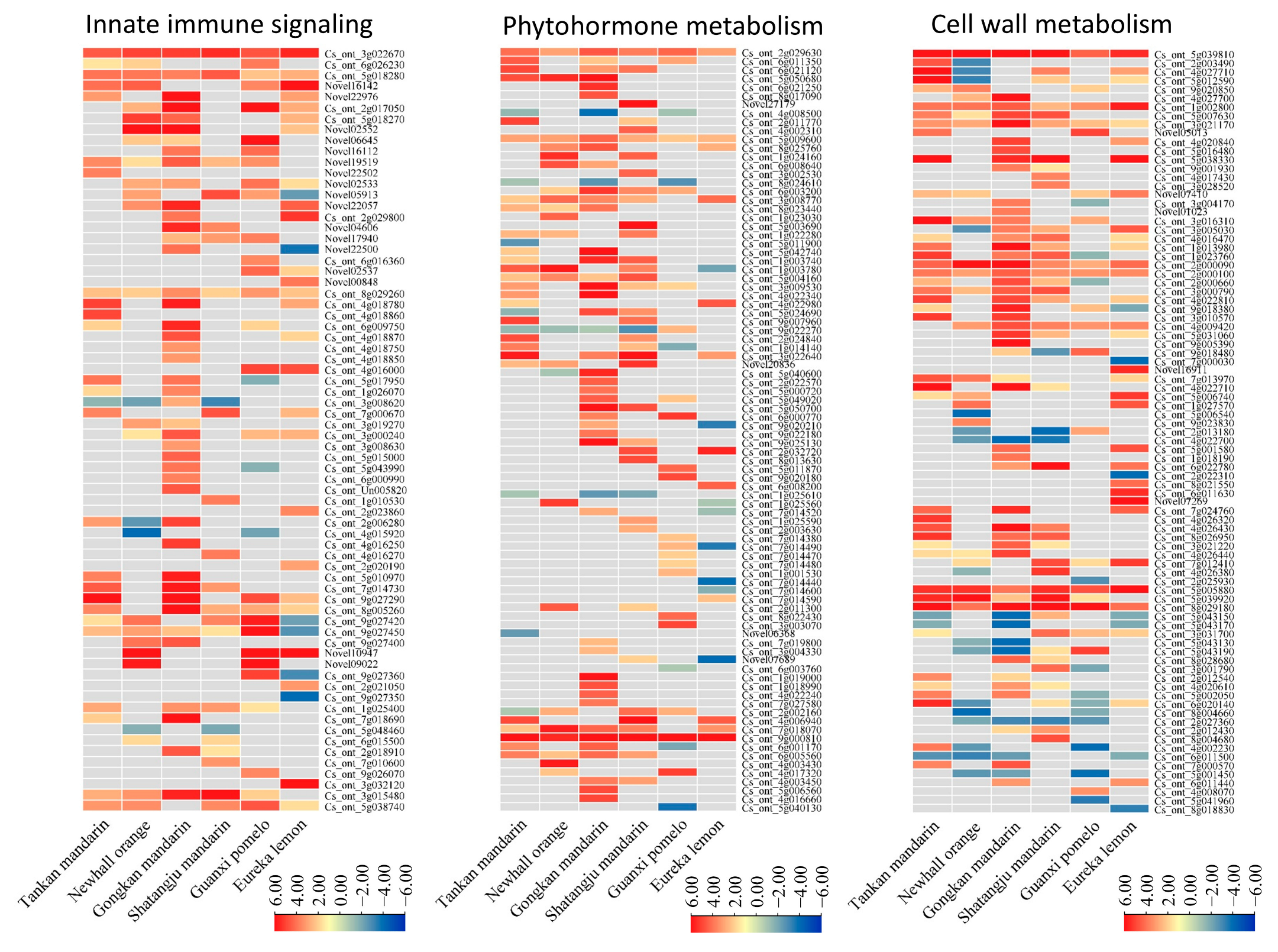
2.7. Innate Immune Signaling
2.8. Phytohormone Metabolism
2.9. Cell Wall Metabolism
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. DNA and RNA Extraction
4.3. RNA Sequencing and Transcriptomic Analysis cDNA Library Production
4.4. CLas Quantification
4.5. Gene Expression Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Zheng, Z.; Chen, J.C.; Deng, X.L. Historical perspectives, management, and current research of citrus HLB in Guangdong Province of China, where the disease has been endemic for over a hundred years. Phytopathology 2018, 108, 1224–1236. [Google Scholar] [CrossRef]
- Jagoueix, S.; Bové, J.M.; Garnier, M. The phloem-limited bacterium of greening disease of citrus is a member of the α subdivision of the Proteobacteria. Int. J. Syst. Evol. Microbiol. 1994, 44, 379–386. [Google Scholar] [CrossRef]
- Teixeira, D.D.C.; Saillard, C.; Eveillard, S.; Danet, J.L.; Costa, P.I.D.; Ayres, A.J.; Bové, J.M. ‘Candidatus Liberibacter americanus’, associated with citrus huanglongbing (greening disease) in São Paulo State, Brazil. Int. J. Syst. Evol. Microbiol. 2005, 55, 1857–1862. [Google Scholar] [CrossRef] [PubMed]
- Folimonova, S.Y.; Robertson, C.J.; Garnsey, S.M.; Gowda, S.; Dawson, W.O. Examination of the responses of different genotypes of citrus to huanglongbing (citrus greening) under different conditions. Phytopathology 2009, 99, 1346–1354. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Li, C.; Li, Z.; Liu, Y.; Li, J.; Guo, J.; Mao, J.; Fang, F.; Wang, C.; Deng, X.; et al. Comparative transcriptome profiling of susceptible and tolerant citrus species at early and late stage of infection by “Candidatus Liberibacter asiaticus”. Front. Plant Sci. 2023, 14, 1191029. [Google Scholar] [CrossRef]
- Miles, G.P.; Stover, E.; Ramadugu, C.; Keremane, M.L.; Lee, R.F. Apparent tolerance to huanglongbing in citrus and citrus-related germplasm. HortScience 2017, 52, 31–39. [Google Scholar] [CrossRef]
- Hu, B.; Yuan, T.; Lu, Z.; Huang, R.; He, J.; Yang, K.; Wu, Q.; Ai, W.; Zhang, W.; Zheng, W.; et al. “Candidatus Liberibacter asiaticus” Infection Induces Citric Acid Accumulation and Immune Responses Mediated by the Transcription Factor CitPH4. Mol. Plant Pathol. 2025, 26, e70062. [Google Scholar] [CrossRef]
- Huang, G.; Hu, Y.; Li, F.; Zuo, X.; Wang, X.; Li, F.; Li, R. Genome-wide characterization of heavy metal-associated isoprenylated plant protein gene family from Citrus sinensis in response to huanglongbing. Front. Plant Sci. 2024, 15, 1369883. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, X.; Huang, H.; Deng, X.; Zhang, B.; Meng, Y.; Wu, L.; Chen, H.; Zhong, Y.; Chen, W. Genome-Wide Identification of the DnaJ Gene Family in Citrus and Functional Characterization of ClDJC24 in Response to Citrus Huanglongbing. Int. J. Mol. Sci. 2024, 25, 11967. [Google Scholar] [CrossRef]
- Zhao, P.; Yang, H.; Sun, Y.; Zhang, J.; Gao, K.; Wu, J.; Zhu, C.; Yin, C.; Chen, X.; Liu, Q.; et al. Targeted MYC2 stabilization confers citrus Huanglongbing resistance. Science 2025, 388, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Mira, A.; Yu, Q.; Gmitter, F.G., Jr. The mechanism of citrus host defense response repression at early stages of infection by feeding of Diaphorina citri transmitting Candidatus Liberibacter asiaticus. Front. Plant Sci. 2021, 12, 635153. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Pang, Z.; Huang, X.; Xu, J.; Pandey, S.S.; Li, J.; Achor, D.S.; Vasconcelos, F.N.C.; Hendrich, C.; Huang, Y.; et al. Citrus Huanglongbing is a pathogen-triggered immune disease that can be mitigated with antioxidants and gibberellin. Nat. Commun. 2022, 13, 529. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.; Xu, J.; Hendrich, C.; Pandey, S.S.; Yu, Q.; Gmitter, F.G., Jr.; Wang, N. Seasonal transcriptome profiling of susceptible and tolerant citrus cultivars to citrus Huanglongbing. Phytopathology 2023, 113, 286–298. [Google Scholar] [CrossRef]
- Balan, B.; Ibáñez, A.M.; Dandekar, A.M.; Caruso, T.; Martinelli, F. Identifying host molecular features strongly linked with responses to huanglongbing disease in citrus leaves. Front. Plant Sci. 2018, 9, 277. [Google Scholar] [CrossRef]
- Schneider, H. Anatomy of greening-disease sweet orange shoots. Phytopathology 1968, 58, 1155–1160. [Google Scholar]
- Schaffer, A.A.; Liu, K.C.; Goldschmidt, E.E.; Boyer, C.D.; Goren, R. Citrus leaf chlorosis induced by sink removal: Starch, nitrogen, and chloroplast ultrastructure. J. Plant Physiol. 1986, 124, 111–121. [Google Scholar] [CrossRef]
- Fan, J.; Chen, C.; Yu, Q.; Khalaf, A.; Achor, D.S.; Brlansky, R.H.; Moore, G.A.; Li, Z.; Gmitter, F.G., Jr. Comparative transcriptional and anatomical analyses of tolerant rough lemon and susceptible sweet orange in response to ‘Candidatus Liberibacter asiaticus’ infection. Mol. Plant-Microbe Interact. 2012, 25, 1396–1407. [Google Scholar] [CrossRef]
- Wu, Y.; Su, H.N.; Huang, A.J.; Zhou, Y.; Li, Z.A.; Liu, J.X.; Zhou, C.Y. Effects of Candidatus Liberibacter asiaticus infection on sugar metabolism in sweet orange leaves. Nongye Kexue 2015, 48, 63–72. [Google Scholar] [CrossRef]
- Iwase, A.; Matsui, K.; Ohme-Takagi, M. Manipulation of plant metabolic pathways by transcription factors. Plant Biotechnol. 2009, 26, 29–38. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Deslandes, L.; Olivier, J.; Theulières, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Naik, J.; Misra, P.; Trivedi, P.K.; Pandey, A. Molecular components associated with the regulation of flavonoid biosynthesis. Plant Sci. 2022, 317, 111196. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, S.; Yu, Y.; Cui, N.; Yu, G.; Zhao, H.; Meng, X.; Fan, H. The pivotal role of MYB transcription factors in plant disease resistance. Planta 2023, 258, 16. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar] [CrossRef]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.S.; Paul, G.; Yu, Z.; Xu, L.; Cheng, T.; Cheng, B.; Aslam, M.N.; Baig, A.; Zhao, H. Comparative Transcriptome and sRNAome Analysis Suggest Coordinated Citrus Immune Responses against Huanglongbing Disease. Plants 2024, 13, 1496. [Google Scholar] [CrossRef]
- Bredow, M.; Monaghan, J. Regulation of plant immune signaling by calcium-dependent protein kinases. Mol. Plant-Microbe Interact. 2019, 32, 6–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, L.; Wang, Y.; Li, B.; Li, X.; Ye, Z.; Zhang, J. A calcium-dependent protein kinase regulates the defense response in Citrus sinensis. Mol. Plant-Microbe Interact. 2024, 37, 459–466. [Google Scholar] [CrossRef]
- Moeder, W.; Urquhart, W.; Ung, H.; Yoshioka, K. The role of cyclic nucleotide-gated ion channels in plant immunity. Mol. Plant 2011, 4, 442–452. [Google Scholar] [CrossRef]
- Mardi, M.; Farsad, L.K.; Gharechahi, J.; Salekdeh, G.H. In-depth transcriptome sequencing of Mexican lime trees infected with Candidatus Phytoplasma aurantifolia. PLoS ONE 2015, 10, e0130425. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Lund, S.T.; Stall, R.E.; Klee, H.J. Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 1998, 10, 371–382. [Google Scholar] [CrossRef]
- Panter, S.N.; Jones, D.A. Age-related resistance to plant pathogens. Adv. Bot. Res. 2002, 38, 251. [Google Scholar] [CrossRef]
- Vidhyasekaran, P. Salicylic acid signaling in plant innate immunity. In Plant Hormone Signaling Systems in Plant Innate Immunity; Vidhyasekaran, P., Ed.; Springer: Dordrecht, The Netherlands, 2015; pp. 27–122. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467. [Google Scholar] [CrossRef]
- Juge, N. Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 2006, 11, 359–367. [Google Scholar] [CrossRef]
- Taylor, N.G.; Laurie, S.; Turner, S.R. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 2000, 12, 2529–2539. [Google Scholar] [CrossRef]
- Hu, Y.; Zhong, X.; Liu, X.; Lou, B.; Zhou, C.; Wang, X. Comparative transcriptome analysis unveils the tolerance mechanisms of Citrus hystrix in response to ‘Candidatus Liberibacter asiaticus’ infection. PLoS ONE 2017, 12, e0189229. [Google Scholar] [CrossRef]
- Bethke, G.; Grundman, R.E.; Sreekanta, S.; Truman, W.; Katagiri, F.; Glazebrook, J. Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae. Plant Physiol. 2014, 164, 1093–1107. [Google Scholar] [CrossRef]
- Xiong, Y.; Ren, X.X.; Zhu, H.; Ren, X.Y.; Pan, Z.Y.; Su, T. Recent Progress in the Functional Research of PMEIs. China Biotechnol. 2022, 42, 99–108. [Google Scholar] [CrossRef]
- Nigro, F.; Ippolito, A.; Salerno, M.G. Mal secco disease of citrus: A journey through a century of research. J. Plant Pathol. 2011, 93, 523–560. [Google Scholar]
- Ben-Hamo, M.; Ezra, D.; Krasnov, H.; Blank, L. Spatial and temporal dynamics of Mal Secco disease spread in lemon orchards in Israel. Phytopathology 2020, 110, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.; Caruso, M.; Arlotta, C.; Lo Piero, A.R.; Nicolosi, E.; Di Silvestro, S. Identification of field tolerance and resistance to mal secco disease in a citrus germplasm collection in sicily. Agronomy 2020, 10, 1806. [Google Scholar] [CrossRef]
- Abbate, L.; Mercati, F.; Del Bosco, S.F. An overview on citrus mal secco disease: Approaches and strategies to select tolerant genotypes in C. limon. Crop Breed. Genet. Genom. 2019, 1, e190018. [Google Scholar] [CrossRef]
- Wang, L.; Huang, Y.; Liu, Z.; He, J.; Jiang, X.; He, F.; Lu, Z.; Yang, S.; Chen, P.; Yu, H.; et al. Somatic variations led to the selection of acidic and acidless orange cultivars. Nat. Plants 2021, 7, 954–965. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.L.; Zheng, Z.; Sun, X.A.; Chen, J.C.; Deng, X.L. Enhancing PCR capacity to detect ‘Candidatus Liberibacter asiaticus’ utilizing whole genome sequence information. Plant Dis. 2020, 104, 527–532. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Fang, F.; Chen, T.; Wu, J.; Zheng, Z.; Deng, X. Transcriptomic Reprogramming and Key Molecular Pathways Underlying Huanglongbing Tolerance and Susceptibility in Six Citrus Cultivars. Int. J. Mol. Sci. 2025, 26, 7359. https://doi.org/10.3390/ijms26157359
Chen X, Fang F, Chen T, Wu J, Zheng Z, Deng X. Transcriptomic Reprogramming and Key Molecular Pathways Underlying Huanglongbing Tolerance and Susceptibility in Six Citrus Cultivars. International Journal of Molecular Sciences. 2025; 26(15):7359. https://doi.org/10.3390/ijms26157359
Chicago/Turabian StyleChen, Xiaohong, Fang Fang, Tingting Chen, Jinghua Wu, Zheng Zheng, and Xiaoling Deng. 2025. "Transcriptomic Reprogramming and Key Molecular Pathways Underlying Huanglongbing Tolerance and Susceptibility in Six Citrus Cultivars" International Journal of Molecular Sciences 26, no. 15: 7359. https://doi.org/10.3390/ijms26157359
APA StyleChen, X., Fang, F., Chen, T., Wu, J., Zheng, Z., & Deng, X. (2025). Transcriptomic Reprogramming and Key Molecular Pathways Underlying Huanglongbing Tolerance and Susceptibility in Six Citrus Cultivars. International Journal of Molecular Sciences, 26(15), 7359. https://doi.org/10.3390/ijms26157359




