Recent Advances in Engineering the Unfolded Protein Response in Recombinant Chinese Hamster Ovary Cell Lines
Abstract
1. Introduction
2. An Overview of the UPR
3. An Optimized UPR Is Necessary for High-Producing CHO Cell Lines
4. The Context Dependency of UPR Engineering
5. Bioreactor Operations Elicit Different ER Stress Responses
5.1. Batch Processes
5.2. Fed-Batch and Perfusion Processes
5.3. Feeds
5.4. Temperature Downshift
6. Controlling the UPR Using Chemical Additives and Cell Line Development
6.1. Chemical Additives
6.2. Cell Line Development
7. Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Monoclonal Antibodies Market Size Set to Hit USD 679.03 Bn by 2033. Available online: https://www.visionresearchreports.com/monoclonal-antibodies-market/38040 (accessed on 17 May 2025).
- Gupta, S.K.; Srivastava, S.K.; Sharma, A.; Nalage, V.H.H.; Salvi, D.; Kushwaha, H.; Chitnis, N.B.; Shukla, P. Metabolic Engineering of CHO Cells for the Development of a Robust Protein Production Platform. PLoS ONE 2017, 12, e0181455. [Google Scholar] [CrossRef]
- Butler, M.; Spearman, M. The Choice of Mammalian Cell Host and Possibilities for Glycosylation Engineering. Curr. Opin. Biotechnol. 2014, 30, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Omasa, T. Optimization of Cell Line Development in the GS-CHO Expression System Using a High-Throughput, Single Cell-Based Clone Selection System. J. Biosci. Bioeng. 2015, 120, 323–329. [Google Scholar] [CrossRef]
- FDALabel. Available online: https://nctr-crs.fda.gov/fdalabel/ui/search (accessed on 17 May 2025).
- Oslowski, C.M.; Urano, F. Measuring ER Stress and the Unfolded Protein Response Using Mammalian Tissue Culture System. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2011; Volume 490, pp. 71–92. [Google Scholar] [CrossRef]
- Hiramatsu, N.; Joseph, V.T.; Lin, J.H. Monitoring and Manipulating Mammalian Unfolded Protein Response. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2011; Volume 491, pp. 183–198. [Google Scholar] [CrossRef]
- Sicari, D.; Delaunay-Moisan, A.; Combettes, L.; Chevet, E.; Igbaria, A. A Guide to Assessing Endoplasmic Reticulum Homeostasis and Stress in Mammalian Systems. FEBS J. 2020, 287, 27–42. [Google Scholar] [CrossRef]
- Torres, M.; Hussain, H.; Dickson, A.J. The Secretory Pathway—The Key for Unlocking the Potential of Chinese Hamster Ovary Cell Factories for Manufacturing Therapeutic Proteins. Crit. Rev. Biotechnol. 2023, 43, 628–645. [Google Scholar] [CrossRef]
- Hussain, H.; Maldonado-Agurto, R.; Dickson, A.J. The Endoplasmic Reticulum and Unfolded Protein Response in the Control of Mammalian Recombinant Protein Production. Biotechnol. Lett. 2014, 36, 1581–1593. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Raju, R.; Alves, C.; Gilbert, A. Debottlenecking Protein Secretion and Reducing Protein Aggregation in the Cellular Host. Curr. Opin. Biotechnol. 2018, 53, 151–157. [Google Scholar] [CrossRef]
- Chevallier, V.; Andersen, M.R.; Malphettes, L. Oxidative Stress-alleviating Strategies to Improve Recombinant Protein Production in CHO Cells. Biotechnol. Bioengr. 2020, 117, 1172–1186. [Google Scholar] [CrossRef]
- Gutiérrez-González, M.; Latorre, Y.; Zúñiga, R.; Aguillón, J.C.; Molina, M.C.; Altamirano, C. Transcription Factor Engineering in CHO Cells for Recombinant Protein Production. Crit. Rev. Biotechnol. 2019, 39, 665–679. [Google Scholar] [CrossRef]
- Hansen, H.G.; Pristovšek, N.; Kildegaard, H.F.; Lee, G.M. Improving the Secretory Capacity of Chinese Hamster Ovary Cells by Ectopic Expression of Effector Genes: Lessons Learned and Future Directions. Biotechnol. Adv. 2017, 35, 64–76. [Google Scholar] [CrossRef]
- Templeton, N.; Young, J.D. Biochemical and Metabolic Engineering Approaches to Enhance Production of Therapeutic Proteins in Animal Cell Cultures. Biochem. Eng. J. 2018, 136, 40–50. [Google Scholar] [CrossRef]
- Henry, M.N.; MacDonald, M.A.; Orellana, C.A.; Gray, P.P.; Gillard, M.; Baker, K.; Nielsen, L.K.; Marcellin, E.; Mahler, S.; Martínez, V.S. Attenuating Apoptosis in Chinese Hamster Ovary Cells for Improved Biopharmaceutical Production. Biotechnol. Bioengr. 2020, 117, 1187–1203. [Google Scholar] [CrossRef]
- Desmurget, C.; Perilleux, A.; Souquet, J.; Borth, N.; Douet, J. Molecular Biomarkers Identification and Applications in CHO Bioprocessing. J. Biotechnol. 2024, 392, 11–24. [Google Scholar] [CrossRef]
- Sommeregger, W.; Mayrhofer, P.; Steinfellner, W.; Reinhart, D.; Henry, M.; Clynes, M.; Meleady, P.; Kunert, R. Proteomic Differences in Recombinant CHO Cells Producing Two Similar Antibody Fragments. Biotechnol. Bioeng. 2016, 113, 1902–1912. [Google Scholar] [CrossRef] [PubMed]
- Johari, Y.B.; Estes, S.D.; Alves, C.S.; Sinacore, M.S.; James, D.C. Integrated Cell and Process Engineering for Improved Transient Production of a “difficult-to-express” Fusion Protein by CHO Cells. Biotechnol. Bioeng. 2015, 112, 2527–2542. [Google Scholar] [CrossRef] [PubMed]
- Le Fourn, V.; Girod, P.-A.; Buceta, M.; Regamey, A.; Mermod, N. CHO Cell Engineering to Prevent Polypeptide Aggregation and Improve Therapeutic Protein Secretion. Metab. Eng. 2014, 21, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Aviram, N.; Schuldiner, M. Targeting and Translocation of Proteins to the Endoplasmic Reticulum at a Glance. J. Cell Sci. 2017, 130, 4079–4085. [Google Scholar] [CrossRef]
- Vincenz-Donnelly, L.; Hipp, M.S. The Endoplasmic Reticulum: A Hub of Protein Quality Control in Health and Disease. Free. Radic. Biol. Med. 2017, 108, 383–393. [Google Scholar] [CrossRef]
- Hwang, J.; Qi, L. Quality Control in the Endoplasmic Reticulum: Crosstalk between ERAD and UPR Pathways. Trends Biochem. Sci. 2018, 43, 593–605. [Google Scholar] [CrossRef]
- Hetz, C.; Papa, F.R. The Unfolded Protein Response and Cell Fate Control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef]
- Prashad, K.; Mehra, S. Dynamics of Unfolded Protein Response in Recombinant CHO Cells. Cytotechnology 2015, 67, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Jäger, R.; Bertrand, M.J.M.; Gorman, A.M.; Vandenabeele, P.; Samali, A. The Unfolded Protein Response at the Crossroads of Cellular Life and Death during Endoplasmic Reticulum Stress. Biol. Cell 2012, 104, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Dorner, A.J.; Wasley, L.C.; Kaufman, R.J. Overexpression of GRP78 Mitigates Stress Induction of Glucose Regulated Proteins and Blocks Secretion of Selective Proteins in Chinese Hamster Ovary Cells. EMBO J. 1992, 11, 1563–1571. [Google Scholar] [CrossRef] [PubMed]
- Pieper, L.A.; Strotbek, M.; Wenger, T.; Olayioye, M.A.; Hausser, A. ATF6β-based Fine-tuning of the Unfolded Protein Response Enhances Therapeutic Antibody Productivity of Chinese Hamster Ovary Cells. Biotechnol. Bioeng. 2017, 114, 1310–1318. [Google Scholar] [CrossRef]
- Thuerauf, D.J.; Marcinko, M.; Belmont, P.J.; Glembotski, C.C. Effects of the Isoform-Specific Characteristics of ATF6α and ATF6β on Endoplasmic Reticulum Stress Response Gene Expression and Cell Viability. J. Biol. Chem. 2007, 282, 22865–22878. [Google Scholar] [CrossRef]
- Kokame, K.; Kato, H.; Miyata, T. Identification of ERSE-II, a New Cis-Acting Element Responsible for the ATF6-Dependent Mammalian Unfolded Protein Response. J. Biol. Chem. 2001, 276, 9199–9205. [Google Scholar] [CrossRef]
- Yoshida, H.; Haze, K.; Yanagi, H.; Yura, T.; Mori, K. Identification of the Cis-Acting Endoplasmic Reticulum Stress Response Element Responsible for Transcriptional Induction of Mammalian Glucose-Regulated Proteins. J. Biol. Chem. 1998, 273, 33741–33749. [Google Scholar] [CrossRef]
- Yoshida, H.; Okada, T.; Haze, K.; Yanagi, H.; Yura, T.; Negishi, M.; Mori, K. ATF6 Activated by Proteolysis Binds in the Presence of NF-Y (CBF) Directly to the Cis -Acting Element Responsible for the Mammalian Unfolded Protein Response. Mol. Cell. Biol. 2000, 20, 6755–6767. [Google Scholar] [CrossRef]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA Is Induced by ATF6 and Spliced by IRE1 in Response to ER Stress to Produce a Highly Active Transcription Factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Hillary, R.F.; FitzGerald, U. A Lifetime of Stress: ATF6 in Development and Homeostasis. J. Biomed. Sci. 2018, 25, 48. [Google Scholar] [CrossRef]
- Adachi, Y.; Yamamoto, K.; Okada, T.; Yoshida, H.; Harada, A.; Mori, K. ATF6 Is a Transcription Factor Specializing in the Regulation of Quality Control Proteins in the Endoplasmic Reticulum. Cell Struct. Funct. 2008, 33, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K. Differential Contributions of ATF6 and XBP1 to the Activation of Endoplasmic Reticulum Stress-Responsive Cis-Acting Elements ERSE, UPRE and ERSE-II. J. Biochem. 2004, 136, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Sato, T.; Matsui, T.; Sato, M.; Okada, T.; Yoshida, H.; Harada, A.; Mori, K. Transcriptional Induction of Mammalian ER Quality Control Proteins Is Mediated by Single or Combined Action of ATF6α and XBP1. Dev. Cell 2007, 13, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Brandizzi, F. IRE1: ER Stress Sensor and Cell Fate Executor. Trends Cell Biol. 2013, 23, 547–555. [Google Scholar] [CrossRef]
- Lee, A.-H.; Iwakoshi, N.N.; Glimcher, L.H. XBP-1 Regulates a Subset of Endoplasmic Reticulum Resident Chaperone Genes in the Unfolded Protein Response. Mol. Cell. Biol. 2003, 23, 7448–7459. [Google Scholar] [CrossRef]
- Shoulders, M.D.; Ryno, L.M.; Genereux, J.C.; Moresco, J.J.; Tu, P.G.; Wu, C.; Yates, J.R.; Su, A.I.; Kelly, J.W.; Wiseman, R.L. Stress-Independent Activation of XBP1s and/or ATF6 Reveals Three Functionally Diverse ER Proteostasis Environments. Cell Rep. 2013, 3, 1279–1292. [Google Scholar] [CrossRef]
- Bommiasamy, H.; Back, S.H.; Fagone, P.; Lee, K.; Meshinchi, S.; Vink, E.; Sriburi, R.; Frank, M.; Jackowski, S.; Kaufman, R.J.; et al. ATF6α Induces XBP1-Independent Expansion of the Endoplasmic Reticulum. J. Cell Sci. 2009, 122, 1626–1636. [Google Scholar] [CrossRef]
- Budge, J.D.; Knight, T.J.; Povey, J.; Roobol, J.; Brown, I.R.; Singh, G.; Dean, A.; Turner, S.; Jaques, C.M.; Young, R.J.; et al. Engineering of Chinese Hamster Ovary Cell Lipid Metabolism Results in an Expanded ER and Enhanced Recombinant Biotherapeutic Protein Production. Metab. Eng. 2020, 57, 203–216. [Google Scholar] [CrossRef]
- Görlach, A.; Klappa, P.; Kietzmann, D.T. The Endoplasmic Reticulum: Folding, Calcium Homeostasis, Signaling, and Redox Control. Antioxid. Redox Signal. 2006, 8, 1391–1418. [Google Scholar] [CrossRef]
- Romine, I.C.; Wiseman, R.L. PERK Signaling Regulates Extracellular Proteostasis of an Amyloidogenic Protein During Endoplasmic Reticulum Stress. Sci. Rep. 2019, 9, 410. [Google Scholar] [CrossRef]
- Ait Ghezala, H.; Jolles, B.; Salhi, S.; Castrillo, K.; Carpentier, W.; Cagnard, N.; Bruhat, A.; Fafournoux, P.; Jean-Jean, O. Translation Termination Efficiency Modulates ATF4 Response by Regulating ATF4 mRNA Translation at 5′ Short ORFs. Nucleic Acids Res. 2012, 40, 9557–9570. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An Integrated Stress Response Regulates Amino Acid Metabolism and Resistance to Oxidative Stress. Mol. Cell 2003, 11, 619–633. [Google Scholar] [CrossRef]
- Han, J.; Kaufman, R.J. The Role of ER Stress in Lipid Metabolism and Lipotoxicity. J. Lipid Res. 2016, 57, 1329–1338. [Google Scholar] [CrossRef]
- Coelho, D.S.; Domingos, P.M. Physiological Roles of Regulated Ire1 Dependent Decay. Front. Genet. 2014, 5, 76. [Google Scholar] [CrossRef] [PubMed]
- Harreither, E.; Hackl, M.; Pichler, J.; Shridhar, S.; Auer, N.; Łabaj, P.P.; Scheideler, M.; Karbiener, M.; Grillari, J.; Kreil, D.P.; et al. Microarray Profiling of Preselected CHO Host Cell Subclones Identifies Gene Expression Patterns Associated with In-creased Production Capacity. Biotechnol. J. 2015, 10, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Lund, A.M.; Kaas, C.S.; Brandl, J.; Pedersen, L.E.; Kildegaard, H.F.; Kristensen, C.; Andersen, M.R. Network Reconstruction of the Mouse Secretory Pathway Applied on CHO Cell Transcriptome Data. BMC Syst. Biol. 2017, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Chromikova, V.; Mader, A.; Steinfellner, W.; Kunert, R. Evaluating the Bottlenecks of Recombinant IgM Production in Mammalian Cells. Cytotechnology 2015, 67, 343–356. [Google Scholar] [CrossRef]
- Sebastião, M.J.; Hoffman, M.; Escandell, J.; Tousi, F.; Zhang, J.; Figueroa, B.; DeMaria, C.; Gomes-Alves, P. Identification of Mispairing Omic Signatures in Chinese Hamster Ovary (CHO) Cells Producing a Tri-Specific Antibody. Biomedicines 2023, 11, 2890. [Google Scholar] [CrossRef]
- Talbot, N.E.; Mead, E.J.; Davies, S.A.; Uddin, S.; Smales, C.M. Application of ER Stress Biomarkers to Predict Formulated Monoclonal Antibody Stability. Biotechnol. J. 2019, 14, 1900024. [Google Scholar] [CrossRef]
- Mathias, S.; Wippermann, A.; Raab, N.; Zeh, N.; Handrick, R.; Gorr, I.; Schulz, P.; Fischer, S.; Gamer, M.; Otte, K. Unraveling What Makes a Monoclonal Antibody Difficult-to-express: From Intracellular Accumulation to Incomplete Folding and Degradation via ERAD. Biotechnol. Bioeng. 2020, 117, 5–16. [Google Scholar] [CrossRef]
- Tang, D.; Sandoval, W.; Lam, C.; Haley, B.; Liu, P.; Xue, D.; Roy, D.; Patapoff, T.; Louie, S.; Snedecor, B.; et al. UBR E3 Ligases and the PDIA3 Protease Control Degradation of Unfolded Antibody Heavy Chain by ERAD. J. Cell Biol. 2020, 219, e201908087. [Google Scholar] [CrossRef]
- Sulaj, E.; Schwaigerlehner, L.; Sandell, F.L.; Dohm, J.C.; Marzban, G.; Kunert, R. Quantitative Proteomics Reveals Cellular Responses to Individual mAb Expression and Tunicamycin in CHO Cells. Appl. Microbiol. Biotechnol. 2024, 108, 381. [Google Scholar] [CrossRef]
- Albrecht, S.; Kaisermayer, C.; Reinhart, D.; Ambrose, M.; Kunert, R.; Lindeberg, A.; Bones, J. Multiple Reaction Monitoring Targeted LC-MS Analysis of Potential Cell Death Marker Proteins for Increased Bioprocess Control. Anal. Bioanal. Chem. 2018, 410, 3197–3207. [Google Scholar] [CrossRef]
- Park, S.-Y.; Egan, S.; Cura, A.J.; Aron, K.L.; Xu, X.; Zheng, M.; Borys, M.; Ghose, S.; Li, Z.; Lee, K. Untargeted Proteomics Reveals Upregulation of Stress Response Pathways during CHO-Based Monoclonal Antibody Manufacturing Process Leading to Disulfide Bond Reduction. mAbs 2021, 13, 1963094. [Google Scholar] [CrossRef] [PubMed]
- Kretz, R.; Walter, L.; Raab, N.; Zeh, N.; Gauges, R.; Otte, K.; Fischer, S.; Stoll, D. Spatial Proteomics Reveals Differences in the Cellular Architecture of Antibody-Producing CHO and Plasma Cell–Derived Cells. Mol. Cell. Proteom. 2022, 21, 100278. [Google Scholar] [CrossRef]
- Henry, M.; Gallagher, C.; Kelly, R.M.; Frye, C.C.; Osborne, M.D.; Brady, C.P.; Barron, N.; Clynes, M.; Meleady, P. Clonal Variation in Productivity and Proteolytic Clipping of an Fc-Fusion Protein in CHO Cells: Proteomic Analysis Suggests a Role for Defective Protein Folding and the UPR. J. Biotechnol. 2018, 281, 21–30. [Google Scholar] [CrossRef]
- Carlage, T.; Kshirsagar, R.; Zang, L.; Janakiraman, V.; Hincapie, M.; Lyubarskaya, Y.; Weiskopf, A.; Hancock, W.S. Analysis of Dynamic Changes in the Proteome of a Bcl-XL Overexpressing Chinese Hamster Ovary Cell Culture during Exponential and Stationary Phases. Biotechnol. Prog. 2012, 28, 814–823. [Google Scholar] [CrossRef]
- Tung, M.; Tang, D.; Wang, S.; Zhan, D.; Kiplinger, K.; Pan, S.; Jing, Y.; Shen, A.; Ahyow, P.; Snedecor, B.; et al. High Intracellular Seed Train BiP Levels Correlate With Poor Production Culture Performance in CHO Cells. Biotechnol. J. 2018, 13, 1700746. [Google Scholar] [CrossRef]
- Hausmann, R.; Chudobová, I.; Spiegel, H.; Schillberg, S. Proteomic Analysis of CHO Cell Lines Producing High and Low Quantities of a Recombinant Antibody before and after Selection with Methotrexate. J. Biotechnol. 2018, 265, 65–69. [Google Scholar] [CrossRef]
- Huhn, S.; Chang, M.; Kumar, A.; Liu, R.; Jiang, B.; Betenbaugh, M.; Lin, H.; Nyberg, G.; Du, Z. Chromosomal Instability Drives Convergent and Divergent Evolution toward Advantageous Inherited Traits in Mammalian CHO Bioproduction Lineages. iScience 2022, 25, 104074. [Google Scholar] [CrossRef]
- Pérez-Rodriguez, S.; Wulff, T.; Voldborg, B.G.; Altamirano, C.; Trujillo-Roldán, M.A.; Valdez-Cruz, N.A. Compartmentalized Proteomic Profiling Outlines the Crucial Role of the Classical Secretory Pathway during Recombinant Protein Production in Chinese Hamster Ovary Cells. ACS Omega 2021, 6, 12439–12458. [Google Scholar] [CrossRef]
- Chakrabarti, L.; Chaerkady, R.; Wang, J.; Weng, S.H.S.; Wang, C.; Qian, C.; Cazares, L.; Hess, S.; Amaya, P.; Zhu, J.; et al. Mitochondrial Membrane Potential-Enriched CHO Host: A Novel and Powerful Tool for Improving Biomanufacturing Capability. mAbs 2022, 14, 2020081. [Google Scholar] [CrossRef]
- Avello, V.; Torres, M.; Vergara, M.; Berrios, J.; Valdez-Cruz, N.A.; Acevedo, C.; Molina Sampayo, M.; Dickson, A.J.; Altamirano, C. Enhanced Recombinant Protein Production in CHO Cell Continuous Cultures under Growth-Inhibiting Conditions Is Associated with an Arrested Cell Cycle in G1/G0 Phase. PLoS ONE 2022, 17, e0277620. [Google Scholar] [CrossRef] [PubMed]
- Kaas, C.S. Characterization of Chinese Hamster Ovary Cells Producing Coagulation Factor VIII Using Multi-Omics Tools. Ph.D. Thesis, Technical University of Denmark, Lyngby, Denmark, 2015. [Google Scholar]
- Orlova, N.A.; Kovnir, S.V.; Gabibov, A.G.; Vorobiev, I.I. Stable High-Level Expression of Factor VIII in Chinese Hamster Ovary Cells in Improved Elongation Factor-1 Alpha-Based System. BMC Biotechnol. 2017, 17, 33. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Inoue, K.; Okubo, J.; Ueda, Y.; Kawaguchi, K.; Sakurai, H.; Wada, I.; Morita, M.; Imanaka, T. Endoplasmic Reticulum Stress Response and Mutant Protein Degradation in CHO Cells Accumulating Antithrombin (C95R) in Russell Bodies. Biol. Pharm. Bull. 2015, 38, 1980–1984. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Kawaguchi, K.; Ueda, Y.; Arai, S.; Morita, M.; Imanaka, T.; Wada, I. Characterization of Russell Bodies Accumulating Mutant Antithrombin Derived from the Endoplasmic Reticulum. Biol. Pharm. Bull. 2015, 38, 852–861. [Google Scholar] [CrossRef]
- Maldonado-Agurto, R.; Dickson, A.J. Multiplexed Digital mRNA Expression Analysis Profiles System-Wide Changes in mRNA Abundance and Responsiveness of UPR-Specific Gene Expression Changes During Batch Culture of Recombinant Chinese Hamster Ovary Cells. Biotechnol. J. 2018, 13, e1700429. [Google Scholar] [CrossRef]
- Coats, M.T.; Bydlinski, N.; Maresch, D.; Diendorfer, A.; Klanert, G.; Borth, N. mRNA Transfection into CHO-Cells Reveals Production Bottlenecks. Biotechnol. J. 2020, 15, 1900198. [Google Scholar] [CrossRef]
- Poulain, A.; Mullick, A.; Massie, B.; Durocher, Y. Reducing Recombinant Protein Expression during CHO Pool Selection Enhances Frequency of High-Producing Cells. J. Biotechnol. 2019, 296, 32–41. [Google Scholar] [CrossRef]
- Sinharoy, P.; Aziz, A.H.; Majewska, N.I.; Ahuja, S.; Handlogten, M.W. Perfusion Reduces Bispecific Antibody Aggregation via Mitigating Mitochondrial Dysfunction-Induced Glutathione Oxidation and ER Stress in CHO Cells. Sci. Rep. 2020, 10, 16620. [Google Scholar] [CrossRef]
- McFarland, K.S.; Hegadorn, K.; Betenbaugh, M.J.; Handlogten, M.W. Elevated Endoplasmic Reticulum pH Is Associated with High Growth and bisAb Aggregation in CHO Cells. Biotechnol. Bioeng. 2025, 122, 137–148. [Google Scholar] [CrossRef]
- Ku, S.C.Y.; Ng, D.T.W.; Yap, M.G.S.; Chao, S. Effects of Overexpression of X-box Binding Protein 1 on Recombinant Protein Production in Chinese Hamster Ovary and NS0 Myeloma Cells. Biotechnol. Bioeng. 2008, 99, 155–164. [Google Scholar] [CrossRef]
- Chandrawanshi, V.; Kulkarni, R.; Prabhu, A.; Mehra, S. Enhancing Titers and Productivity of rCHO Clones with a Combination of an Optimized Fed-Batch Process and ER-Stress Adaptation. J. Biotechnol. 2020, 311, 49–58. [Google Scholar] [CrossRef]
- Romanova, N.; Schelletter, L.; Hoffrogge, R.; Noll, T. Hyperosmolality in CHO Cell Culture: Effects on the Proteome. Appl. Microbiol. Biotechnol. 2022, 106, 2569–2586. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dai, S.; Bones, J.; Ray, S.; Cha, S.; Karger, B.L.; Li, J.J.; Wilson, L.; Hinckle, G.; Rossomando, A. A Quantitative Proteomic Analysis of Cellular Responses to High Glucose Media in Chinese Hamster Ovary Cells. Biotechnol. Prog. 2015, 31, 1026–1038. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.S.; Raju, R.; Kshirsagar, R.; Ivanov, A.R.; Gilbert, A.; Zang, L.; Karger, B.L. Multi-Omics Study on the Impact of Cysteine Feed Level on Cell Viability and mAb Production in a CHO Bioprocess. Biotechnol. J. 2019, 14, e1800352. [Google Scholar] [CrossRef]
- Ali, A.S.; Chen, R.; Raju, R.; Kshirsagar, R.; Gilbert, A.; Zang, L.; Karger, B.L.; Ivanov, A.R. Multi-Omics Reveals Impact of Cysteine Feed Concentration and Resulting Redox Imbalance on Cellular Energy Metabolism and Specific Productivity in CHO Cell Bioprocessing. Biotechnol. J. 2020, 15, e1900565. [Google Scholar] [CrossRef] [PubMed]
- Komuczki, D.; Stadermann, A.; Bentele, M.; Unsoeld, A.; Grillari, J.; Mueller, M.M.; Paul, A.; Fischer, S. High Cysteine Concentrations in Cell Culture Media Lead to Oxidative Stress and Reduced Bioprocess Performance of Recombinant CHO Cells. Biotechnol. J. 2022, 17, e2200029. [Google Scholar] [CrossRef]
- Handlogten, M.W.; Lee-O’Brien, A.; Roy, G.; Levitskaya, S.V.; Venkat, R.; Singh, S.; Ahuja, S. Intracellular Response to Process Optimization and Impact on Productivity and Product Aggregates for a High-titer CHO Cell Process. Biotechnol. Bioeng. 2018, 115, 126–138. [Google Scholar] [CrossRef]
- Torres, M.; Dickson, A.J. Combined Gene and Environmental Engineering Offers a Synergetic Strategy to Enhance R-protein Production in Chinese Hamster Ovary Cells. Biotechnol. Bioeng. 2022, 119, 550–565. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Yi, D.; Zhang, C.; Ning, B.; Fu, Y.; Jia, Y.; Wang, T.; Wang, X. Synergistic Promotion of Transient Transgene Expression in CHO Cells by PDI/XBP-1s Co-Transfection and Mild Hypothermia. Bioprocess. Biosyst. Eng. 2024, 47, 557–565. [Google Scholar] [CrossRef]
- Latorre, Y.; Torres, M.; Vergara, M.; Berrios, J.; Sampayo, M.M.; Gödecke, N.; Wirth, D.; Hauser, H.; Dickson, A.J.; Altamirano, C. Engineering of Chinese Hamster Ovary Cells for Co-Overexpressing MYC and XBP1s Increased Cell Proliferation and Recombinant EPO Production. Sci. Rep. 2023, 13, 1482. [Google Scholar] [CrossRef]
- Gulis, G.; Simi, K.C.R.; De Toledo, R.R.; Maranhao, A.Q.; Brigido, M.M. Optimization of Heterologous Protein Production in Chinese Hamster Ovary Cells under Overexpression of Spliced Form of Human X-Box Binding Protein. BMC Biotechnol. 2014, 14, 26. [Google Scholar] [CrossRef]
- Hu, D.; Sun, Y.; Liu, X.; Liu, J.; Zhang, X.; Zhao, L.; Wang, H.; Tan, W.-S.; Fan, L. Understanding the Intracellular Effects of Yeast Extract on the Enhancement of Fc-Fusion Protein Production in Chinese Hamster Ovary Cell Culture. Appl. Microbiol. Biotechnol. 2015, 99, 8429–8440. [Google Scholar] [CrossRef] [PubMed]
- Bedoya-López, A.; Estrada, K.; Sanchez-Flores, A.; Ramírez, O.T.; Altamirano, C.; Segovia, L.; Miranda-Ríos, J.; Trujillo-Roldán, M.A.; Valdez-Cruz, N.A. Effect of Temperature Downshift on the Transcriptomic Responses of Chinese Hamster Ovary Cells Using Recombinant Human Tissue Plasminogen Activator Production Culture. PLoS ONE 2016, 11, e0151529. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Zúñiga, R.; Gutierrez, M.; Vergara, M.; Collazo, N.; Reyes, J.; Berrios, J.; Aguillon, J.C.; Molina, M.C.; Altamirano, C. Mild Hypothermia Upregulates Myc and Xbp1s Expression and Improves Anti-TNFα Production in CHO Cells. PLoS ONE 2018, 13, e0194510. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Akhtar, S.; McKenzie, E.A.; Dickson, A.J. Temperature Down-Shift Modifies Expression of UPR-/ERAD-Related Genes and Enhances Production of a Chimeric Fusion Protein in CHO Cells. Biotechnol. J. 2021, 16, e2000081. [Google Scholar] [CrossRef]
- Baik, J.Y.; Lee, M.S.; An, S.R.; Yoon, S.K.; Joo, E.J.; Kim, Y.H.; Park, H.W.; Lee, G.M. Initial Transcriptome and Proteome Analyses of Low Culture Temperature-induced Expression in CHO Cells Producing Erythropoietin. Biotechnol. Bioeng. 2006, 93, 361–371. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, T.; Chen, J.; Liu, C.; Tang, J.; Xie, Q. The Effect of Culture Temperature on the Aggregation of Recombinant TNFR-Fc Is Regulated by the PERK-eIF2a Pathway in CHO Cells. Protein Peptide Lett. 2018, 25, 570–579. [Google Scholar] [CrossRef]
- Tossolini, I.; López-Díaz, F.J.; Kratje, R.; Prieto, C.C. Characterization of Cellular States of CHO-K1 Suspension Cell Culture through Cell Cycle and RNA-Sequencing Profiling. J. Biotechnol. 2018, 286, 56–67. [Google Scholar] [CrossRef]
- Baek, E.; Lee, J.S.; Lee, G.M. Untangling the Mechanism of 3-methyladenine in Enhancing the Specific Productivity: Transcriptome Analysis of Recombinant Chinese Hamster Ovary Cells Treated with 3-methyladenine. Biotechnol. Bioeng. 2018, 115, 2243–2254. [Google Scholar] [CrossRef]
- Ha, T.K.; Hansen, A.H.; Kol, S.; Kildegaard, H.F.; Lee, G.M. Baicalein Reduces Oxidative Stress in CHO Cell Cultures and Improves Recombinant Antibody Productivity. Biotechnol. J. 2018, 13, e1700425. [Google Scholar] [CrossRef]
- Ha, T.K.; Hansen, A.H.; Kildegaard, H.F.; Lee, G.M. BiP Inducer X: An ER Stress Inhibitor for Enhancing Recombinant Antibody Production in CHO Cell Culture. Biotechnol. J. 2019, 14, e1900130. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Treiber, D.; McCarter, J.D.; Fomina-Yadlin, D.; Saleem, R.A.; McCoy, R.E.; Zhang, Y.; Tharmalingam, T.; Leith, M.; Follstad, B.D.; et al. Use of a Small Molecule Cell Cycle Inhibitor to Control Cell Growth and Improve Specific Productivity and Product Quality of Recombinant Proteins in CHO Cell Cultures. Biotechnol. Bioeng. 2015, 112, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Huhn, S.; Nelson, L.; Betenbaugh, M.; Du, Z. Significant Impact of mTORC1 and ATF4 Pathways in CHO Cell Recombinant Protein Production Induced by CDK4/6 Inhibitor. Biotechnol. Bioeng. 2022, 119, 1189–1206. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Schneider, J.W.; Robinson, A.S. Rosmarinic Acid Enhances CHO Cell Productivity and Proliferation through Activation of the Unfolded Protein Response and the mTOR Pathway. Biotechnol. J. 2024, 19, e2300397. [Google Scholar] [CrossRef]
- Selvaprakash, K.; Sideri, C.; Henry, M.; Efeoglu, E.; Ryan, D.; Meleady, P. Characterization of the Ubiquitin-Modified Proteome of Recombinant Chinese Hamster Ovary Cells in Response to Endoplasmic Reticulum Stress. Biotechnol. J. 2024, 19, e202400413. [Google Scholar] [CrossRef]
- Mortazavi, M.; Shokrgozar, M.A.; Sardari, S.; Azadmanesh, K.; Mahdian, R.; Kaghazian, H.; Hosseini, S.N.; Hedayati, M.H. Using Chemical Chaperones to Increase Recombinant Human Erythropoietin Secretion in CHO Cell Line. Prep. Biochem. Biotechnol. 2019, 49, 535–544. [Google Scholar] [CrossRef]
- Lim, J.; Lim, J.-H.; Lee, J.-H.; Cheon, S.-H.; Lee, G.; Kim, Z.-H.; Kim, D.-I. Effect of 4-Phenylbutyrate Addition Timing on Titer of Fc-Fusion Protein in Chinese Hamster Ovary Cell Cultures. Biotechnol. Bioprocess Eng. 2024, 29, 712–720. [Google Scholar] [CrossRef]
- Kumar, S.; Dhara, V.G.; Orzolek, L.D.; Hao, H.; More, A.J.; Lau, E.C.; Betenbaugh, M.J. Elucidating the Impact of Cottonseed Hydrolysates on CHO Cell Culture Performance through Transcriptomic Analysis. Appl. Microbiol. Biotechnol. 2021, 105, 271–285. [Google Scholar] [CrossRef]
- Segar, K.P.; Chandrawanshi, V.; Mehra, S. Activation of Unfolded Protein Response Pathway Is Important for Valproic Acid Mediated Increase in Immunoglobulin G Productivity in Recombinant Chinese Hamster Ovary Cells. J. Biosci. Bioeng. 2017, 124, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Mahameed, M.; Tirosh, B. Engineering CHO Cells with an Oncogenic KIT Improves Cells Growth, Resilience to Stress, and Productivity. Biotechnol. Bioeng. 2017, 114, 2560–2570. [Google Scholar] [CrossRef]
- Mohan, C.; Lee, G.M. Calnexin Overexpression Sensitizes Recombinant CHO Cells to Apoptosis Induced by Sodium Butyrate Treatment. Cell Stress Chaperones 2009, 14, 49–60. [Google Scholar] [CrossRef]
- Ryan, D.; Sideri, C.-K.; Henry, M.; Efeoglu, E.; Meleady, P. Label-Free Quantitative Proteomics Analysis of Producer and Non-Producer Chinese Hamsters Ovary (CHO) Cells under ER Stress Conditions. Curr. Res. Biotechnol. 2023, 6, 100141. [Google Scholar] [CrossRef]
- Becker, E.; Florin, L.; Pfizenmaier, K.; Kaufmann, H. An XBP-1 Dependent Bottle-Neck in Production of IgG Subtype Antibodies in Chemically Defined Serum-Free Chinese Hamster Ovary (CHO) Fed-Batch Processes. J. Biotechnol. 2008, 135, 217–223. [Google Scholar] [CrossRef]
- Cain, K.; Peters, S.; Hailu, H.; Sweeney, B.; Stephens, P.; Heads, J.; Sarkar, K.; Ventom, A.; Page, C.; Dickson, A. A CHO Cell Line Engineered to Express XBP1 and ERO1-Lα Has Increased Levels of Transient Protein Expression. Biotechnol. Prog. 2013, 29, 697–706. [Google Scholar] [CrossRef]
- Ohya, T.; Hayashi, T.; Kiyama, E.; Nishii, H.; Miki, H.; Kobayashi, K.; Honda, K.; Omasa, T.; Ohtake, H. Improved Production of Recombinant Human Antithrombin III in Chinese Hamster Ovary Cells by ATF4 Overexpression. Biotechnol. Bioeng. 2008, 100, 317–324. [Google Scholar] [CrossRef]
- Campos-da-Paz, M.; Costa, C.S.; Quilici, L.S.; Simões, I.D.C.; Kyaw, C.M.; Maranhão, A.Q.; Brigido, M.M. Production of Recombinant Human Factor VIII in Different Cell Lines and the Effect of Human XBP1 Co-Expression. Mol. Biotechnol. 2008, 39, 155–158. [Google Scholar] [CrossRef] [PubMed]
- Rahimpour, A. Engineering the Cellular Protein Secretory Pathway for Enhancement of Recombinant Tissue Plasminogen Activator Expression in Chinese Hamster Ovary Cells: Effects of CERT and XBP1s Genes. J. Microbiol. Biotechnol. 2013, 23, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Rahimpour, A.; Ahani, R.; Najaei, A.; Adeli, A.; Barkhordari, F.; Mahboudi, F. Development of Genetically Modified Chinese Hamster Ovary Host Cells for the Enhancement of Recombinant Tissue Plasminogen Activator Expression. Malays. J. Med. Sci. 2016, 23, 6–13. [Google Scholar]
- Raab, N.; Zeh, N.; Kretz, R.; Weiß, L.; Stadermann, A.; Lindner, B.; Fischer, S.; Stoll, D.; Otte, K. Nature as Blueprint: Global Phenotype Engineering of CHO Production Cells Based on a Multi-Omics Comparison with Plasma Cells. Metab. Eng. 2024, 83, 110–122. [Google Scholar] [CrossRef]
- Torres, M.; Dickson, A.J. Overexpression of Transcription Factor BLIMP1/Prdm1 Leads to Growth Inhibition and Enhanced Secretory Capacity in Chinese Hamster Ovary Cells. Metab. Eng. 2021, 67, 237–249. [Google Scholar] [CrossRef]
- Torres, M.; Dickson, A.J. Reprogramming of Chinese Hamster Ovary Cells towards Enhanced Protein Secretion. Metab. Eng. 2022, 69, 249–261. [Google Scholar] [CrossRef]
- Kim, S.H.; Baek, M.; Park, S.; Shin, S.; Lee, J.S.; Lee, G.M. Improving the Secretory Capacity of CHO Producer Cells: The Effect of Controlled Blimp1 Expression, a Master Transcription Factor for Plasma Cells. Metab. Eng. 2022, 69, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, A.L.; Shapiro-Shelef, M.; Iwakoshi, N.N.; Lee, A.-H.; Qian, S.-B.; Zhao, H.; Yu, X.; Yang, L.; Tan, B.K.; Rosenwald, A.; et al. XBP1, Downstream of Blimp-1, Expands the Secretory Apparatus and Other Organelles, and Increases Protein Synthesis in Plasma Cell Differentiation. Immunity 2004, 21, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Castellano, B.M.; Tang, D.; Marsters, S.; Lam, C.; Liu, P.; Rose, C.M.; Sandoval, W.; Ashkenazi, A.; Snedecor, B.; Misaghi, S. Activation of the PERK Branch of the Unfolded Protein Response during Production Reduces Specific Productivity in CHO Cells via Downregulation of PDGFRa and IRE1a Signaling. Biotechnol. Prog. 2023, 39, e3354. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.C.Y.; Toh, P.C.; Lee, Y.Y.; Chusainow, J.; Yap, M.G.S.; Chao, S. Regulation of XBP-1 Signaling during Transient and Stable Recombinant Protein Production in CHO Cells. Biotechnol. Prog. 2010, 26, 517–526. [Google Scholar] [CrossRef]
- Becker, E.; Florin, L.; Pfizenmaier, K.; Kaufmann, H. Evaluation of a Combinatorial Cell Engineering Approach to Overcome Apoptotic Effects in XBP-1(s) Expressing Cells. J. Biotechnol. 2010, 146, 198–206. [Google Scholar] [CrossRef]
- Berger, A.; Le Fourn, V.; Masternak, J.; Regamey, A.; Bodenmann, I.; Girod, P.; Mermod, N. Overexpression of Transcription Factor Foxa1 and Target Genes Remediate Therapeutic Protein Production Bottlenecks in Chinese Hamster Ovary Cells. Biotechnol. Bioeng. 2020, 117, 1101–1116. [Google Scholar] [CrossRef]
- Borth, N.; Mattanovich, D.; Kunert, R.; Katinger, H. Effect of Increased Expression of Protein Disulfide Isomerase and Heavy Chain Binding Protein on Antibody Secretion in a Recombinant CHO Cell Line. Biotechnol. Prog. 2008, 21, 106–111. [Google Scholar] [CrossRef]
- Mohan, C.; Park, S.H.; Chung, J.Y.; Lee, G.M. Effect of Doxycycline-regulated Protein Disulfide Isomerase Expression on the Specific Productivity of Recombinant CHO Cells: Thrombopoietin and Antibody. Biotechnol. Bioeng. 2007, 98, 611–615. [Google Scholar] [CrossRef]
- Kirimoto, Y.; Yamano-Adachi, N.; Koga, Y.; Omasa, T. Effect of Co-Overexpression of the Cargo Receptor ERGIC-53/MCFD2 on Antibody Production and Intracellular IgG Secretion in Recombinant Chinese Hamster Ovary Cells. J. Biosci. Bioeng. 2023, 136, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Nishimiya, D.; Mano, T.; Miyadai, K.; Yoshida, H.; Takahashi, T. Overexpression of CHOP Alone and in Combination with Chaperones Is Effective in Improving Antibody Production in Mammalian Cells. Appl. Microbiol. Biotechnol. 2013, 97, 2531–2539. [Google Scholar] [CrossRef]
- Zhang, C.; Fu, Y.; Zheng, W.; Chang, F.; Shen, Y.; Niu, J.; Wang, Y.; Ma, X. Enhancing the Antibody Production Efficiency of Chinese Hamster Ovary Cells through Improvement of Disulfide Bond Folding Ability and Apoptosis Resistance. Cells 2024, 13, 1481. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, J.F.; Arnall, C.L.; Patel, Y.D.; Barber, N.O.W.; Lovelady, C.S.; Rosignoli, G.; Harris, C.L.; Dunn, S.; Field, R.P.; Dean, G.; et al. A Platform for Context-Specific Genetic Engineering of Recombinant Protein Production by CHO Cells. J. Biotechnol. 2020, 312, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Rives, D.; Peak, C.; Blenner, M.A. RNASeq Highlights ATF6 Pathway Regulators for CHO Cell Engineering with Different Impacts of ATF6β and WFS1 Knockdown on Fed-Batch Production of IgG1. Sci. Rep. 2024, 14, 14141. [Google Scholar] [CrossRef]
- Davis, R.; Schooley, K.; Rasmussen, B.; Thomas, J.; Reddy, P. Effect of PDI Overexpression on Recombinant Protein Secretion in CHO Cells. Biotechnol. Prog. 2000, 16, 736–743. [Google Scholar] [CrossRef]
- Roobol, A.; Roobol, J.; Smith, M.E.; Carden, M.J.; Hershey, J.W.B.; Willis, A.E.; Smales, C.M. Engineered Transient and Stable Overexpression of Translation Factors eIF3i and eIF3c in CHOK1 and HEK293 Cells Gives Enhanced Cell Growth Associated with Increased C-Myc Expression and Increased Recombinant Protein Synthesis. Metab. Eng. 2020, 59, 98–105. [Google Scholar] [CrossRef]
- Haredy, A.M.; Nishizawa, A.; Honda, K.; Ohya, T.; Ohtake, H.; Omasa, T. Improved Antibody Production in Chinese Hamster Ovary Cells by ATF4 Overexpression. Cytotechnology 2013, 65, 993–1002. [Google Scholar] [CrossRef]
- Jiang, Q.; Sun, Y.; Guo, Z.; Fu, M.; Wang, Q.; Zhu, H.; Lei, P.; Shen, G. Overexpression of GRP78 Enhances Survival of CHO Cells in Response to Serum Deprivation and Oxidative Stress. Eng. Life Sci. 2017, 17, 107–116. [Google Scholar] [CrossRef]
- Komatsu, K.; Kumon, K.; Arita, M.; Onitsuka, M.; Omasa, T.; Yohda, M. Effect of the Disulfide Isomerase PDIa4 on the Antibody Production of Chinese Hamster Ovary Cells. J. Biosci. Bioeng. 2020, 130, 637–643. [Google Scholar] [CrossRef]
- Omasa, T.; Takami, T.; Ohya, T.; Kiyama, E.; Hayashi, T.; Nishii, H.; Miki, H.; Kobayashi, K.; Honda, K.; Ohtake, H. Overexpression of GADD34 Enhances Production of Recombinant Human Antithrombin III in Chinese Hamster Ovary Cells. J. Biosci. Bioeng. 2008, 106, 568–573. [Google Scholar] [CrossRef]
- Majors, B.S.; Betenbaugh, M.J.; Pederson, N.E.; Chiang, G.G. Enhancement of Transient Gene Expression and Culture Viability Using Chinese Hamster Ovary Cells Overexpressing Bcl-xL. Biotechnol. Bioeng. 2008, 101, 567–578. [Google Scholar] [CrossRef]
- Onitsuka, M.; Kinoshita, Y.; Nishizawa, A.; Tsutsui, T.; Omasa, T. Enhanced IgG1 Production by Overexpression of Nuclear Factor Kappa B Inhibitor Zeta (NFKBIZ) in Chinese Hamster Ovary Cells. Cytotechnology 2018, 70, 675–685. [Google Scholar] [CrossRef]
- Samy, A.; Kaneyoshi, K.; Omasa, T. Improvement of Intracellular Traffic System by Overexpression of KDEL Receptor 1 in Antibody-Producing CHO Cells. Biotechnol. J. 2020, 15, e1900352. [Google Scholar] [CrossRef] [PubMed]
- Budge, J.D.; Knight, T.J.; Povey, J.; Roobol, J.; Brown, I.R.; Singh, G.; Dean, A.; Turner, S.; Jaques, C.M.; Young, R.J.; et al. Data for Engineering Lipid Metabolism of Chinese Hamster Ovary (CHO) Cells for Enhanced Recombinant Protein Production. Data Brief. 2020, 29, 105217. [Google Scholar] [CrossRef]
- Florin, L.; Pegel, A.; Becker, E.; Hausser, A.; Olayioye, M.A.; Kaufmann, H. Heterologous Expression of the Lipid Transfer Protein CERT Increases Therapeutic Protein Productivity of Mammalian Cells. J. Biotechnol. 2009, 141, 84–90. [Google Scholar] [CrossRef]
- Tigges, M.; Fussenegger, M. Xbp1-Based Engineering of Secretory Capacity Enhances the Productivity of Chinese Hamster Ovary Cells. Metab. Eng. 2006, 8, 264–272. [Google Scholar] [CrossRef]
- Barzadd, M.M.; Lundqvist, M.; Harris, C.; Malm, M.; Volk, A.-L.; Thalén, N.; Chotteau, V.; Grassi, L.; Smith, A.; Abadi, M.L.; et al. Autophagy and Intracellular Product Degradation Genes Identified by Systems Biology Analysis Reduce Aggregation of Bispecific Antibody in CHO Cells. New Biotechnol. 2022, 68, 68–76. [Google Scholar] [CrossRef]
- Hwang, S.O.; Chung, J.Y.; Lee, G.M. Effect of Doxycycline-Regulated ERp57 Expression on Specific Thrombopoietin Productivity of Recombinant CHO Cells. Biotechnol. Prog. 2003, 19, 179–184. [Google Scholar] [CrossRef]
- Pieper, L.A.; Strotbek, M.; Wenger, T.; Gamer, M.; Olayioye, M.A.; Hausser, A. Secretory Pathway Optimization of CHO Producer Cells by Co-Engineering of the mitosRNA-1978 Target Genes CerS2 and Tbc1D20. Metab. Eng. 2017, 40, 69–79. [Google Scholar] [CrossRef]
- Pybus, L.P.; Dean, G.; West, N.R.; Smith, A.; Daramola, O.; Field, R.; Wilkinson, S.J.; James, D.C. Model-Directed Engineering of “Difficult-to-Express” Monoclonal Antibody Production by Chinese Hamster Ovary Cells. Biotechnol. Bioengr. 2014, 111, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Malm, M.; Kuo, C.-C.; Barzadd, M.M.; Mebrahtu, A.; Wistbacka, N.; Razavi, R.; Volk, A.-L.; Lundqvist, M.; Kotol, D.; Tegel, H.; et al. Harnessing Secretory Pathway Differences between HEK293 and CHO to Rescue Production of Difficult to Express Proteins. Metab. Eng. 2022, 72, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.; Sathyamurthy, M.; Lee, G.M. A Role of GADD153 in ER Stress-Induced Apoptosis in Recombinant Chinese Hamster Ovary Cells. Biotechnol. Bioprocess Eng. 2012, 17, 446–455. [Google Scholar] [CrossRef]
- Brown, A.J.; Gibson, S.J.; Hatton, D.; Arnall, C.L.; James, D.C. Whole Synthetic Pathway Engineering of Recombinant Protein Production. Biotechnol. Bioengr. 2019, 116, 375–387. [Google Scholar] [CrossRef]
- Bryan, L.; Henry, M.; Barron, N.; Gallagher, C.; Kelly, R.M.; Frye, C.C.; Osborne, M.D.; Clynes, M.; Meleady, P. Differential Expression of miRNAs and Functional Role of Mir-200a in High and Low Productivity CHO Cells Expressing an Fc Fusion Protein. Biotechnol. Lett. 2021, 43, 1551–1563. [Google Scholar] [CrossRef]
- Vito, D.; Eriksen, J.C.; Skjødt, C.; Weilguny, D.; Rasmussen, S.K.; Smales, C.M. Defining lncRNAs Correlated with CHO Cell Growth and IgG Productivity by RNA-Seq. iScience 2020, 23, 100785. [Google Scholar] [CrossRef]
- Costello, A.; Coleman, O.; Lao, N.T.; Henry, M.; Meleady, P.; Barron, N.; Clynes, M. Depletion of Endogenous miRNA-378-3p Increases Peak Cell Density of CHO DP12 Cells and Is Correlated with Elevated Levels of Ubiquitin Carboxyl-Terminal Hydrolase 14. J. Biotechnol. 2018, 288, 30–40. [Google Scholar] [CrossRef]
- Kober, L.; Zehe, C.; Bode, J. Development of a Novel ER Stress Based Selection System for the Isolation of Highly Productive Clones. Biotechnol. Bioengr. 2012, 109, 2599–2611. [Google Scholar] [CrossRef]
- Tanemura, H.; Masuda, K.; Okumura, T.; Takagi, E.; Kajihara, D.; Kakihara, H.; Nonaka, K.; Ushioda, R. Development of a Stable Antibody Production System Utilizing an Hspa5 Promoter in CHO Cells. Sci. Rep. 2022, 12, 7239. [Google Scholar] [CrossRef]
- Kyeong, M.; Lee, J.S. Endogenous BiP Reporter System for Simultaneous Identification of ER Stress and Antibody Production in Chinese Hamster Ovary Cells. Metab. Eng. 2022, 72, 35–45. [Google Scholar] [CrossRef]
- Roy, G.; Zhang, S.; Li, L.; Higham, E.; Wu, H.; Marelli, M.; Bowen, M.A. Development of a Fluorescent Reporter System for Monitoring ER Stress in Chinese Hamster Ovary Cells and Its Application for Therapeutic Protein Production. PLoS ONE 2017, 12, e0183694. [Google Scholar] [CrossRef]
- Willmott, J.A. Developing Novel Biosensors for Monitoring Antibody Production in Chinese Hamster Ovary (CHO) Cells. Ph.D. Thesis, University of Sheffield, Sheffield, UK, 2022. [Google Scholar]
- Du, Z.; Treiber, D.; McCoy, R.E.; Miller, A.K.; Han, M.; He, F.; Domnitz, S.; Heath, C.; Reddy, P. Non-invasive UPR Monitoring System and Its Applications in CHO Production Cultures. Biotechnol. Bioengr. 2013, 110, 2184–2194. [Google Scholar] [CrossRef]
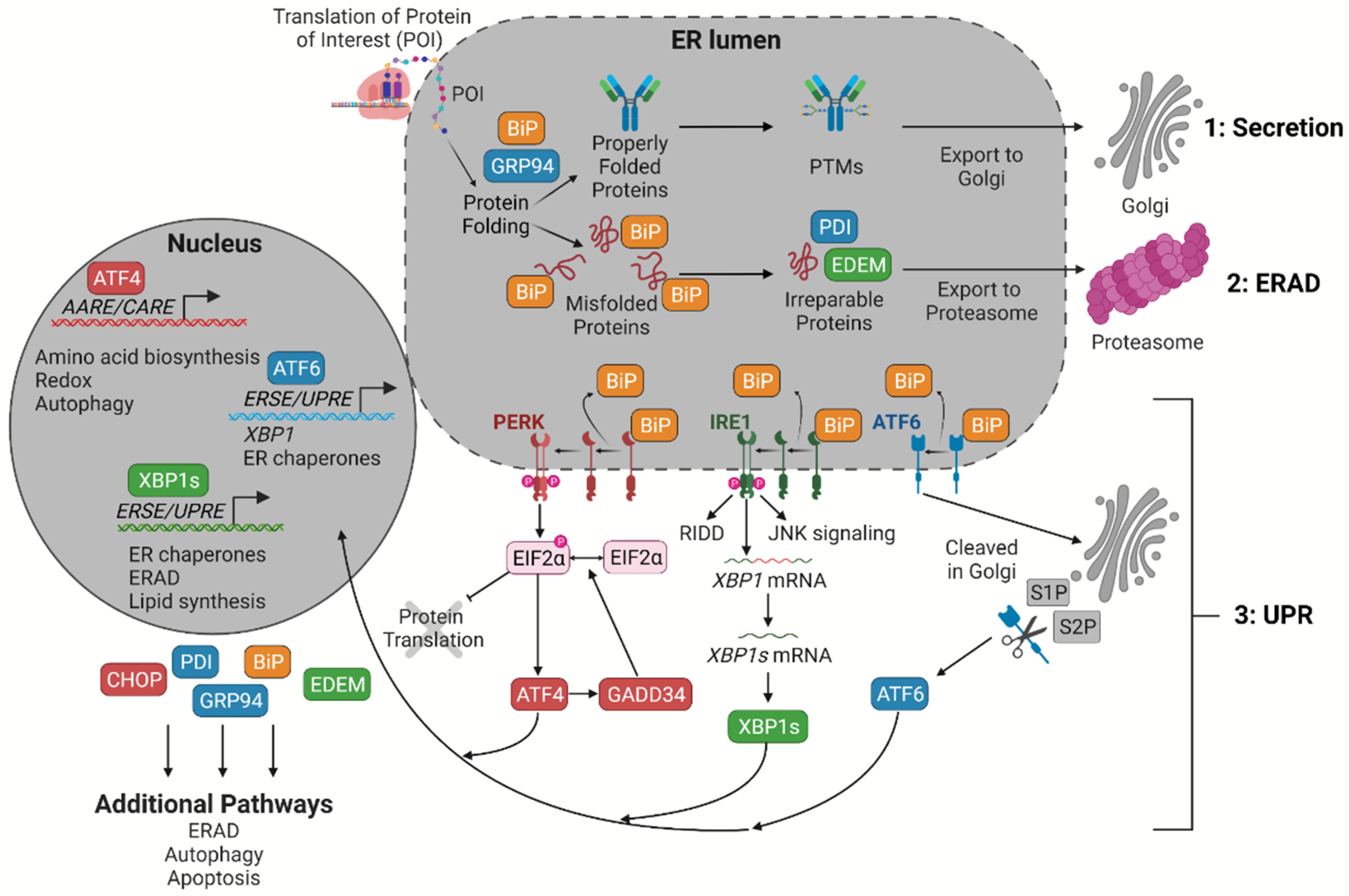
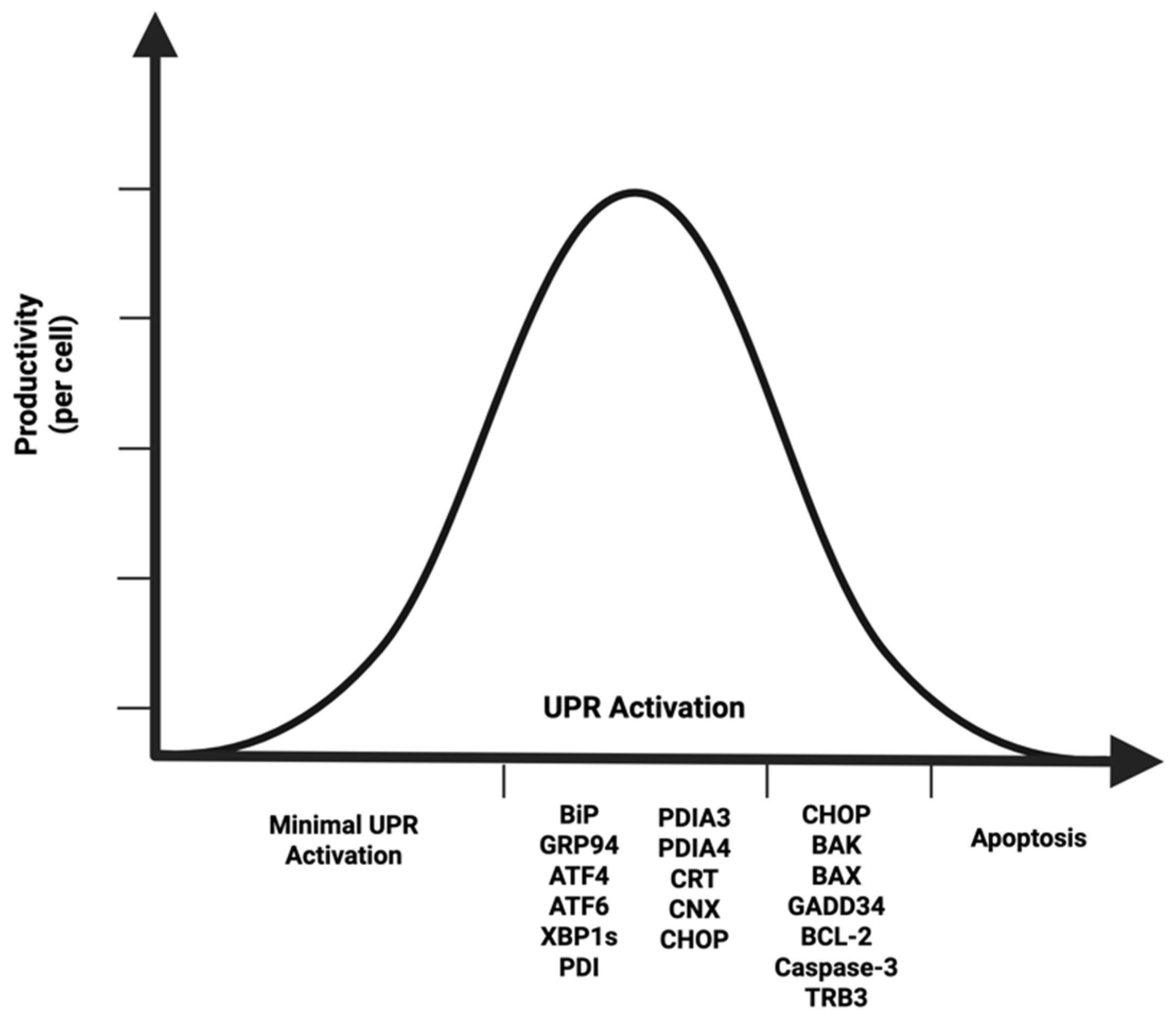

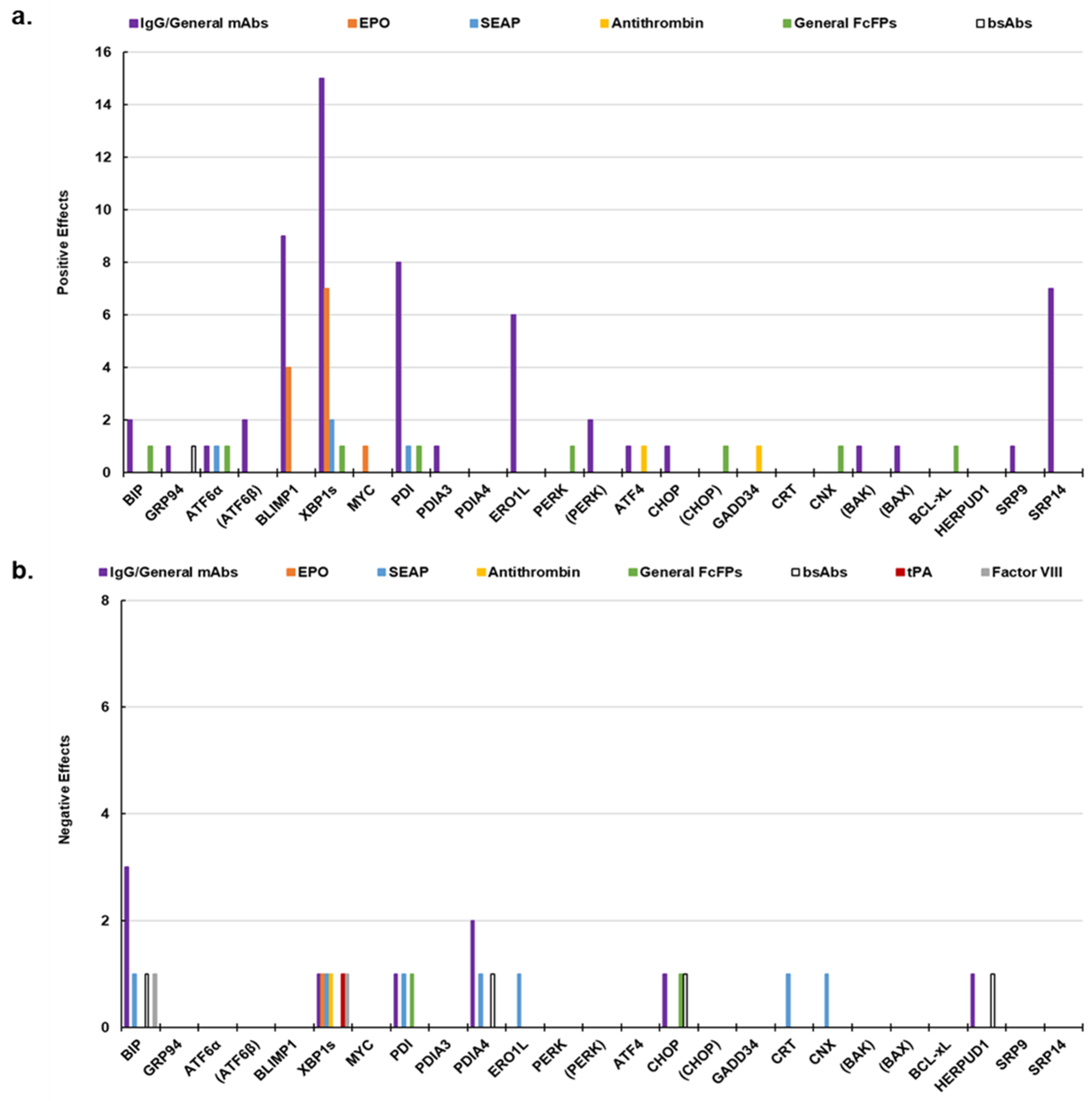
| Marker * | Role |
|---|---|
| HSPA5/GRP78/BIP | UPR initiator; chaperone |
| HSP90B1/GRP94 | Chaperone |
| ATF6c/ATF6α | UPR initiator; transcription factor |
| ERN1/IRE1 | UPR initiator; endoribonuclease |
| XBP1s | Transcription factor |
| P4HB/ERP59/PDIA1/PDI | Isomerase; chaperone |
| ERP57/PDIA3 | Isomerase; chaperone |
| ERP72/PDIA4 | Isomerase; chaperone |
| ERO1L | ER oxidoreductase |
| JNK | Kinase |
| PERK | UPR initiator; kinase |
| EIF2α | Translation |
| ATF4 | Transcription factor |
| GADD153/DDIT3/CHOP | Transcription factor |
| PPP1R15A/GADD34 | Translation initiation; apoptosis |
| EDEM1, EDEM2, EDEM3 | ERAD; mannosidases |
| DERL2, DERL3 | ERAD |
| HSPA8 | Heat shock protein; chaperone; ERAD |
| HSP70 | Heat shock protein; chaperone |
| CALR/CRT | Calcium-dependent chaperone |
| CANX/CNX | Calcium-dependent chaperone |
| BAK | Apoptosis |
| BAX | Apoptosis |
| BCL2 | Apoptosis |
| Caspase-3 | Apoptosis |
| TRB3 | Apoptosis |
| HERPUD1 | ERAD |
| HYOU1 | Hypoxia |
| Product | Markers Identified by Omics/Profiling * | Reference |
|---|---|---|
| IgG1 ** | CHOP, ATF4, BIP, GRP94, HERPUD1, PDIA3, BCL-XL, PRDX1, USP14, SOD1, SOD2, BCL2L11, PDIA4, PDI, PDIA6, RAGC, RPN1, CRT, CNX, ERDJ4, ERO1α, XBP1s, UGGT1-V1, UGGT2, GADD34, NRF2, HYOU1, SIL1, DNAJC1, DNAJC3, DNAJC10, DNJC11, FKBP9, HSPE1, PRDX1, (CREDL1), (SELENBP1) | [25,49,50,53,54,55,56,57,58,59] |
| IgG2 | ATF4, BIP, RAGC, RPN1, CHAC1, DERL3, HSP70 CRT, HERPUD1, HSPA9, RAGC, RPN1 | [53] |
| IgG4 | UGGT1, HSP90AB1, WFS1, GRP94, BIP, HYOU1, PDIA5, PDIA4, ERP29 | [60] |
| IgM | - | [51] |
| General mAbs *** | (PDIA3) FK506-binding proteins 7 and 14, calumenin, NCK1, PRKRA, BIP, PERK, CHOP, ATF6, XBP1s, PDI, GRP94, PDIA4, CNX, SEC61, HSP90, DNAJB9, DNAJB11, PDIA2, PDIA3, EDEM1, EDEM3, UGGT1, KDELR1, (CLCC1), (DNAJC3), (EMC7), (OS9), (MINPP1), (TMED4), (UFC1), (PRKCD), (PITPNM1), (SURF4) | [61,62,63,64,65,66] |
| tPA | HSPA8 | [67] |
| Factor VIII | BIP, XBP1s, CRT, CNX, PDIA3, PDIA4, PDIA6, EDEM1, EDEM2, DERL2, HERPUD1, PRDX1 | [68,69] |
| Antithrombin (AT(C95R)) | BIP, GRP97, PDI | [70,71] |
| EPO | CHKB, CHKA, CEPT, HERPUD1, SYVN1, SELS, EDEM3, SQSTM1, XBP1, PDI, GRP94, BIP, BIRC5, ODZ4, ERO1L, TRB3, CHOP, ATF5, ATF4 | [72,73] |
| General FcFPs **** | Cathepsin B, PDIA3, CRT, PDIA4, DNAJC7, PDI, PDIA6, GRP94, GRPLE1, p-EIF2α, EI5FA, EIF4A1, XBP1s, BIP, PRDX1, CAT, HSP90AB1 | [18,74] |
| bsAbs | BIP, ATF6, PDI, PERK, CHOP | [66,75,76] |
| tsAbs | (PDI), (DNAJA3), (DNAJC1), (XBP1s), (ATF4), (ATF6), (CEBPA), (CEBPB), (CEBPD), (CEBPG), (IRE1), (INSIG1), (MAP2K7), (MAPK8), (NRF2), (PDI), (ATF5), (RPL28), (SCAP), (SREBF1), (NUPR1), (UBXN4) CEBPZ, DNAJC7, DNAJC21, HSPA9 | [52] |
| Target * | Cell Line ** | Recombinant Product | Effects *** | Impact on Quality (Y/N/U) **** | Reference |
|---|---|---|---|---|---|
| XBP1s | DG44 | IgG | Increased yield, qp | N | [110] |
| XBP1s/XIAP | DG44 | IgG | Increased yield, qp | N | [110,123] |
| ERP27 | CHO-K1d | ETE Trastuzumab (Tras) | Increased titer | U | [124] |
| ERP27/PDIA3 | DTE Infliximab (Infli) | Increased titer, VCD, viability | U | ||
| ERP27/PDIA3 | DTE Etanercept | Increased titer, VCD, viability | U | ||
| (PERK) | CHO-K1 | mAb2 | Increased titer, qp, decreased viability | N | [121] |
| (PERK/Bax/Bak) | CHO-K1 | mAb3 | Increased titer, qp, IVCC, viability | N | |
| (ATF6β) | DG44 | IgG | Increased titer, VCD | N | [28] |
| (ATF6β) | CHO-K1d | IgG1 | Decreased VCD, no change in titer, increased qp | U | [131] |
| (WFS1) | Decreased titer, no change in growth | U | |||
| BIP | CHO DHFR- | humAb 2F5 IgG | Decreased production rate | U | [125] |
| PDI | Increased production rate | U | |||
| BIP/PDI | Decreased production rate | U | |||
| XBP1s | CHO-S | Multiple mAbs | Increased mAb expression levels | U | [111] |
| ERO1a | CHO-K1 | Multiple mAbs | Increased mAb expression levels | U | |
| XBP1s/ERO1a | CHO-S | Multiple mAbs | Increased mAb titers | N | |
| XBP1s | CHO-K1 | Human Factor VIII | No improvement in production | U | [113] |
| XBP1s | CHO-K1 | Tissue Plasminogen Activator (t-Pa) | No improvement in titer | U | [114,115] |
| PDI | CHO-DUKX B-11 | TNFR:Fc | Decreased secretion | U | [132] |
| PDI | IL-15 | None | U | ||
| BIP | CHO-DUKX B-11 | von Willebrand Factor | Decreased secretion | U | [27] |
| BIP | Mutant Factor VIII | Decreased secretion | U | ||
| BIP | M-CSF | None | U | ||
| eIF3c | CHO-K1 | cap- and IRES-Dependent Recombinant Protein | Improved recombinant protein synthesis, cell count | U | [133] |
| XBP1s | CHO-K1 | IgG | Increased qp, ER size | N | [88] |
| ATF4 | CHO-DP12 SF | anti-IL-8 IgG | Increased qp | U | [134] |
| BIP | - | TfR-Ab | Increased titer, viability | N | [135] |
| (PDIA4) | CHO-HcD6 (CHO-K1d) | ETE Trastuzumab (Tras) | Decrease in secreted antibody | U | [136] |
| PDIA4 | None | U | |||
| XBP1s | CHO DG44 | mAb | No improvement in titer | U | [77] |
| CHO DHFR- | Interferon γ (IFNγ) | No improvement in titer | U | ||
| CHO-K1 | EPO | No improvement in titer | U | ||
| XBP1s | CHO-K1 | EPO | Increase in titer is dependent on product/XBP1s dosage levels | U | [77,122] |
| (XBP1s) | Decreased product titer | U | |||
| MYC | CHO-K1d | EPO | Increased IVCC | U | [87] |
| XBP1S | Increased titer, qp | U | |||
| MYC/XBP1s | Increased IVCC, specific growth rate, titer, qp | U | |||
| PDI | - | Thrombopoietin (TPO) | No increase in qp | U | [126] |
| CHO DG44 | mAb | Slight increased qp | U | ||
| BLIMP1 | DG44 | mAb | Increased titer, qp | U | [116] |
| DNAJC3 | U | ||||
| SYVN1 | U | ||||
| SELENOF | U | ||||
| HSPA8 | U | ||||
| BLIMP1 | CHO-K1 | IgG and DTE Doppelmab | Increased titer | U | |
| SYVN1 | U | ||||
| DNAJC3 | U | ||||
| ATF4 | CHO DXB11 | Antithrombin III (AT-III) | Increased qp | U | [112] |
| XBP1s | No improvement in qp | U | |||
| GADD34 | CHO DXB11 | Antithrombin III (AT-III) | Decreased VCD, Increased qp | N | [137] |
| BCL-xL | CHO DG44 | Fusion Protein (FP) | Increased qp | N | [138] |
| NFKBIZ | CHO-HcD6 | IgG1 | Increased qp | N | [139] |
| PDI/XBP1s | CHO-S | Adalimumab | Increased titer, qp | U | [86] |
| SEAP | Increased product expression | U | |||
| KDEL receptor 1 | CHO-K1 | IgG | Increased qp | N | [140] |
| BLIMP1 | CHO-K1 | IgG1 | Decreased VCDs, prolonged viability, Increased titers, qp | U | [85,117,118] |
| EPO-Fc | U | ||||
| CHO-S | IgG1 | U | |||
| BLIMP1 | CHO-K1 | EPO-Fc | Decreased VCD, increased titer, qp | U | |
| CHO-S | IgG1 | U | |||
| XBP1s | CHO-K1 | IgG1 | Prolonged viability, increased titer | U | |
| EPO-Fc | U | ||||
| BLIMP1/XBP1s | IgG1 | Decreased VCD, prolonged viability, increased titer, qp | U | ||
| EPO-Fc | U | ||||
| XBP1s | CHO-S | IgG1 | Prolonged viability, increased titer | U | |
| EPO-Fc | U | ||||
| BLIMP1/XBP1s | IgG1 | Decreased VCD, prolonged viability, increased titer, qp | U | ||
| EPO-Fc | U | ||||
| BLIMP1α | CHO DG44 | DTE Human Bone Morphogenetic Protein-4 (rhBMP-4) | Increased qp | U | [119] |
| BLIMP1β | Increased qp, yields | U | |||
| CHO-K1 | ETE Rituximab | Decreased specific growth rate, increased titer, qp | U | ||
| SCD1 | CHO-K1d | cB72.3, FcFP, DTE IgG1 | Increased titers | U | [42,141] |
| SREBF1 | U | ||||
| PERK | CHO DG44 | TNFR-Fc | Decreased aggregates | N | [94] |
| CERT | CHO DG44 | Human Serum Albumin (HSA) | Increased titers, qp | U | [142] |
| IgGs | Increased secretion | U | |||
| XBP1s | CHO-K1/CHO-K1d | Secreted Alkaline Phosphatase (SEAP) | Increased production | U | [143] |
| Bacillus stearothermophilus-derived a-amylase (SAMY) | U | ||||
| Vascular Endothelial Growth Factor 121 (VEGF121) | U | ||||
| SRP14 | CHO-K1 | ETE Trastuzumab (Tras) | Prolonged viability, increased qp | U | [20] |
| DTE Infliximab (Infli) | Increased qp | U | |||
| SRP14/SRP9/SRP54/SR | U | ||||
| SRP14/SR/Translocon | U | ||||
| BIP | CHO-S | DTE Sp35Fc | Dose-dependent; Decreased IVCD, increased titer, qp | N | [19] |
| PDI | Increased titer, qp, product aggregation | Y | |||
| CypB | Increased IVCD, titer, decreased product aggregation | N | |||
| ATF6α | Dose-dependent; decreased IVCD, increased titer, qp | N | |||
| XBP1s | Dose-dependent; decreased IVCD, increased titer, qp | N | |||
| PDIA4 | CAT-S/CHO-K1d | BsAb1 | None | U | [144] |
| UBXN8 | Decreased titer | U | |||
| DNAJB9 | None | U | |||
| BIP | Decreased titer | U | |||
| GRP94 | Decreased product aggregation | N | |||
| DNAJC3 | None | U | |||
| CHOP | Decreased product aggregation, titer | N | |||
| HERPUD1 | Decreased titer | U | |||
| PDIA4 | CHO-Sd | ETE Trastuzumab (Tras) | None | U | |
| UBXN8 | None | U | |||
| DNAJB9 | None | U | |||
| BIP | None | U | |||
| GRP94 | Increased titer | U | |||
| DNAJC3 | Increased titer | U | |||
| CHOP | None | U | |||
| HERPUD1 | None | U | |||
| PDIA3 | CHO-DUKX B-11 | Thrombopoietin (TPO) | Increased titer, qp | U | [145] |
| ERGIC-53 | CHO-HcD6 (CHO-K1d) | IgG1 | Increased VCD, titer, qp | N | [127] |
| ERGIC-53/MCFD2 | Decreased VCD, increased titer, qp | N | |||
| (CerS2/Tbc1D20) | CHO DG44 | Human Serum Albumin (HSA) and IgG | Increased titer, qp | N | [146] |
| CHOP | CHO-S | hTRA-8 | Increased titer | N | [128] # |
| BIP | CHO-K1d | Multiple IgG1-type mAbs | Increased titer, qp for one mAb | U | [147] |
| CypB | Increased cell growth, titer, decreased qp | U | |||
| PDI | Increased titer, qp for one mAb | U | |||
| ATF6α | Increased titer, qp dependent on expression level | U | |||
| XBP1s | U | ||||
| (UBR4/UBR5) | - | IgG | Increased titer | U | [55] |
| EIF2AK2 | CHO-S | DTE Thrombospondin 4 (THBS4) | Decreased titer | U | [148] |
| HSPA1B | None | U | |||
| TBC1D9 | None | U | |||
| HSPA4L | None | U | |||
| RAB11FIP1 | Decreased titer | U | |||
| MYO5B | None | U | |||
| MGAT3 | Decreased titer | U | |||
| SNAP25 | Decreased titer | U | |||
| AGAP2 | None | U | |||
| RAB6B | None | U | |||
| DERL3 | Decreased titer | U | |||
| SVIP1 | Decreased titer | U | |||
| GALNT18 | Decreased titer | U | |||
| JUN | Increased titer | U | |||
| PDIA4 | None | U | |||
| ATF4 | Increased titer | U | |||
| SRP9 | Increased titer | U | |||
| HSPA8 | None | U | |||
| PDIA3 | None | U | |||
| RAB31 | None | U | |||
| RAB43 | None | U | |||
| HSPA1B | DTE Artemin (ARTN) | None | U | ||
| ATF4 | Increased titer | U | |||
| SRP9 | None | U | |||
| PDIA3 | Increased titer | U | |||
| RAB43 | Decreased titer | U | |||
| HSPA8 | Increased titer | U | |||
| HsQSOX1b/Survivin | CHO-K1 | Pembrolizumab (PAb) | Increased titer, qp | N | [129] |
| (CHOP) | - | TNFR-Fc | Decreased percentage of non-viable/apoptotic cells under ER stress conditions | U | [149] |
| CHOP | Increased percentage of non-viable/apoptotic cells under ER stress conditions | U | |||
| Onco-tyrosine kinase receptor (KIT) | CHO-K1 | Green Fluorescent Protein (GFP)-Fc | Increased titer | U | [107] |
| XBP1s | CHO-K1d | mAb-transient | Increased titer, qp | U | [130] ## |
| Light Chain/XBP1s | Increased titer, qp | U | |||
| CRELD2 | Increased titer, qp | U | |||
| Light Chain/CRELD2 | Increased titer, qp | U | |||
| XBP1s/CRELD2 | Increased titer, qp | U | |||
| Light Chain/XBP1s/CRELD2 | Increased titer, qp | U | |||
| PDI | mAb-stable | Increased titer, qp | U | ||
| ERO1α | Increased titer, qp | U | |||
| PDI/ERO1a | Increased titer, qp | U | |||
| SRP14 | Increased titer, qp | U | |||
| PDI/SRP14 | Increased titer, qp | U | |||
| ERO1α/SRP14 | Increased titer, qp | U | |||
| PDI/ERO1α/SRP14 | Increased titer, qp | U | |||
| CNX | - | TNFR-Fc | Increased qp | U | [108] |
| ATF6α | CHO-S | SEAP | Increased yield, qp | U | [150] |
| XBP1s | No increase in yield | U | |||
| CypB | No increase in yield | U | |||
| ERO1α | No increase in yield | U | |||
| PDI | No increase in yield | U | |||
| PDIA4 | No increase in yield | U | |||
| BIP | No increase in yield | U | |||
| CRT | No increase in yield | U | |||
| CNX | No increase in yield | U | |||
| HSPA1A | No increase in yield | U | |||
| TOR1A | No increase in yield | U | |||
| CERT | No increase in yield | U |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rives, D.; Richbourg, T.; Gurtler, S.; Martone, J.; Blenner, M.A. Recent Advances in Engineering the Unfolded Protein Response in Recombinant Chinese Hamster Ovary Cell Lines. Int. J. Mol. Sci. 2025, 26, 7189. https://doi.org/10.3390/ijms26157189
Rives D, Richbourg T, Gurtler S, Martone J, Blenner MA. Recent Advances in Engineering the Unfolded Protein Response in Recombinant Chinese Hamster Ovary Cell Lines. International Journal of Molecular Sciences. 2025; 26(15):7189. https://doi.org/10.3390/ijms26157189
Chicago/Turabian StyleRives, Dyllan, Tara Richbourg, Sierra Gurtler, Julia Martone, and Mark A. Blenner. 2025. "Recent Advances in Engineering the Unfolded Protein Response in Recombinant Chinese Hamster Ovary Cell Lines" International Journal of Molecular Sciences 26, no. 15: 7189. https://doi.org/10.3390/ijms26157189
APA StyleRives, D., Richbourg, T., Gurtler, S., Martone, J., & Blenner, M. A. (2025). Recent Advances in Engineering the Unfolded Protein Response in Recombinant Chinese Hamster Ovary Cell Lines. International Journal of Molecular Sciences, 26(15), 7189. https://doi.org/10.3390/ijms26157189





