Machine Learning Approach to Select Small Compounds in Plasma as Predictors of Alzheimer’s Disease
Abstract
1. Introduction
2. Results
2.1. Correlations Between Data from Patients and Controls
2.1.1. Dataset Overview and Data Cleaning Outcome
2.1.2. Selection of the Metabolite_c for Each Biochemical Family
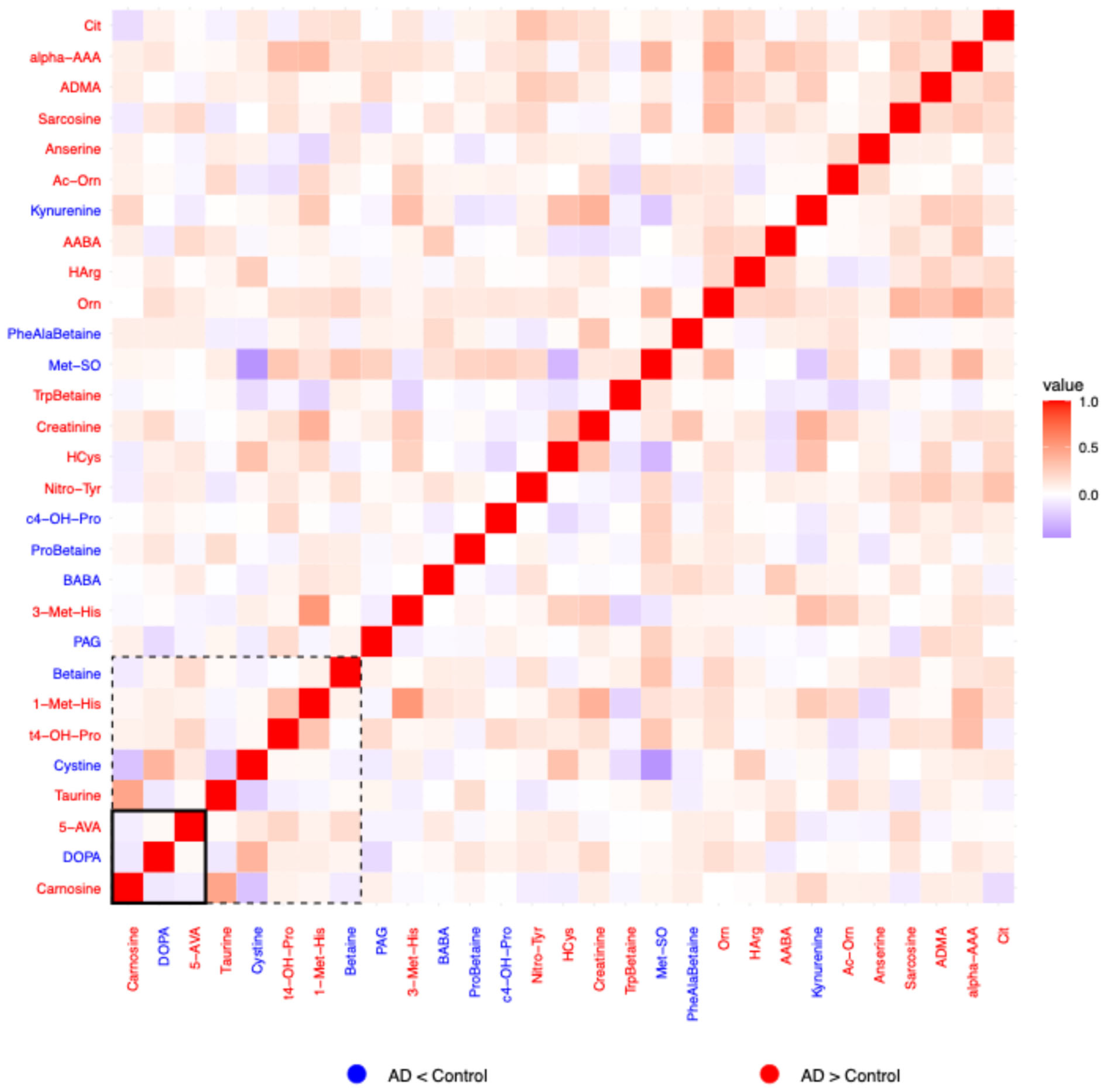
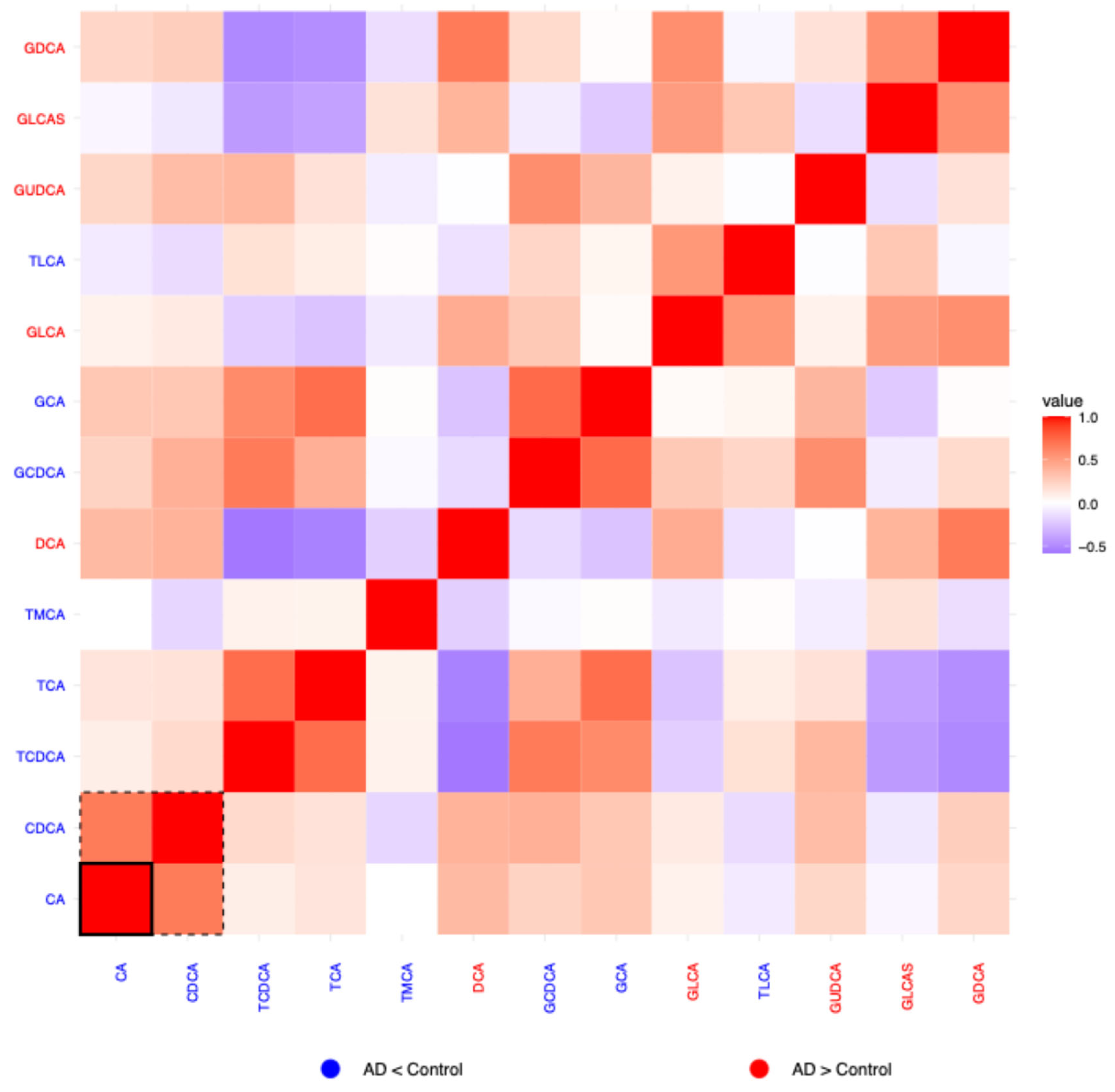
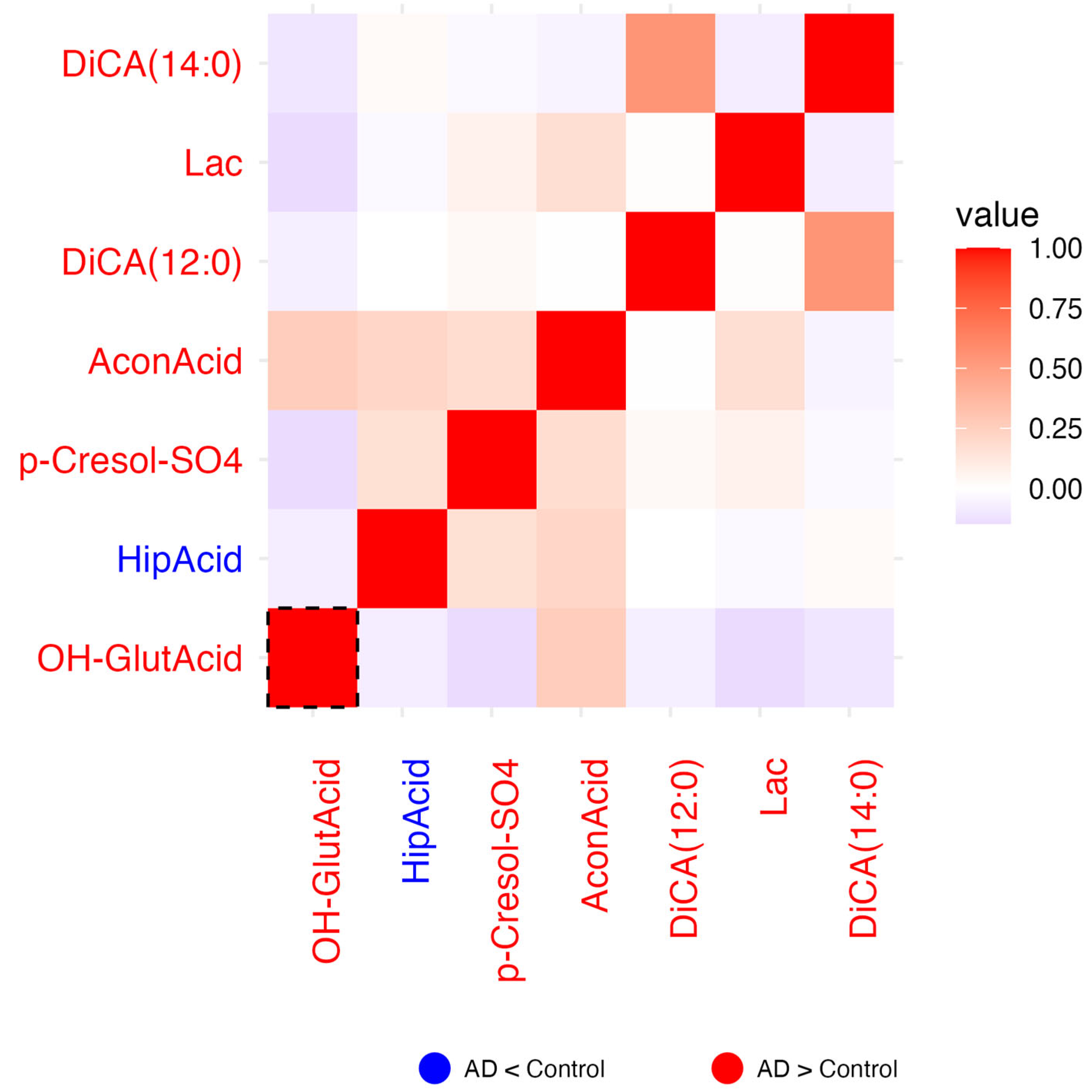
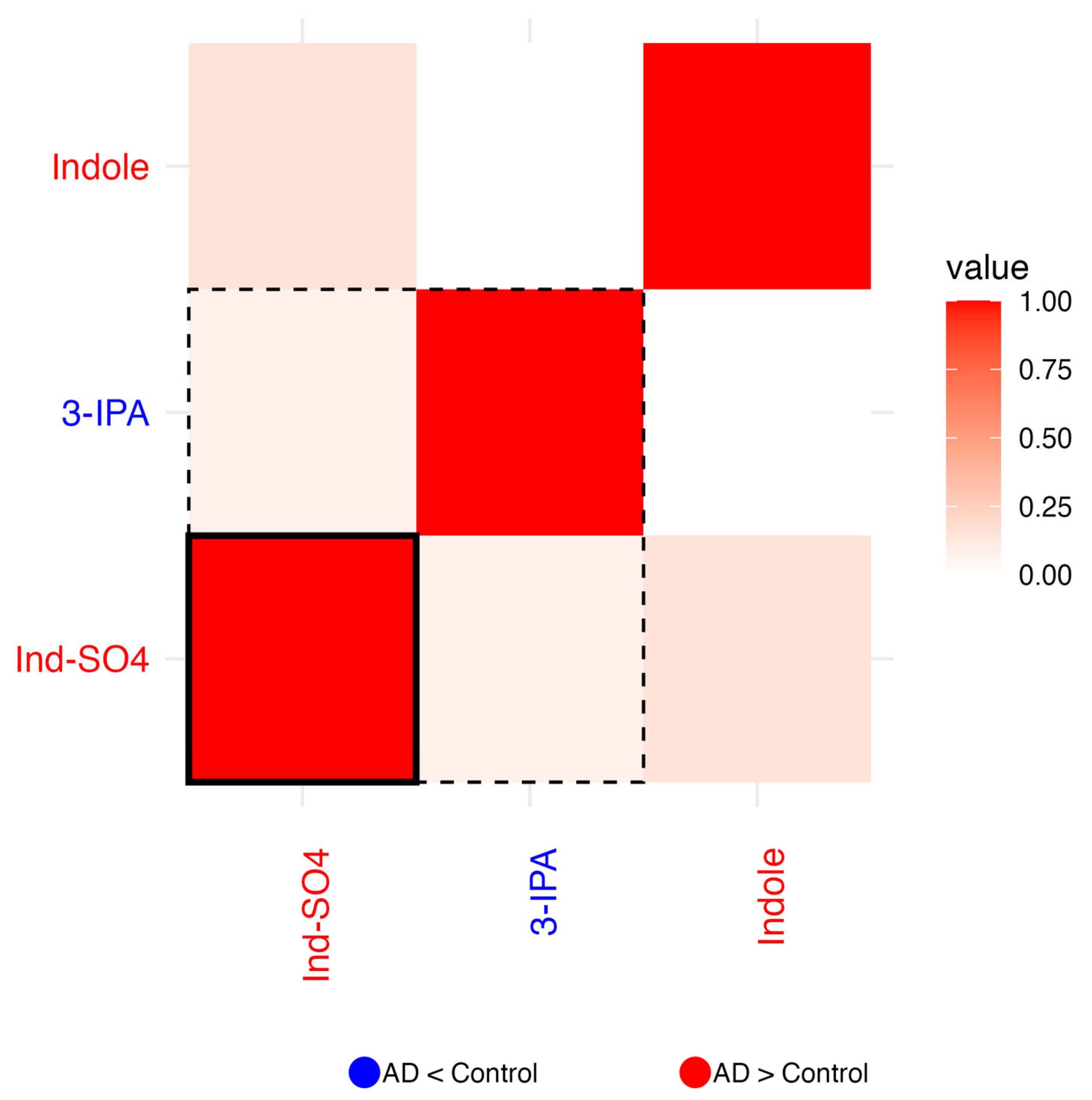
2.1.3. Residual Correlation Heatmaps
2.1.4. Amino Acid Related
2.1.5. Bile Acids
2.1.6. Biogenic Amines
2.1.7. Carboxylic Acids
2.1.8. Indoles
2.1.9. Fatty Acids
2.2. Classification Performance via LDA
2.2.1. Univariate Analysis
2.2.2. Multivariate Analysis: Four Metabolite Combinations
2.2.3. Multivariate Analysis: Five Metabolite Combinations
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. Data Handling
4.3. Statistical Analysis
4.4. Determination of the Concomitant Metabolite for Each Biochemical Family
4.5. Residual Correlation Heatmaps
4.6. Linear Discriminant Analysis (LDA)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chételat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef]
- Long, J.M.; Holtzman, D.M. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 2019, 179, 312–339. [Google Scholar] [CrossRef]
- Avila, J.; Pérez, M.; Avila-Villanueva, M.; Santa-María, I.; Hernández, F. Looking at the periphery—New hypothesis to look for new targets for Alzheimer’s disease therapy. Explor. Neuroprotective Ther. 2023, 3, 151–163. [Google Scholar] [CrossRef]
- Whiley, L.; Sen, A.; Heaton, J.; Proitsi, P.; García-Gómez, D.; Leung, R.; Smith, N.; Thambisetty, M.; Kloszewska, I.; Mecocci, P.; et al. Evidence of altered phosphatidylcholine metabolism in Alzheimer’s disease. Neurobiol. Aging 2014, 35, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Kalecký, K.; German, D.C.; Montillo, A.A.; Bottiglieri, T. Targeted Metabolomic Analysis in Alzheimer’s Disease Plasma and Brain Tissue in Non-Hispanic Whites. J. Alzheimer’s Dis. 2022, 86, 1875–1895. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Marín, J.; Marin, S.; Iglesias, A.; Lillo, J.; Garrigós, C.; Capó, T.; Reyes-Resina, I.; Alkozi, H.A.; Cascante, M.; Sánchez-Navés, J.; et al. Novel protocol for metabolomics data normalization and biomarker discovery in human tears. Clin. Chem. Lab. Med. 2025, 63, 1599–1609. [Google Scholar] [CrossRef]
- Stefanini, E.; Serrano-Marín, J.; Iglesias, A.; Fernández, M.G.; Sánchez-Navés, J.; Alkozi, H.A.; Bernal-Casas, D.; Franco, R. CE(20:4) and CE(22:5) cholesteryl ester levels are elevated in the plasma of Alzheimer’s disease patients with ε3/ε4 genotype. Explor. Neuroprotective Ther. 2025, 5, 100497. [Google Scholar] [CrossRef]
- Passaro, A.; Al Bakir, M.; Hamilton, E.G.; Diehn, M.; André, F.; Roy-Chowdhuri, S.; Mountzios, G.; Wistuba, I.I.; Swanton, C.; Peters, S. Cancer biomarkers: Emerging trends and clinical implications for personalized treatment. Cell 2024, 187, 1617–1635. [Google Scholar] [CrossRef]
- Schöll, M.; Verberk, I.M.W.; del Campo, M.; Delaby, C.; Therriault, J.; Chong, J.R.; Palmqvist, S.; Alcolea, D. Challenges in the practical implementation of blood biomarkers for Alzheimer’s disease. Lancet Healthy Longev. 2024, 5, 100630. [Google Scholar] [CrossRef]
- Serrano-Marín, J.; Marin, S.; Bernal-Casas, D.; Lillo, A.; González-Subías, M.; Navarro, G.; Cascante, M.; Sánchez-Navés, J.; Franco, R. A metabolomics study in aqueous humor discloses altered arginine metabolism in Parkinson’s disease. Fluids Barriers CNS 2023, 20, 90. [Google Scholar] [CrossRef]
- Bernal-Casas, D.; Serrano-Marín, J.; Sánchez-Navés, J.; Oller, J.M.; Franco, R. Advancing Personalized Medicine by Analytical Means: Selection of Three Metabolites That Allows Discrimination between Glaucoma, Diabetes, and Controls. Metabolites 2024, 14, 149. [Google Scholar] [CrossRef]
- Lillo, A.; Marin, S.; Serrano-Marín, J.; Binetti, N.; Navarro, G.; Cascante, M.; Sánchez-Navés, J.; Franco, R. Targeted Metabolomics Shows That the Level of Glutamine, Kynurenine, Acyl-Carnitines and Lysophosphatidylcholines Is Significantly Increased in the Aqueous Humor of Glaucoma Patients. Front. Med. 2022, 9, 2082. [Google Scholar] [CrossRef]
- Adler, C.H.; Beach, T.G.; Hentz, J.G.; Shill, H.A.; Caviness, J.N.; Driver-Dunckley, E.; Sabbagh, M.N.; Sue, L.I.; Jacobson, S.A.; Belden, C.M.; et al. Low clinical diagnostic accuracy of early vs. advanced Parkinson disease: Clinicopathologic study. Neurology 2014, 83, 406. [Google Scholar] [CrossRef]
- Beach, T.G.; Adler, C.H. Importance of low diagnostic Accuracy for early Parkinson’s disease. Mov. Disord. 2018, 33, 1551–1554. [Google Scholar] [CrossRef]
- Vlaar, A.M.M.; van Kroonenburgh, M.J.P.G.; Kessels, A.G.H.; Weber, W.E.J. Meta-analysis of the literature on diagnostic accuracy of SPECT in parkinsonian syndromes. BMC Neurol. 2007, 7, 27. [Google Scholar] [CrossRef]
- Hacohen, Y.; Brownlee, W.; Mankad, K.; Chong, W.K.K.; Thompson, A.; Lim, M.; Wassmer, E.; Hemingway, C.; Barkhof, F.; Ciccarelli, O. Improved performance of the 2017 McDonald criteria for diagnosis of multiple sclerosis in children in a real-life cohort. Mult. Scler. J. 2020, 26, 1372–1380. [Google Scholar] [CrossRef]
- Yao, X.; Li, X.; Ye, Q.; Huang, Y.; Cheng, Q.; Zhang, G.Q. A robust deep learning approach for automatic classification of seizures against non-seizures. Biomed. Signal Process. Control 2021, 64, 102215. [Google Scholar] [CrossRef]
- Ahmed, H.; Leyrolle, Q.; Koistinen, V.; Kärkkäinen, O.; Layé, S.; Delzenne, N.; Hanhineva, K. Microbiota-derived metabolites as drivers of gut-brain communication. Gut Microbes 2022, 14, 2102878. [Google Scholar] [CrossRef]
- Caruso, G.; Caraci, F.; Jolivet, R.B. Pivotal role of carnosine in the modulation of brain cells activity: Multimodal mechanism of action and therapeutic potential in neurodegenerative disorders. Prog. Neurobiol. 2019, 175, 35–53. [Google Scholar] [CrossRef]
- MahmoudianDehkordi, S.; Arnold, M.; Nho, K.; Ahmad, S.; Jia, W.; Xie, G.; Louie, G.; Kueider-Paisley, A.; Moseley, M.A.; Thompson, J.W.; et al. Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease-An emerging role for gut microbiome. Alzheimer’s Dement. 2019, 15, 76–92. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Wang, M.C.; Wang, Y.N.; Hu, H.H.; Liu, Q.Q.; Liu, H.J.; Zhao, Y.Y. Redox signaling and Alzheimer’s disease: From pathomechanism insights to biomarker discovery and therapy strategy. Biomark. Res. 2020, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.C.; Lu, K.C.; Huang, C.L.; Wang, J.J.; Yeh, T.Y.; Lin, S.M.; Chung, Y.L.; Hou, Y.C.; Huang, Y.S. Indoxyl sulfate is associated with cognitive impairment in ESRD patients by activating the extrinsic apoptosis in the neuronal cells during differentiating process. Int. J. Med. Sci. 2025, 22, 1736–1749. [Google Scholar] [CrossRef] [PubMed]
- Pais, M.V.; Forlenza, O.V.; Diniz, B.S. Plasma Biomarkers of Alzheimer’s Disease: A Review of Available Assays, Recent Developments, and Implications for Clinical Practice. J. Alzheimer’s Dis. Rep. 2023, 7, 355–380. [Google Scholar] [CrossRef] [PubMed]
- O’Bryant, S.E.; Waring, S.C.; Cullum, C.M.; Hall, J.; Lacritz, L.; Massman, P.J.; Lupo, P.J.; Reisch, J.S.; Doody, R. Staging dementia using Clinical Dementia Rating Scale Sum of Boxes scores: A Texas Alzheimer’s research consortium study. Arch. Neurol. 2008, 65, 1091–1095. [Google Scholar] [CrossRef]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef]
| Group | Model a | R2 | F Test-p-Value |
|---|---|---|---|
| AA-related | log10(metabolite)predicted ~ Group + Age * Sex + Fasting_time + log10(SDMA) | 0.121 | 0.099 |
| Bile Acids | log10(metabolite)predicted ~ Group + Age * Sex + Fasting_time + log10(TDCA) | 0.309 | 0.013 |
| Biogenic Amines | log10(metabolite)predicted ~ Group + Age * Sex + Fasting_time + log10(Spermine) | 0.112 | 0.101 |
| Carboxylic Acids | log10(metabolite)predicted ~ Group + Age * Sex + Fasting_time + log10(Succinic) | 0.108 | 0.134 |
| Indoles | log10(metabolite)predicted ~ Group + Age * Sex + Fasting_time + log10(3-IAA) | 0.054 | 0.188 |
| Fatty Acids | log10(metabolite)predicted ~ Group + Age * Sex + Fasting_time + log10(DHA) | 0.294 | 0.014 |
| AA-Related | Bile Acids | Biogenic Amines | Carboxylic Acids | Indoles | Fatty Acids | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| 5-AVA, Carnosine | CA, CDCA | 0.76 | 0.74 | 0.79 | ||||
| 5-AVA, Carnosine, c4−OH−Pro | CA | 0.76 | 0.78 | 0.74 | ||||
| 5-AVA, Carnosine,1−Met−His | CA | 0.76 | 0.76 | 0.76 | ||||
| 5-AVA | CA | Serotonin | Ind-SO4 | 0.75 | 0.8 | 0.69 | ||
| Carnosine, Orn | CA | Hypoxanthine | 0.75 | 0.73 | 0.79 | |||
| 5-AVA, Carnosine | CA | Lactic | 0.75 | 0.74 | 0.77 | |||
| Carnosine, PAG | TCA | TMAO | 0.75 | 0.74 | 0.77 | |||
| 5-AVA, Carnosine, Nitro-Tyr | Hypoxanthine | 0.75 | 0.74 | 0.77 | ||||
| Carnosine | CA | Hypoxanthine | Ind-SO4 | 0.75 | 0.78 | 0.72 | ||
| 5-AVA, Carnosine | CA | Hippuric | 0.75 | 0.78 | 0.72 |
| AA-Related | Bile Acids | Biogenic Amines | Carboxylic Acids | Indoles | Fatty Acids | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| Carnosine, Nitro-Tyr | Hypoxanthine | Ind-SO4 | 0.74 | 0.76 | 0.71 | |||
| 5-AVA, Carnosine | CA | Lac | 0.74 | 0.74 | 0.74 | |||
| Carnosine, HCys | Hypoxanthine | Ind-SO4 | 0.73 | 0.78 | 0.67 | |||
| 5-AVA, Carnosine | CA, TMCA | 0.73 | 0.73 | 0.72 | ||||
| 5-AVA, Carnosine, 3−Met−His | CA | 0.73 | 0.73 | 0.72 | ||||
| 5-AVA | CA | Serotonin | Ind-SO4 | 0.73 | 0.78 | 0.65 | ||
| 5-AVA, Carnosine | CA, GUDCA | 0.73 | 0.75 | 0.69 | ||||
| 5-AVA, Carnosine, Nitro-Tyr | Hypoxanthine | 0.73 | 0.72 | 0.74 | ||||
| Carnosine, PAG | TCA | TMAO | 0.73 | 0.73 | 0.72 | |||
| 5-AVA, Carnosine,1−Met−His | CA | 0.73 | 0.75 | 0.69 |
| AA-Related | Bile Acids | Biogenic Amines | Carboxylic Acids | Indoles | Fatty Acids | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| 5-AVA, Carnosine | CA | Hypoxanthine | 3-IPA | 0.78 | 0.78 | 0.79 | ||
| 5-AVA, Carnosine, c4−OH−Pro | CA | FA(18:2) | 0.78 | 0.8 | 0.76 | |||
| 5-AVA, Carnosine | CA | FA(20:3), FA(20:1) | 0.78 | 0.76 | 0.81 | |||
| Carnosine, Nitro-Tyr, Harg | Hypoxanthine | Ind-SO4 | 0.78 | 0.81 | 0.72 | |||
| 5-AVA, Carnosine, c4−OH−Pro | CA | FA(18:1) | 0.78 | 0.8 | 0.76 | |||
| Carnosine, Orn, ADMA | CA | Hypoxanthine | 0.78 | 0.75 | 0.81 | |||
| 5-AVA, Carnosine, c4−OH−Pro,PAG | CA | 0.78 | 0.79 | 0.76 | ||||
| Carnosine, Nitro-Tyr, Cit | Hypoxanthine | Ind-SO4 | 0.78 | 0.79 | 0.76 | |||
| 5-AVA, Carnosine, Nitro-Tyr, ProBetaine | CA | 0.78 | 0.78 | 0.77 | ||||
| 5-AVA, 3−Met−His, t4−OH−Pro | CA | Xanthine | 0.78 | 0.79 | 0.76 |
| AA-Related | Bile Acids | Biogenic Amines | Carboxylic Acids | Indoles | Fatty Acids | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| 5-AVA, Carnosine, c4−OH−Pro | CA | FA(18:2) | 0.76 | 0.78 | 0.74 | |||
| 5-AVA, Met−SO | CA | Serotonin | Ind-SO4 | 0.76 | 0.79 | 0.71 | ||
| Carnosine, Orn, alpha-AAA | CA | Hypoxanthine | 0.76 | 0.75 | 0.76 | |||
| Carnosine | CA | Hypoxanthine | DiCA(14:0) | Ind-SO4 | 0.76 | 0.79 | 0.71 | |
| 5-AVA, Carnosine | CA | Hypoxanthine | 3-IPA | 0.76 | 0.73 | 0.79 | ||
| Carnosine, Nitro-Tyr | Hypoxanthine | Ind-SO4 | FA(20:3) | 0.76 | 0.79 | 0.71 | ||
| Carnosine, Orn, PAG | CA | Hypoxanthine | 0.76 | 0.75 | 0.76 | |||
| 5-AVA, Carnosine, c4−OH−Pro | CA | DiCA(14:0) | 0.75 | 0.78 | 0.71 | |||
| 5-AVA, Carnosine, PAG | CA | OH−GlutAcid | 0.75 | 0.75 | 0.74 | |||
| Carnosine, Nitro-Tyr, c4−OH−Pro | Hypoxanthine | Ind-SO4 | 0.75 | 0.78 | 0.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefanini, E.; Iglesias, A.; Serrano-Marín, J.; Sánchez-Navés, J.; Alkozi, H.A.; Pallàs, M.; Griñán-Ferré, C.; Bernal-Casas, D.; Franco, R. Machine Learning Approach to Select Small Compounds in Plasma as Predictors of Alzheimer’s Disease. Int. J. Mol. Sci. 2025, 26, 6991. https://doi.org/10.3390/ijms26146991
Stefanini E, Iglesias A, Serrano-Marín J, Sánchez-Navés J, Alkozi HA, Pallàs M, Griñán-Ferré C, Bernal-Casas D, Franco R. Machine Learning Approach to Select Small Compounds in Plasma as Predictors of Alzheimer’s Disease. International Journal of Molecular Sciences. 2025; 26(14):6991. https://doi.org/10.3390/ijms26146991
Chicago/Turabian StyleStefanini, Eleonora, Alberto Iglesias, Joan Serrano-Marín, Juan Sánchez-Navés, Hanan A. Alkozi, Mercè Pallàs, Christian Griñán-Ferré, David Bernal-Casas, and Rafael Franco. 2025. "Machine Learning Approach to Select Small Compounds in Plasma as Predictors of Alzheimer’s Disease" International Journal of Molecular Sciences 26, no. 14: 6991. https://doi.org/10.3390/ijms26146991
APA StyleStefanini, E., Iglesias, A., Serrano-Marín, J., Sánchez-Navés, J., Alkozi, H. A., Pallàs, M., Griñán-Ferré, C., Bernal-Casas, D., & Franco, R. (2025). Machine Learning Approach to Select Small Compounds in Plasma as Predictors of Alzheimer’s Disease. International Journal of Molecular Sciences, 26(14), 6991. https://doi.org/10.3390/ijms26146991









