Reconstruction of a Genome-Scale Metabolic Model for Aspergillus oryzae Engineered Strain: A Potent Computational Tool for Enhancing Cordycepin Production
Abstract
1. Introduction
2. Results
2.1. GSMM of the Cordycepin-Producing A. oryzae Strain
2.2. The GSMM Biomass Formulation
2.3. Scenario GSMM Validation by Optimizing Growth and Synthetic Cordycepin Production
2.4. In Silico Identification of Gene Amplification Targets for Improving Cordycepin Production
2.5. Optimizing Nutrients and the C:N Ratio Toward Rational Design of Synthetic Media for Cordycepin Overproduction in A. oryzae
3. Discussion
4. Materials and Methods
4.1. Fungal Strain and Cultivation
4.2. Determination of Cell Growth, Proximate Compositions, and Cordycepin Production of A. oryzae Strains
4.3. Reconstruction of GSMM for Cordycepin-Producing A. oryzae Strain
4.4. Formulation of Biomass Reactions
4.5. GSMM Simulation, Validation, and Analysis
4.6. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AICAR | 5-amino-1-(5-phospho-d-ribosyl)imidazole-4-carboxamide |
| DCW | dry cell weight |
| EC | enzyme commission |
| FBA | flux balance analysis |
| FSEOF | flux scanning based on enforced objective flux |
| GPR | gene-protein-reaction |
| GSMM | genome-scale metabolic model |
| PPP | pentose phosphate pathway |
| PRFICA | 5-formamido-1-(5-phospho-d-ribosyl)imidazole-4-carboxamide |
References
- Tuli, H.S.; Sandhu, S.S.; Sharma, A.K. Pharmacological and Therapeutic Potential of Cordyceps with Special Reference to Cordycepin. 3 Biotech 2014, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Radhi, M.; Ashraf, S.; Lawrence, S.; Tranholm, A.A.; Wellham, P.A.D.; Hafeez, A.; Khamis, A.S.; Thomas, R.; McWilliams, D.; De Moor, C.H. A Systematic Review of the Biological Effects of Cordycepin. Molecules 2021, 26, 5886. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Lee, S.; Kwon, J.; Moon, S.; Lee, S.; Lee, C.-K.; Cho, K.; Ha, N.-J.; Kim, K. Cordycepin Suppresses Expression of Diabetes Regulating Genes by Inhibition of Lipopolysaccharide-Induced Inflammation in Macrophages. Immune Netw. 2009, 9, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yan, H.; Zeng, B.; Hu, Z. Research Progress on Cordycepin Synthesis and Methods for Enhancement of Cordycepin Production in Cordyceps militaris. Bioengineering 2022, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Hansske, F.; Robins, M.J. Regiospecific and Stereoselective Conversion of Ribonucleosides to 3′-Deoxynucleosides. A High Yield Three-Stage Synthesis of Cordycepin from Adenosine. Tetrahedron Lett. 1985, 26, 4295–4298. [Google Scholar] [CrossRef]
- Aman, S.; Anderson, D.J.; Connolly, T.J.; Crittall, A.J.; Ji, G. From Adenosine to 3′-Deoxyadenosine: Development and Scale Up. Org. Process Res. Dev. 2000, 4, 601–605. [Google Scholar] [CrossRef]
- Duan, X.; Yang, H.; Wang, C.; Liu, H.; Lu, X.; Tian, Y. Microbial Synthesis of Cordycepin, Current Systems and Future Perspectives. Trends Food Sci. Technol. 2023, 132, 162–170. [Google Scholar] [CrossRef]
- Lou, H.; Lin, J.; Guo, L.; Wang, X.; Tian, S.; Liu, C.; Zhao, Y.; Zhao, R. Advances in Research on Cordyceps militaris Degeneration. Appl. Microbiol. Biotechnol. 2019, 103, 7835–7841. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, R.; Wang, S.; Li, C.; Xu, Y.; Li, S.; Li, Q.; Wang, L. Prospects for Cordycepin Biosynthesis in Microbial Cell Factories. Front. Chem. Eng. 2024, 6, 1446454. [Google Scholar] [CrossRef]
- Xia, Y.; Luo, F.; Shang, Y.; Chen, P.; Lu, Y.; Wang, C. Fungal Cordycepin Biosynthesis Is Coupled with the Production of the Safeguard Molecule Pentostatin. Cell Chem. Biol. 2017, 24, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.-Y.; Tian, Y.; Song, Z.-Q.; Song, L.-P.; Lin, W.-B.; Wang, C.; Yang, H.; Lu, X.-Y.; Ji, X.-J.; Liu, H.-H. High-Level de Novo Biosynthesis of Cordycepin by Systems Metabolic Engineering in Yarrowia lipolytica. Bioresour. Technol. 2022, 363, 127862. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Wang, L.; Wang, H.; Cheng, Y.; Li, X.; Wan, H.; Liu, C.; Liu, T.; Li, Q. Engineering Komagataella phaffii to Biosynthesize Cordycepin from Methanol Which Drives Global Metabolic Alterations at the Transcription Level. Synth. Syst. Biotechnol. 2023, 8, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Jeennor, S.; Anantayanon, J.; Panchanawaporn, S.; Chutrakul, C.; Vongsangnak, W.; Laoteng, K. Efficient de Novo Production of Bioactive Cordycepin by Aspergillus oryzae Using a Food-Grade Expression Platform. Microb. Cell Fact. 2023, 22, 253. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Kim, G.B.; Kim, W.J.; Kim, H.U.; Lee, S.Y. Current Status and Applications of Genome-Scale Metabolic Models. Genome Biol. 2019, 20, 121. [Google Scholar] [CrossRef] [PubMed]
- Antonakoudis, A.; Barbosa, R.; Kotidis, P.; Kontoravdi, C. The Era of Big Data: Genome-Scale Modelling Meets Machine Learning. Comput. Struct. Biotechnol. J. 2020, 18, 3287–3330. [Google Scholar] [CrossRef] [PubMed]
- Raethong, N.; Wang, H.; Nielsen, J.; Vongsangnak, W. Optimizing Cultivation of Cordyceps militaris for Fast Growth and Cordycepin Overproduction Using Rational Design of Synthetic Media. Comput. Struct. Biotechnol. J. 2020, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vongsangnak, W.; Olsen, P.; Hansen, K.; Krogsgaard, S.; Nielsen, J. Improved Annotation through Genome-Scale Metabolic Modeling of Aspergillus oryzae. BMC Genomics 2008, 9, 245. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Feizi, A.; Österlund, T.; Hjort, C.; Nielsen, J. Genome-Scale Analysis of the High-Efficient Protein Secretion System of Aspergillus oryzae. BMC Syst. Biol. 2014, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Thammarongtham, C.; Nookaew, I.; Vorapreeda, T.; Srisuk, T.; Land, M.L.; Jeennor, S.; Laoteng, K. Genome Characterization of Oleaginous Aspergillus oryzae BCC7051: A Potential Fungal-Based Platform for Lipid Production. Curr. Microbiol. 2018, 75, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Shuvalov, O.; Petukhov, A.; Daks, A.; Fedorova, O.; Vasileva, E.; Barlev, N.A. One-Carbon Metabolism and Nucleotide Biosynthesis as Attractive Targets for Anticancer Therapy. Oncotarget 2017, 8, 23955–23977. [Google Scholar] [CrossRef] [PubMed]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Weikert, T.; Niehues, A.; Cord-Landwehr, S.; Hellmann, M.J.; Moerschbacher, B.M. Reassessment of Chitosanase Substrate Specificities and Classification. Nat. Commun. 2017, 8, 1698. [Google Scholar] [CrossRef] [PubMed]
- Synowiecki, J.; Al-Khateeb, N.A. Production, Properties, and Some New Applications of Chitin and Its Derivatives. Crit. Rev. Food Sci. Nutr. 2003, 43, 145–171. [Google Scholar] [CrossRef] [PubMed]
- Rinaudo, M. Chitin and Chitosan: Properties and Applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Anantayanon, J.; Chamkhuy, W.; Rattanaphan, N.; Panchanawaporn, S.; Laoteng, K.; Jeennor, S. Enhancing Bioactive Cordycepin Production via Precision Fermentation with an Engineered Aspergillus oryzae. Fermentation 2025, 11, 32. [Google Scholar] [CrossRef]
- Shih, I.; Chang, S.; Chen, Y. Cultivation of Cordyceps militaris in Solid and Liquid Culture. J. Am. Diet. Assoc. 2010, 110, A51. [Google Scholar] [CrossRef]
- Chutrakul, C.; Panchanawaporn, S.; Vorapreeda, T.; Jeennor, S.; Anantayanon, J.; Laoteng, K. The Exploring Functional Role of Ammonium Transporters of Aspergillus oryzae in Nitrogen Metabolism: Challenges towards Cell Biomass Production. Int. J. Mol. Sci. 2022, 23, 7567. [Google Scholar] [CrossRef] [PubMed]
- Hadadi, N.; Pandey, V.; Chiappino-Pepe, A.; Morales, M.; Gallart-Ayala, H.; Mehl, F.; Ivanisevic, J.; Sentchilo, V.; Meer, J.R. van der Mechanistic Insights into Bacterial Metabolic Reprogramming from Omics-Integrated Genome-Scale Models. NPJ Syst. Biol. Appl. 2020, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; van Pelt-KleinJan, E.; van Olst, B.; Douwenga, S.; Boeren, S.; Bachmann, H.; Molenaar, D.; Nielsen, J.; Teusink, B. Proteome Constraints Reveal Targets for Improving Microbial Fitness in Nutrient-rich Environments. Mol. Syst. Biol. 2021, 17, e10093. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Cai, G.; He, Y.; Tong, G. Separation of Cordycepin from Cordyceps militaris Fermentation Supernatant Using Preparative HPLC and Evaluation of Its Antibacterial Activity as an NAD+-Dependent DNA Ligase Inhibitor. Exp. Ther. Med. 2016, 12, 1812–1816. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Marcišauskas, S.; Sánchez, B.J.; Domenzain, I.; Hermansson, D.; Agren, R.; Nielsen, J.; Kerkhoven, E.J. RAVEN 2.0: A Versatile Toolbox for Metabolic Network Reconstruction and a Case Study on Streptomyces coelicolor. PLoS Comput. Biol. 2018, 14, e1006541. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc Database of Metabolic Pathways and Enzymes—A 2019 Update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Agren, R.; Liu, L.; Shoaie, S.; Vongsangnak, W.; Nookaew, I.; Nielsen, J. The RAVEN Toolbox and Its Use for Generating a Genome-Scale Metabolic Model for Penicillium chrysogenum. PLoS Comput. Biol. 2013, 9, e1002980. [Google Scholar] [CrossRef] [PubMed]
- David, H.; Özçelik, İ.Ş.; Hofmann, G.; Nielsen, J. Analysis of Aspergillus nidulans Metabolism at the Genome-Scale. BMC Genomics 2008, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Dreyfuss, J.M.; Zucker, J.D.; Hood, H.M.; Ocasio, L.R.; Sachs, M.S.; Galagan, J.E. Reconstruction and Validation of a Genome-Scale Metabolic Model for the Filamentous Fungus Neurospora crassa Using FARM. PLoS Comput. Biol. 2013, 9, e1003126. [Google Scholar] [CrossRef] [PubMed]
- Vorapreeda, T.; Khongto, B.; Thammarongtham, C.; Srisuk, T.; Laoteng, K. Metabolic Regulation of Sugar Assimilation for Lipid Production in Aspergillus oryzae BCC7051 through Comparative Transcriptome Perspective. Biology 2021, 10, 885. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Worgan, J.T. Lipid Production by Aspergillus oryzae From Starch Substrates. Eur. J. Appl. Microbiol. Biotechnol. 1982, 16, 126–130. [Google Scholar] [CrossRef]
- Lv, G.; Xu, Y.; Tu, Y.; Cheng, X.; Zeng, B.; Huang, J.; He, B. Effects of Nitrogen and Phosphorus Limitation on Fatty Acid Contents in Aspergillus oryzae. Front. Microbiol. 2021, 21, 739569. [Google Scholar] [CrossRef] [PubMed]


| Genomic Characteristics | A. oryzae BCC7051 | A. oryzae RIB40 | C. militaris CM01 |
|---|---|---|---|
| Genome size (Mb) | 38.51 | 37.20 | 32.20 |
| No. of protein-coding genes | 11,456 | 12,096 | 9651 |
| GSMM characteristics | iNR1684 (This study) | iWV1346 | iNR1329 |
| Total genes | 1684 | 1346 | 1329 |
| Total metabolites | 1155 | 810 | 1171 |
| Total reactions | 1947 | 2360 | 1821 |
| - GPR associations (% gap) | 1810 (11) | 2174 (9) | 1684 (14) |
| - Non-GPR associations | 137 | 186 | 137 |
| Compartments | 4 | 4 | 4 |
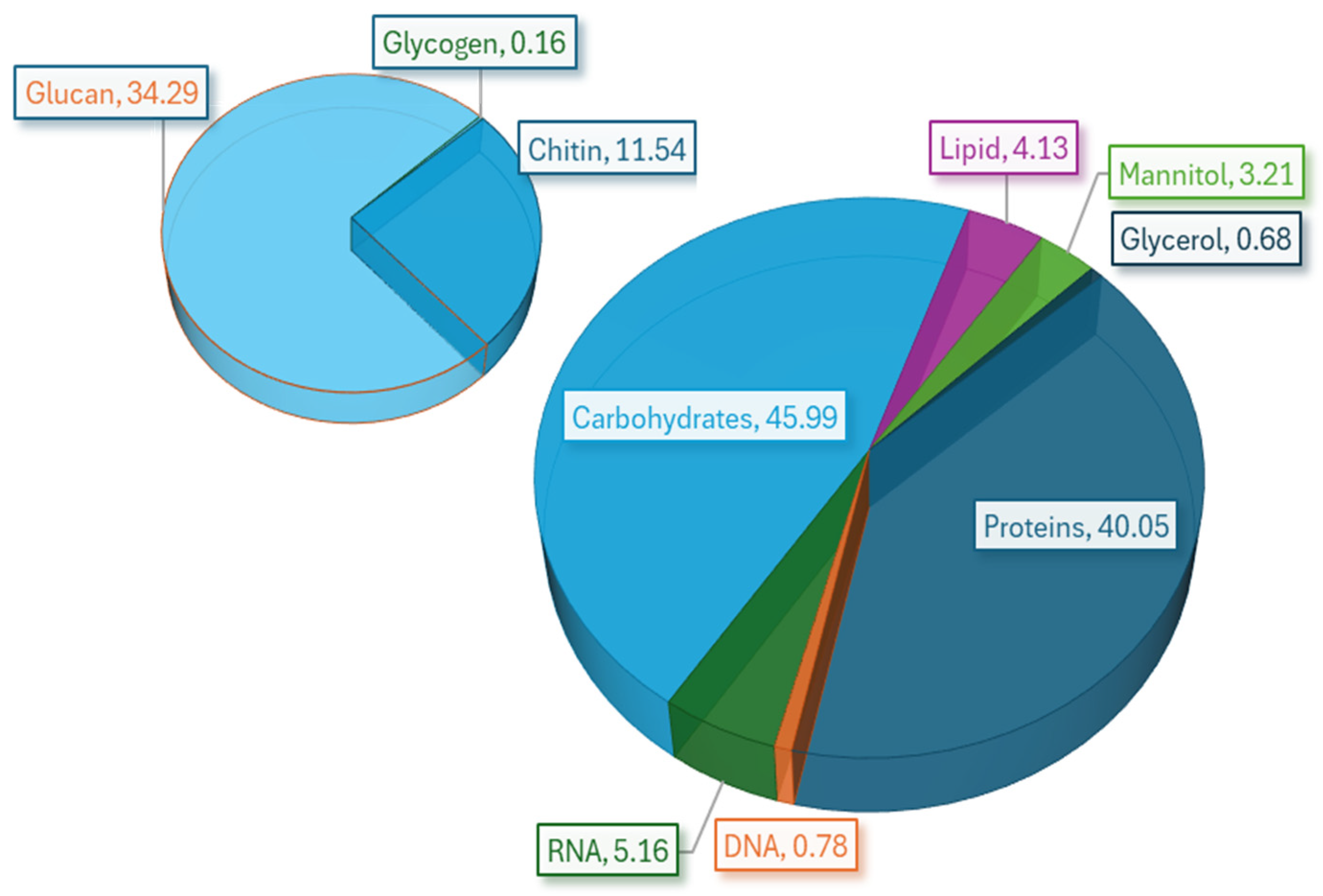
| Biomass Component | Average Molecular Weight (g/mol) | Content (g/100 gDCW) | Stoichiometric Coefficient (mmol/gDCW) |
|---|---|---|---|
| Proteins | 127.03 | 40.05 | 3.153 |
| Carbohydrates | - | - | - |
| Chitin | 203.20 | 11.54 | 0.568 |
| Glucan | 162.10 | 34.29 | 2.115 |
| Glycogen | 666.60 | 0.16 | 0.002 |
| RNA | 495.30 | 5.16 | 0.104 |
| DNA | 482.73 | 0.78 | 0.016 |
| Lipids | - | - | - |
| Triacylglycerol | 821.95 | 1.28 | 0.016 |
| Phosphatidylcholine | 744.76 | 0.15 | 0.002 |
| Phosphatidylethanolamine | 701.67 | 0.06 | 0.001 |
| Palmitic acid (16:0) | 200.32 | 0.17 | 0.009 |
| Oleic acid (18:1 n-9) | 242.40 | 0.15 | 0.006 |
| Stearic acid (18:0) | 239.20 | 0.33 | 0.014 |
| Linoleic acid (18:2 n-6) | 254.41 | 0.63 | 0.025 |
| Arachidic acid (20:0) | 265.30 | 0.01 | 0.0003 |
| Sterol esters | 302.45 | 0.48 | 0.016 |
| Ergosterol | 396.65 | 0.87 | 0.022 |
| Others | - | - | - |
| D-Mannitol | 182.20 | 3.21 | 0.176 |
| Glycerol | 92.10 | 0.68 | 0.074 |
| Parameters | Wild Type | Cordycepin-Producing Strain |
|---|---|---|
| Exponential phase (h) | 24–48 | 24–48 |
| Growth rate, µmax (h−1) | 0.032 ± 0.004 | 0.025 ± 0.002 |
| Biomass production (gDW L−1) | 8.690 ± 0.070 | 5.860 ± 0.170 |
| Sugar uptake rate (mmol gDW−1 h−1) | 0.632 ± 0.042 | 0.549 ± 0.026 |
| Cordycepin production rate (mmol gDW−1 h−1) | - | 0.013 ± 0.001 |
| Cordycepin yield on biomass (mg gDW−1) | - | 81.975 ± 0.005 |
| Cordycepin titer (mg L−1) | - | 479.970 ± 13.590 |
| In silico growth rate (h−1) | 0.032 | 0.025 |
| % error rate | 0.94% | 2.77% |
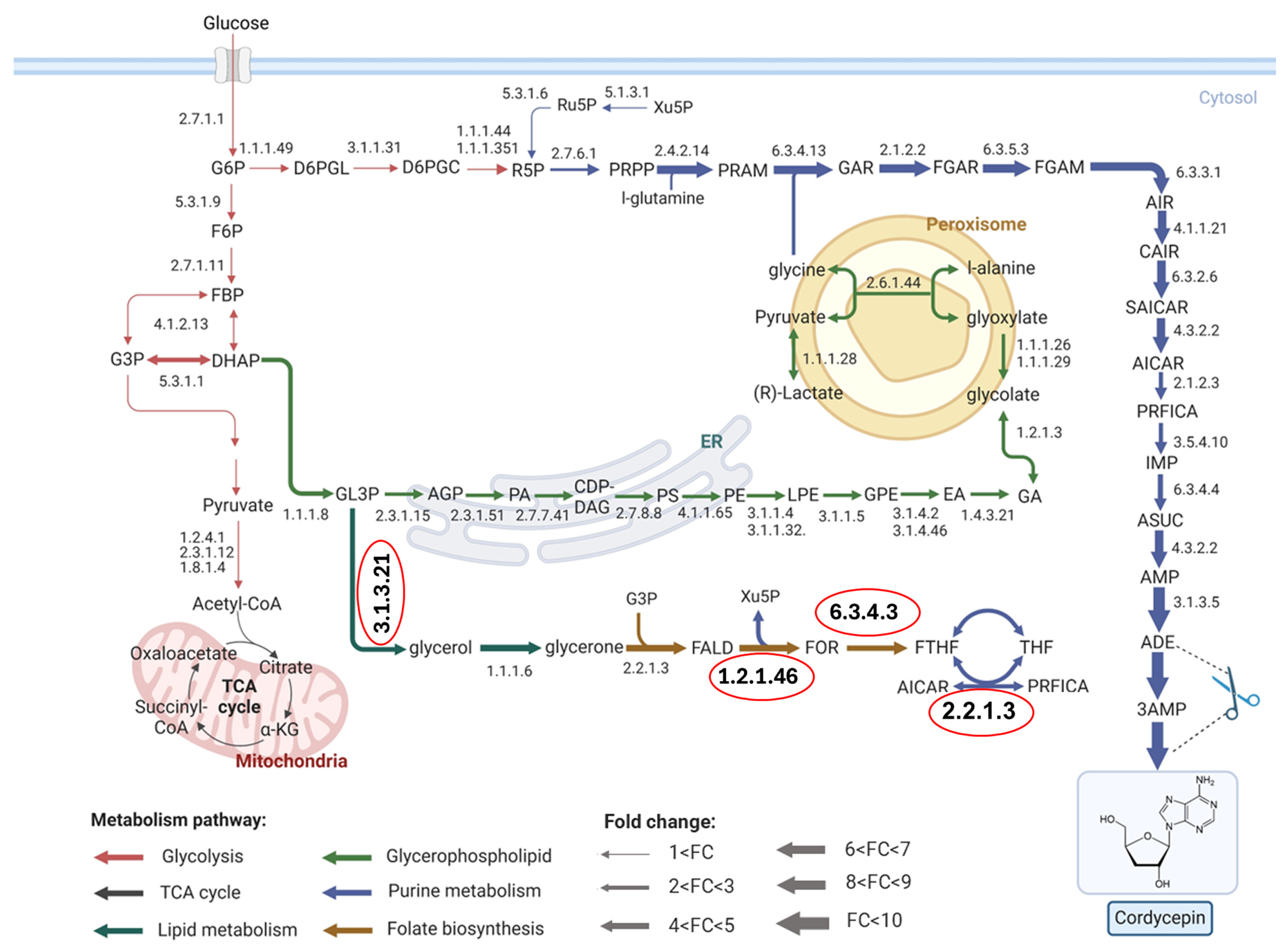
| EC Number | Enzyme Name | GPR Association |
|---|---|---|
| 1.2.1.46 | Formaldehyde dehydrogenase | OAory_01014160 OAory_01017500 OAory_01018710 |
| 2.2.1.3 | Formaldehyde transketolase | OAory_01025450 OAory_01077260 OAory_01105810 |
| 6.3.4.3 | Formate tetrahydrofolate ligase | OAory_01002190 |
| 3.1.3.21 | Glycerol-3-phosphate phosphohydrolase | OAory_01027100 OAory_01104250 |
| Production Data | C:N Ratio | ||||||
|---|---|---|---|---|---|---|---|
| 7.3:1 | 9.7:1 | 11.6:1 | 14.6:1 | 19.4:1 | 29.1:1 | 58.2:1 | |
| Cell biomass (DCW, gL−1) | 20.315 ± 0.276 | 19.544 ± 0.079 | 18.019 ± 0.143 | 16.635 ± 1.557 | 14.097 ± 0.683 | 12.373 ± 0.564 | 9.619 ± 0.284 |
| Biomass productivity (gL−1 h−1) | 0.423 ± 0.006 | 0.407 ± 0.002 | 0.375 ± 0.003 | 0.347 ± 0.032 | 0.294 ± 0.014 | 0.258 ± 0.012 | 0.2 ± 0.006 |
| Cordycepin titer (gL−1) | 1.288 ± 0.017 | 1.366 ± 0.01 | 1.521 ± 0.015 | 1.416 ± 0.025 | 1.35 ± 0.003 | 1.133 ± 0.008 | 0.95 ± 0.003 |
| Cordycepin productivity (gL−1 h−1) | 0.027 ± 0 | 0.028 ± 0 | 0.032 ± 0 | 0.029 ± 0.001 | 0.028 ± 0 | 0.024 ± 0 | 0.02 ± 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raethong, N.; Jeennor, S.; Anantayanon, J.; Wannawilai, S.; Vongsangnak, W.; Laoteng, K. Reconstruction of a Genome-Scale Metabolic Model for Aspergillus oryzae Engineered Strain: A Potent Computational Tool for Enhancing Cordycepin Production. Int. J. Mol. Sci. 2025, 26, 6906. https://doi.org/10.3390/ijms26146906
Raethong N, Jeennor S, Anantayanon J, Wannawilai S, Vongsangnak W, Laoteng K. Reconstruction of a Genome-Scale Metabolic Model for Aspergillus oryzae Engineered Strain: A Potent Computational Tool for Enhancing Cordycepin Production. International Journal of Molecular Sciences. 2025; 26(14):6906. https://doi.org/10.3390/ijms26146906
Chicago/Turabian StyleRaethong, Nachon, Sukanya Jeennor, Jutamas Anantayanon, Siwaporn Wannawilai, Wanwipa Vongsangnak, and Kobkul Laoteng. 2025. "Reconstruction of a Genome-Scale Metabolic Model for Aspergillus oryzae Engineered Strain: A Potent Computational Tool for Enhancing Cordycepin Production" International Journal of Molecular Sciences 26, no. 14: 6906. https://doi.org/10.3390/ijms26146906
APA StyleRaethong, N., Jeennor, S., Anantayanon, J., Wannawilai, S., Vongsangnak, W., & Laoteng, K. (2025). Reconstruction of a Genome-Scale Metabolic Model for Aspergillus oryzae Engineered Strain: A Potent Computational Tool for Enhancing Cordycepin Production. International Journal of Molecular Sciences, 26(14), 6906. https://doi.org/10.3390/ijms26146906







