Selection of Stable Reference Genes for Gene Expression Studies in Activated and Non-Activated PBMCs Under Normoxic and Hypoxic Conditions
Abstract
1. Introduction
2. Results
2.1. Standard Curve and PCR Efficiency
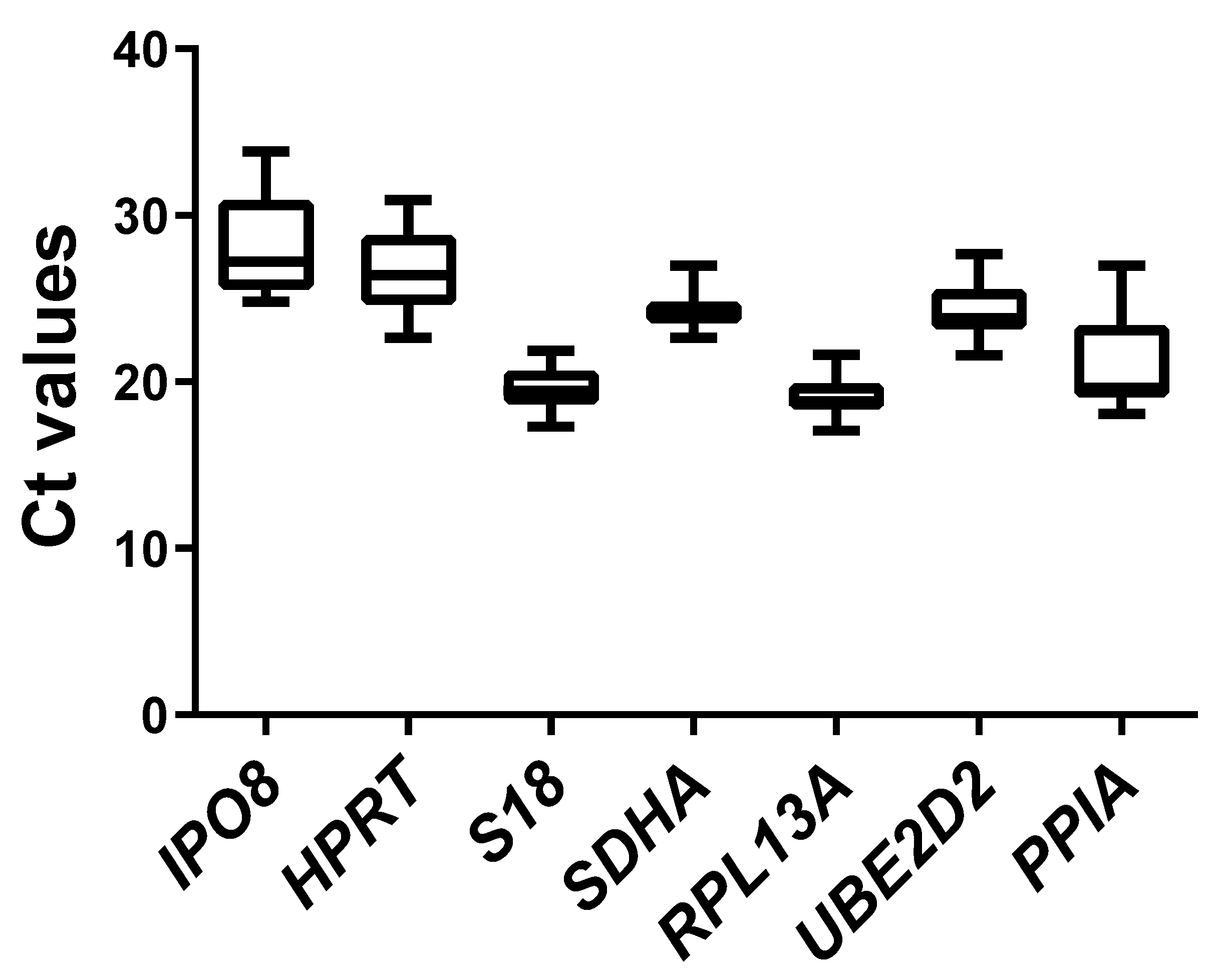
2.2. Expression Profiles of Candidate Reference Genes
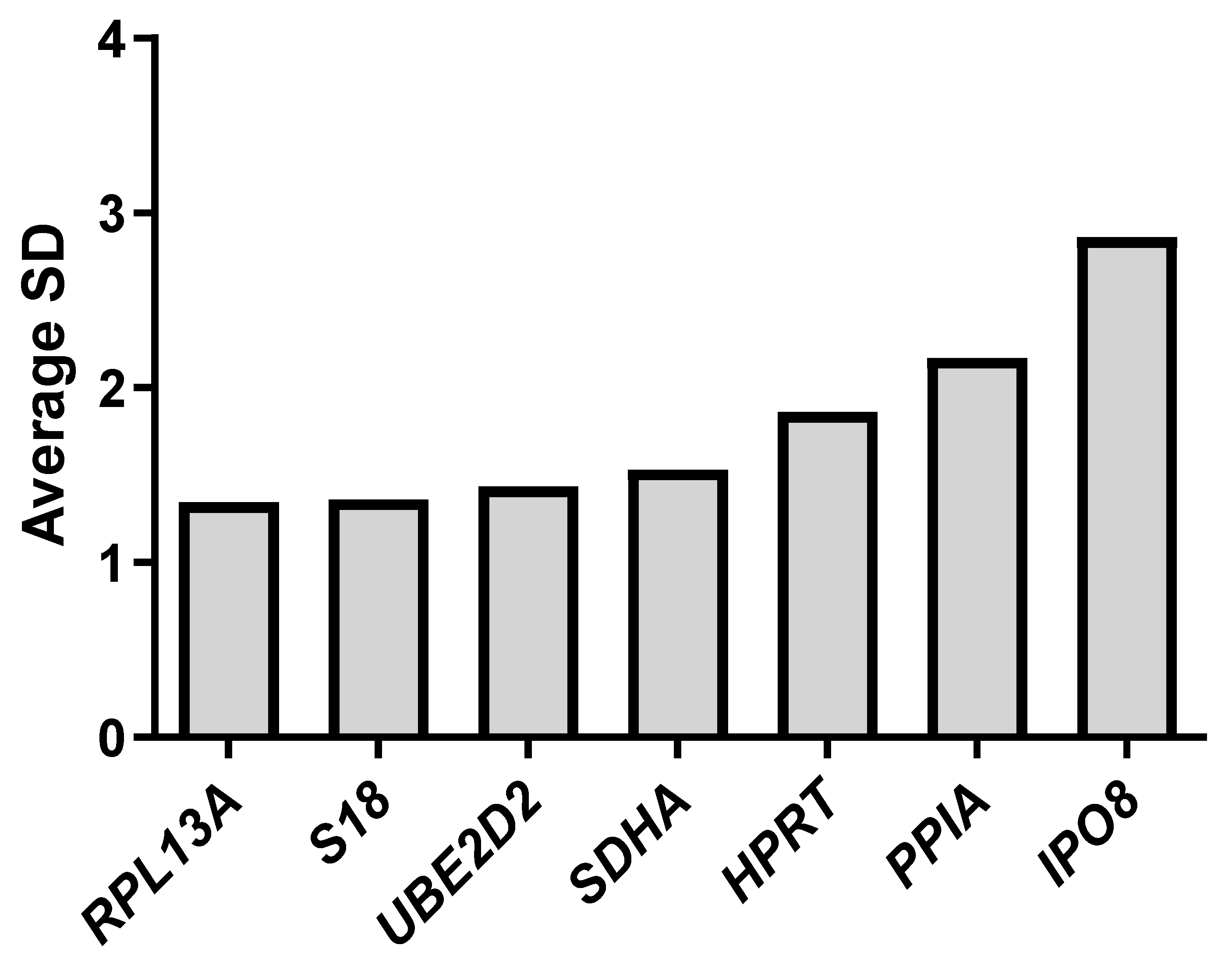
2.3. Evaluation of Gene Expression Stability Using Delta Ct, NormFinder, BestKeeper, GeNorm, and RefFinder
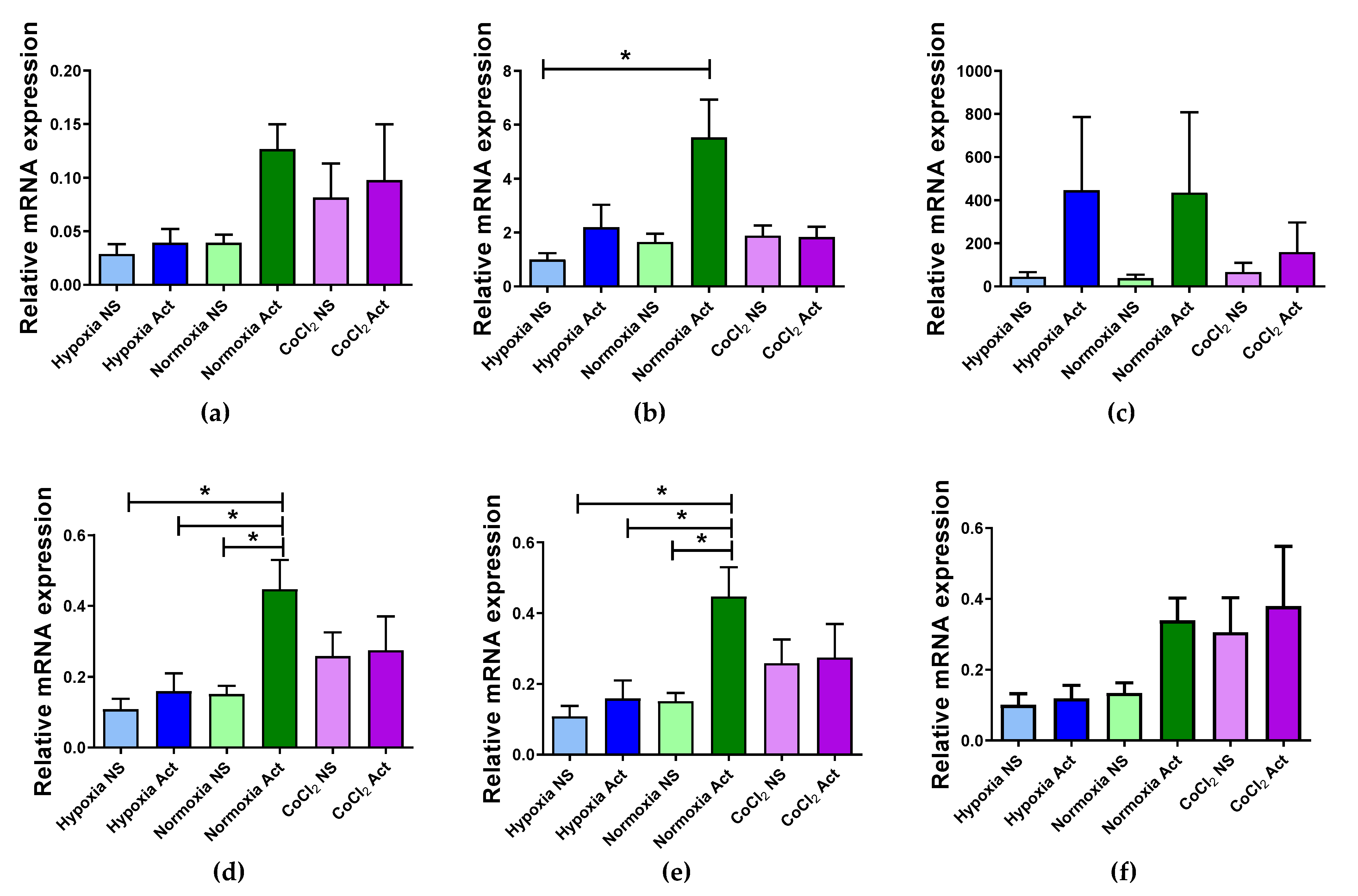
2.4. Validation of Selected Reference Genes
3. Discussion
4. Materials and Methods
4.1. PBMC Isolation
4.2. Cell Culture
4.3. RNA Isolation and cDNA Synthesis
4.4. Primer Design and Quantitative PCR with SYBR Green
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACTB | Actin beta |
| B2M | Beta-2-microglobulin |
| CAR-T | Chimeric antigen receptor T cell |
| CD3/CD28 | Cluster of differentiation 3/Cluster of differentiation 28 |
| CoCl2 | Cobalt (II) chloride |
| COVID-19 | Coronavirus disease 2019 |
| CV | Coefficient of variation |
| DOAJ | Directory of open access journals |
| EEF1A1 | Eukaryotic elongation factor 1 alpha 1 |
| FBS | Fetal bovine serum |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| HIF | Hypoxia-inducible factors |
| HPRT | Hypoxanthine phosphoribosyltransferase 1 |
| IPO8 | Importin 8 |
| IQR | Interquartile range |
| LD | Linear dichroism |
| LPS | Lipopolysaccharide |
| MDPI | Multidisciplinary Digital Publishing Institute |
| NS | Non-stimulated |
| PBL | Peripheral blood lymphocytes |
| PBMCs | Peripheral blood mononuclear cells |
| PBS | Phosphate buffered saline |
| PPIA | Peptidylprolyl isomerase A |
| qPCR | Quantitative polymerase chain reaction |
| RGs | Reference genes |
| RPL13A | Ribosomal protein L13a |
| RPL18 | Ribosomal protein L18 |
| RPL19 | Ribosomal protein L19 |
| RPL27 | Ribosomal protein L27 |
| RPL4 | Ribosomal protein L4 |
| RPLP0 | Ribosomal protein lateral stalk subunit P0 |
| RPS15 | Ribosomal protein S15 |
| RT | Room temperature |
| RT-qPCR | Quantitative reverse transcription polymerase chain reaction |
| S18 | Ribosomal protein S18 |
| SD | Standard deviation |
| SDHA | Succinate dehydrogenase complex flavoprotein subunit A |
| SYBR | SYnergy BRands Green I |
| TME | Tumor microenvironment |
| TPP | Tripeptidyl peptidase |
| UBE2D2 | Ubiquitin conjugating enzyme E2 D2 |
References
- Lahiri, A.; Maji, A.; Potdar, P.D.; Singh, N.; Parikh, P.; Bisht, B.; Mukherjee, A.; Paul, M.K. Lung Cancer Immunotherapy: Progress, Pitfalls, and Promises. Mol. Cancer 2023, 22, 40. [Google Scholar] [CrossRef] [PubMed]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A Guide to Cancer Immunotherapy: From T Cell Basic Science to Clinical Practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, M.; Zhang, D.; Chen, M.; Zhu, D. Clinical Cancer Immunotherapy: Current Progress and Prospects. Front. Immunol. 2022, 13, 961805. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.R.; Wu, X.L.; Sun, Y.L. Therapeutic Targets and Biomarkers of Tumor Immunotherapy: Response versus Non-Response. Signal Transduct. Target. Ther. 2022, 7, 331. [Google Scholar] [CrossRef] [PubMed]
- Desai, I.; Thakur, S.; Pagariya, P. Current Advances in Immunotherapy for Cancer. Oral Oncol. Rep. 2024, 12, 100652. [Google Scholar] [CrossRef]
- Augustin, R.C.; Delgoffe, G.M.; Najjar, Y.G. Characteristics of the Tumor Microenvironment That Influence Immune Cell Functions: Hypoxia, Oxidative Stress, Metabolic Alterations. Cancers 2020, 12, 3802. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.; Gaiteiro, C.; Peixoto, A.; Soares, J.; Neves, M.; Santos, L.L.; Ferreira, J.A. Reference Genes for Addressing Gene Expression of Bladder Cancer Cell Models under Hypoxia: A Step towards Transcriptomic Studies. PLoS ONE 2016, 11, e0166120. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.A.; Peixoto, A.; Neves, M.; Gaiteiro, C.; Reis, C.A.; Assaraf, Y.G.; Santos, L.L. Mechanisms of Cisplatin Resistance and Targeting of Cancer Stem Cells: Adding Glycosylation to the Equation. Drug Resist. Updates 2016, 24, 34–54. [Google Scholar] [CrossRef] [PubMed]
- Chouaib, S.; Noman, M.Z.; Kosmatopoulos, K.; Curran, M.A. Hypoxic Stress: Obstacles and Opportunities for Innovative Immunotherapy of Cancer. Oncogene 2017, 36, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Najafi, M.; Farhood, B.; Mortezaee, K.; Kharazinejad, E.; Majidpoor, J.; Ahadi, R. Hypoxia in Solid Tumors: A Key Promoter of Cancer Stem Cell (CSC) Resistance. J. Cancer Res. Clin. Oncol. 2020, 146, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Challapalli, A.; Carroll, L.; Aboagye, E.O. Molecular Mechanisms of Hypoxia in Cancer. Clin. Transl. Imaging 2017, 5, 225–253. [Google Scholar] [CrossRef] [PubMed]
- Triner, D.; Shah, Y.M. Hypoxia-Inducible Factors: A Central Link between Inflammation and Cancer. J. Clin. Investig. 2016, 126, 3689–3698. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Sharma, A.; Sodhi, M.; Tiwari, M.; Vivek, P.; Kataria, R.S.; Nirajan, S.K.; Bharti, V.K.; Singh, P.; Lathwal, S.S.; et al. Identification of Internal Reference Genes in Peripheral Blood Mononuclear Cells of Cattle Populations Adapted to Hot Arid Normoxia and Cold Arid Hypoxia Environments. Front. Genet. 2022, 12, 730599. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Sodhi, M.; Verma, P.; Vivek, P.; Kataria, R.S.; Niranjan, S.K.; Bharti, V.K.; Masharing, N.; Gujar, G.; Chanda, D.; et al. Selection of Species Specific Panel of Reference Genes in Peripheral Blood Mononuclear Cells of Native Livestock Species Adapted to Trans-Himalayan Region of Leh-Ladakh. Sci. Rep. 2022, 12, 18473. [Google Scholar] [CrossRef] [PubMed]
- Hannemann, J.; Schmidt-Hutten, L.; Hannemann, J.; Kleinsang, F.; Böger, R. Selection of Reference Genes for Normalization of Gene Expression After Exposure of Human Endothelial and Epithelial Cells to Hypoxia. Int. J. Mol. Sci. 2025, 26, 1763. [Google Scholar] [CrossRef] [PubMed]
- Moein, S.; Javanmard, S.; Abedi, M.; Izadpanahi, M.; Gheisari, Y. Identification of Appropriate Housekeeping Genes for Gene Expression Analysis in Long-Term Hypoxia-Treated Kidney Cells. Adv. Biomed. Res. 2017, 6, 15. [Google Scholar] [CrossRef]
- Malcolm, J.R.; Bridge, K.S.; Holding, A.N.; Brackenbury, W.J. Identification of Robust RT-QPCR Reference Genes for Studying Changes in Gene Expression in Response to Hypoxia in Breast Cancer Cell Lines. BMC Genom. 2025, 26, 59. [Google Scholar] [CrossRef] [PubMed]
- De Backer, J.; Maric, D.; Bosman, M.; Dewilde, S.; Hoogewijs, D. A Reliable Set of Reference Genes to Normalize Oxygen-Dependent Cytoglobin Gene Expression Levels in Melanoma. Sci. Rep. 2021, 11, 10879. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.G.; McElhaney, J.E.; Verschoor, C.P. Reliable Reference Genes for the Quantification of MRNA in Human T-Cells and PBMCs Stimulated with Live Influenza Virus. BMC Immunol. 2020, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; He, S.; Wu, Y.; Chen, W.; Song, J.; Zhu, Y.; Chen, H.; Wang, Q.; Wang, S.; Tan, S.; et al. Validation of Reference Genes for the Normalization of the RT-QPCR in Peripheral Blood Mononuclear Cells of Septic Patients. Heliyon 2023, 9, e15269. [Google Scholar] [CrossRef] [PubMed]
- Oturai, D.B.; Søndergaard, H.B.; Börnsen, L.; Sellebjerg, F.; Romme Christensen, J. Identification of Suitable Reference Genes for Peripheral Blood Mononuclear Cell Subset Studies in Multiple Sclerosis. Scand. J. Immunol. 2016, 83, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Miglioranza Scavuzzi, B.; Abcouwer, S.F.; Zacks, D.N. A Simplified Protocol to Induce Hypoxia in a Standard Incubator: A Focus on Retinal Cells. Exp. Eye Res. 2023, 236, 109653. [Google Scholar] [CrossRef] [PubMed]
- Ledderose, C.; Heyn, J.; Limbeck, E.; Kreth, S. Selection of Reliable Reference Genes for Quantitative Real-Time PCR in Human T Cells and Neutrophils. BMC Res. Notes 2011, 4, 427. [Google Scholar] [CrossRef] [PubMed]
- Henig, N.; Avidan, N.; Mandel, I.; Staun-Ram, E.; Ginzburg, E.; Paperna, T.; Pinter, R.Y.; Miller, A. Interferon-Beta Induces Distinct Gene Expression Response Patterns in Human Monocytes versus T Cells. PLoS ONE 2013, 8, e62366. [Google Scholar] [CrossRef] [PubMed]
- Radke, L.; Giese, C.; Lubitz, A.; Hinderlich, S.; Sandig, G.; Hummel, M.; Frohme, M. Reference Gene Stability in Peripheral Blood Mononuclear Cells Determined by QPCR and NanoString. Microchim. Acta 2014, 181, 1733–1742. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, Z.C.; Xu, J.; Ji, A.J.; Luo, H.M.; Song, J.Y.; Sun, C.; Hu, Y.L.; Chen, S.L. Selection and Validation of Reference Genes for Normalization of Quantitative Real-Time Reverse Transcription PCR Analysis in Poria Cocos (Schw.) Wolf (Fuling). Chin. Med. 2016, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chen, J.X.; Yang, S.; Fu, X.P.; Zhang, Z.; Chen, K.H.; Huang, Y.; Li, Y.; Xie, Y.; Mao, Y.M. Selection of Reliable Reference Genes for Gene Expression Study in Nasopharyngeal Carcinoma. Acta Pharmacol. Sin. 2010, 31, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Riedel, G.; Rüdrich, U.; Fekete-Drimusz, N.; Manns, M.P.; Vondran, F.W.R.; Bock, M. An Extended ΔCT-Method Facilitating Normalisation with Multiple Reference Genes Suited for Quantitative RT-PCR Analyses of Human Hepatocyte-like Cells. PLoS ONE 2014, 9, e93031. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lai, Q.; He, C.; Fang, Y.; Yan, Q.; Zhang, Y.; Wang, X.; Gu, C.; Wang, Y.; Ye, L.; et al. RUNX1 Promotes Tumour Metastasis by Activating the Wnt/β-Catenin Signalling Pathway and EMT in Colorectal Cancer. J. Exp. Clin. Cancer Res. 2019, 38, 334. [Google Scholar] [CrossRef] [PubMed]
- Carbonneau, M.; Gagne, L.M.; Lalonde, M.E.; Germain, M.A.; Motorina, A.; Guiot, M.C.; Secco, B.; Vincent, E.E.; Tumber, A.; Hulea, L.; et al. The Oncometabolite 2-Hydroxyglutarate Activates the MTOR Signalling Pathway. Nat. Commun. 2016, 7, 12700. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- El Hadi, H.; Abdellaoui-Maane, I.; Kottwitz, D.; El Amrani, M.; Bouchoutrouch, N.; Qmichou, Z.; Karkouri, M.; ElAttar, H.; Errihani, H.; Fernandez, P.L.; et al. Development and Evaluation of a Novel RT-QPCR Based Test for the Quantification of HER2 Gene Expression in Breast Cancer. Gene 2017, 605, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Svingen, T.; Letting, H.; Hadrup, N.; Hass, U.; Vinggaard, A.M. Selection of Reference Genes for Quantitative RT-PCR (RT-QPCR) Analysis of Rat Tissues under Physiological and Toxicological Conditions. PeerJ 2015, 3, e855. [Google Scholar] [CrossRef] [PubMed]
- Taïhi, I.; Nassif, A.; Berbar, T.; Isaac, J.; Berdal, A.; Gogly, B.; Fournier, B.P. Validation of Housekeeping Genes to Study Human Gingival Stem Cells and Their in Vitro Osteogenic Differentiation Using Real-Time RT-QPCR. Stem Cells Int. 2016, 2016, 6261490. [Google Scholar] [CrossRef] [PubMed]
- Dzaki, N.; Ramli, K.N.; Azlan, A.; Ishak, I.H.; Azzam, G. Evaluation of Reference Genes at Different Developmental Stages for Quantitative Real-Time PCR in Aedes Aegypti. Sci. Rep. 2017, 7, srep43618. [Google Scholar] [CrossRef] [PubMed]
- Joehanes, R.; Johnson, A.D.; Barb, J.J.; Raghavachari, N.; Liu, P.; Woodhouse, K.A.; O’Donnell, C.J.; Munson, P.J.; Levy, D. Gene Expression Analysis of Whole Blood, Peripheral Blood Mononuclear Cells, and Lymphoblastoid Cell Lines from the Framingham Heart Study. Physiol. Genom. 2012, 44, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, T.; Klenow, S.; Borowicki, A.; Gill, C.I.R.; Pool-Zobel, B.L.; Glei, M. Gene Expression Profiles in Human Peripheral Blood Mononuclear Cells as Biomarkers for Nutritional in Vitro and in Vivo Investigations. Genes Nutr. 2010, 5, 309–319. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, C.; Iommelli, F.; De Rosa, V.; Ercolano, G.; Sodano, F.; Tuccillo, C.; Amato, L.; Tirino, V.; Ariano, A.; Cimmino, F.; et al. PBMCs as Tool for Identification of Novel Immunotherapy Biomarkers in Lung Cancer. Biomedicines 2024, 12, 809. [Google Scholar] [CrossRef] [PubMed]
- Lê, H.; Deforges, J.; Hua, G.; Idoux-Gillet, Y.; Ponté, C.; Lindner, V.; Olland, A.; Falcoz, P.E.; Zaupa, C.; Jain, S.; et al. In Vitro Vascularized Immunocompetent Patient-Derived Model to Test Cancer Therapies. iScience 2023, 26, 108094. [Google Scholar] [CrossRef] [PubMed]
- Geigges, M.; Gubser, P.M.; Unterstab, G.; Lecoultre, Y.; Paro, R.; Hess, C. Reference Genes for Expression Studies in Human CD8+ Naïve and Effector Memory T Cells under Resting and Activating Conditions. Sci. Rep. 2020, 10, 9411. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Q.; Bai, J.; Xuan, Y.; Wang, Y. Evaluation of Eight Reference Genes for Quantitative Polymerase Chain Reaction Analysis in Human T Lymphocytes Co-Cultured with Mesenchymal Stem Cells. Mol. Med. Rep. 2015, 12, 7721–7727. [Google Scholar] [CrossRef] [PubMed]
- Curtis, K.M.; Gomez, L.A.; Rios, C.; Garbayo, E.; Raval, A.P.; Perez-Pinzon, M.A.; Schiller, P.C. EF1α and RPL13a Represent Normalization Genes Suitable for RT-QPCR Analysis of Bone Marrow Derived Mesenchymal Stem Cells. BMC Mol. Biol. 2010, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Jeon, R.H.; Lee, W.J.; Son, Y.B.; Bharti, D.; Shivakumar, S.B.; Lee, S.L.; Rho, G.J. PPIA, HPRT1, and YWHAZ Genes Are Suitable for Normalization of MRNA Expression in Long-Term Expanded Human Mesenchymal Stem Cells. Biomed. Res. Int. 2019, 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Röhn, G.; Koch, A.; Krischek, B.; Stavrinou, P.; Goldbrunner, R.; Timmer, M. ACTB and SDHA Are Suitable Endogenous Reference Genes for Gene Expression Studies in Human Astrocytomas Using Quantitative RT-PCR. Technol. Cancer Res. Treat. 2018, 17, 1533033818802318. [Google Scholar] [CrossRef] [PubMed]
- Panina, Y.; Germond, A.; Watanabe, T.M. Analysis of the Stability of 70 Housekeeping Genes during IPS Reprogramming. Sci. Rep. 2020, 10, 21711. [Google Scholar] [CrossRef] [PubMed]
- Iyer, G.; Wang, A.R.; Brennan, S.R.; Bourgeois, S.; Armstrong, E.; Shah, P.; Harari, P.M. Identification of Stable Housekeeping Genes in Response to Ionizing Radiation in Cancer Research. Sci. Rep. 2017, 7, srep43763. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Kreth, S.; Heyn, J.; Grau, S.; Kretzschmar, H.A.; Egensperger, R.; Kreth, F.W. Identification of Valid Endogenous Control Genes for Determining Gene Expression in Human Glioma. Neuro Oncol. 2010, 12, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper—Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Hurtado Del Pozo, C.; Calvo, R.M.; Vesperinas-García, G.; Gómez-Ambrosi, J.; Frühbeck, G.; Corripio-Sánchez, R.; Rubio, M.A.; Obregon, M.J. IPO8 and FBXL10: New Reference Genes for Gene Expression Studies in Human Adipose Tissue. Obesity 2010, 18, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Nguewa, P.A.; Agorreta, J.; Blanco, D.; Lozano, M.D.; Gomez-Roman, J.; Sanchez, B.A.; Valles, I.; Pajares, M.J.; Pio, R.; Rodriguez, M.J.; et al. Identification of Importin 8 (IPO8) as the Most Accurate Reference Gene for the Clinicopathological Analysis of Lung Specimens. BMC Mol. Biol. 2008, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Rácz, G.A.; Nagy, N.; Tóvári, J.; Apáti, Á.; Vértessy, B.G. Identification of New Reference Genes with Stable Expression Patterns for Gene Expression Studies Using Human Cancer and Normal Cell Lines. Sci. Rep. 2021, 11, 19459. [Google Scholar] [CrossRef] [PubMed]
- Tóth, O.; Rácz, G.A.; Oláh, E.; Tóth, M.; Szabó, E.; Várady, G.; Vértessy, B.G.; Nagy, N. Identification of New Reference Genes with Stable Expression Patterns for Cell Cycle Experiments in Human Leukemia Cell Lines. Sci. Rep. 2025, 15, 1052. [Google Scholar] [CrossRef] [PubMed]
- Bantulà, M.; Arismendi, E.; Picado, C.; Mullol, J.; Roca-Ferrer, J.; Tubita, V. Reference Gene Validation for RT–QPCR in PBMCs from Asthmatic Patients with or without Obesity. Methods Protoc. 2022, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Cinar, M.U.; Islam, M.A.; Pröll, M.; Kocamis, H.; Tholen, E.; Tesfaye, D.; Looft, C.; Schellander, K.; Uddin, M.J. Evaluation of Suitable Reference Genes for Gene Expression Studies in Porcine PBMCs in Response to LPS and LTA. BMC Res. Notes 2013, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Miao, Y.; Wang, Y.; Shan, Q. Hprt Serves as an Ideal Reference Gene for QRT-PCR Normalization in Rat DRG Neurons. J. Integr. Neurosci. 2023, 22, 125. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.H.; Patrizi, A. Assessment of Common Housekeeping Genes as Reference for Gene Expression Studies Using RT-QPCR in Mouse Choroid Plexus. Sci. Rep. 2021, 11, 3278. [Google Scholar] [CrossRef] [PubMed]
- Townsend, M.H.; Felsted, A.M.; Ence, Z.E.; Piccolo, S.R.; Robison, R.A.; O’Neill, K.L. Falling from Grace: HPRT Is Not Suitable as an Endogenous Control for Cancer-Related Studies. Mol. Cell. Oncol. 2019, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Drzewiecka, E.M.; Liszewska, E.; Kozłowski, K.; Ciereszko, A.; Słowińska, M. Research Note: Reference Genes Selection for Gene Expression Analyses in Reproductive Turkey (Meleagris Gallopavo) with Yellow Semen Syndrome. Poult. Sci. 2025, 104, 105093. [Google Scholar] [CrossRef] [PubMed]
- Røge, R.; Thorsen, J.; Tørring, C.; Øzbay, A.; Møller, B.K.; Carstens, J. Commonly Used Reference Genes Are Actively Regulated in in Vitro Stimulated Lymphocytes. Scand. J. Immunol. 2007, 65, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Baddela, V.S.; Baufeld, A.; Yenuganti, V.R.; Vanselow, J.; Singh, D. Suitable Housekeeping Genes for Normalization of Transcript Abundance Analysis by Real-Time RT-PCR in Cultured Bovine Granulosa Cells during Hypoxia and Differential Cell Plating Density. Reprod. Biol. Endocrinol. 2014, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Facci, M.R.; Auray, G.; Meurens, F.; Buchanan, R.; van Kessel, J.; Gerdts, V. Stability of Expression of Reference Genes in Porcine Peripheral Blood Mononuclear and Dendritic Cells. Vet. Immunol. Immunopathol. 2011, 141, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Volland, M.; Blasco, J.; Hampel, M. Validation of Reference Genes for RT-QPCR in Marine Bivalve Ecotoxicology: Systematic Review and Case Study Using Copper Treated Primary Ruditapes Philippinarum Hemocytes. Aquat. Toxicol. 2017, 185, 86–94. [Google Scholar] [CrossRef] [PubMed]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference Gene Validation for RT-QPCR, a Note on Different Available Software Packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef] [PubMed]
- Bollinger, T.; Bollinger, A.; Gies, S.; Feldhoff, L.; Solbach, W.; Rupp, J. Transcription Regulates HIF-1α Expression in CD4+ T Cells. Immunol. Cell Biol. 2016, 94, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Rius, J.; Guma, M.; Schachtrup, C.; Akassoglou, K.; Zinkernagel, A.S.; Nizet, V.; Johnson, R.S.; Haddad, G.G.; Karin, M. NF-ΚB Links Innate Immunity to the Hypoxic Response through Transcriptional Regulation of HIF-1α. Nature 2008, 453, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Simon, M.C. Hypoxia-Inducible Factors: Key Regulators of Myeloid Cells during Inflammation. J. Clin. Investig. 2016, 126, 3661–3671. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Ojo, O.A.; Ding, H.; Mullen, L.J.; Xing, C.; Hossain, M.I.; Yassin, A.; Shi, V.Y.; Lewis, Z.; Podgorska, E.; et al. HIF1α-Regulated Glycolysis Promotes Activation-Induced Cell Death and IFN-γ Induction in Hypoxic T Cells. Nat. Commun. 2024, 15, 9394. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.Z.; Wang, R.; Huang, G.; Vogel, P.; Neale, G.; Green, D.R.; Chi, H. HIF1α-Dependent Glycolytic Pathway Orchestrates a Metabolic Checkpoint for the Differentiation of TH17 and Treg Cells. J. Exp. Med. 2011, 208, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Méndez, D.; Huerta, L.; Villarreal, C. Modeling the Effect of Environmental Cytokines, Nutrient Conditions and Hypoxia on CD4+ T Cell Differentiation. Front. Immunol. 2022, 13, 962175. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. MiRDeepFinder: A MiRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]


| Abbreviation | Gene Name | NCBI Accession Number | Gene Function * |
|---|---|---|---|
| HPRT | Hypoxanthine Phosphoribosyltransferase 1 | NM_000194 | Involved in the purine salvage pathway |
| S18 | Ribosomal Protein S18 | NM_022551.3 | Initiation and fidelity of translation as a core component of the small ribosomal subunit |
| IPO8 | Importin 8 | NM_001190995.2 | Nuclear transport receptor mediates the proteins and ribonucleoproteins into the nucleus |
| RPL13A | Ribosomal Protein L13a | NM_001270491.2 | Contributes to protein synthesis by participating in the assembly and function of ribosomes |
| UBE2D2 | Ubiquitin Conjugating Enzyme E2 D2 | NM_181838.2 | Part of the ubiquitination system, acts as an E2 ubiquitin-conjugating enzyme and works with E3 ligases to attach ubiquitin to target proteins/regulates protein turnover, cell cycle progression, and responses to stress |
| PPIA | Peptidylprolyl Isomerase A | NM_001300981.2 | Protein folding, intracellular signaling, inflammation, and viral infection processes |
| SDHA | Succinate Dehydrogenase Complex Flavoprotein Subunit A | NM_001294332.2 | Catalyzes the oxidation of succinate to fumarate and transfers electrons to the electron transport chain |
| Gene | Primer Sequence (5′-3′) | Efficiency [%] | R2 | Slope | Amplicon Size (bp) | Source |
|---|---|---|---|---|---|---|
| HPRT | F: CCTGGCGTCGTGATTAGTGAT R: AGACGTTCAGTCCTGTCCATAA | 113.5 | 0.992 | −3.036 | 131 | de novo |
| S18 | F: TGGTCTGGACAACAAGCTCC R: GAAGTGACGCAGCCCTCTAT | 103.93 | 0.996 | −3.231 | 76 | de novo |
| IPO8 | F: GTGTAAGCTTCGTGAGGGC R: TGTGAGTTGCAGAAGACGGA | 91.13 | 0.996 | −3.555 | 108 | de novo |
| RPL13A | F: AAAAGCGGATGGTGGTTCCT R: GCTGTCACTGCCTGGTACTT | 98.32 | 0.999 | −3.363 | 118 | [21] |
| UBE2D2 | F: ATTGAATGATCTGGCACGGG R: GTCATTTGGCCCCATTATTG | 92.07 | 0.997 | −3.528 | 100 | [25] |
| PPIA | F:TGAGAACTTCATCCTAAAGCATAC R: CATCCAACCACTCAGTCTTG | 113.6 | 0.999 | −3.034 | 116 | [26] |
| SDHA | F: TATATGGAAGGTCTCTGCGA R: GTGTTCTTTGCTCTTATGCG | 99.77 | 0.987 | −3.328 | 145 | [26] |
| IPO8 | HPRT | S18 | SDHA | RPL13A | UBE2D2 | PPIA | |
|---|---|---|---|---|---|---|---|
| Geometric mean [CP] | 28.04 | 26.53 | 19.53 | 24.22 | 19.03 | 24.23 | 20.85 |
| Arithmetic mean [CP] | 28.19 | 26.64 | 19.57 | 24.24 | 19.07 | 24.28 | 20.99 |
| Minimum [CP] | 24.82 | 22.64 | 17.33 | 22.65 | 17.10 | 21.60 | 18.08 |
| Maximum [CP] | 33.86 | 30.90 | 21.89 | 26.99 | 21.63 | 27.67 | 26.99 |
| Standard deviation [±CP] | 2.76 | 1.95 | 1.07 | 0.83 | 0.97 | 1.35 | 2.19 |
| Coefficient variation [%CP] | 9.79 | 7.33 | 5.49 | 3.44 | 5.10 | 5.56 | 10.43 |
| Minimum [x-fold] | −6.88 | −24.38 | −4.99 | −2.95 | −3.71 | −4.98 | −9.65 |
| Maximum [x-fold] | 32.68 | 35.98 | 5.61 | 6.76 | 5.79 | 8.16 | 155.29 |
| Standard deviation [±x-fold] | 5.22 | 3.22 | 1.90 | 1.65 | 1.79 | 2.25 | 3.71 |
| Ranking Order (Better-Good-Average) | |||||||
|---|---|---|---|---|---|---|---|
| Conditions | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| Normoxia | RPL13A | UBE2D2 | SDHA | S18 | HPRT | PPIA | IPO8 |
| Hypoxia | RPL13A | UBE2D2 | SDHA | S18 | HPRT | PPIA | IPO8 |
| Chemical hypoxia | RPL13A | S18 | HPRT | UBE2D2 | SDHA | IPO8 | PPIA |
| Recommended comprehensive ranking | RPL13A | UBE2D2 | S18 | SDHA | HPRT | PPIA | IPO8 |
| Donor No. | Sex | Age | Blood Type | Rh (D) |
|---|---|---|---|---|
| NDA | Male | 44 | B | + |
| NDB | Male | 54 | A | − |
| NDC | Male | 33 | B | + |
| NDD | Male | 33 | 0 | + |
| NDE | Male | 45 | 0 | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wardaszka, A.; Smolarska, A.; Bednarczyk, P.; Bujak, J.K. Selection of Stable Reference Genes for Gene Expression Studies in Activated and Non-Activated PBMCs Under Normoxic and Hypoxic Conditions. Int. J. Mol. Sci. 2025, 26, 6790. https://doi.org/10.3390/ijms26146790
Wardaszka A, Smolarska A, Bednarczyk P, Bujak JK. Selection of Stable Reference Genes for Gene Expression Studies in Activated and Non-Activated PBMCs Under Normoxic and Hypoxic Conditions. International Journal of Molecular Sciences. 2025; 26(14):6790. https://doi.org/10.3390/ijms26146790
Chicago/Turabian StyleWardaszka, Artur, Anna Smolarska, Piotr Bednarczyk, and Joanna Katarzyna Bujak. 2025. "Selection of Stable Reference Genes for Gene Expression Studies in Activated and Non-Activated PBMCs Under Normoxic and Hypoxic Conditions" International Journal of Molecular Sciences 26, no. 14: 6790. https://doi.org/10.3390/ijms26146790
APA StyleWardaszka, A., Smolarska, A., Bednarczyk, P., & Bujak, J. K. (2025). Selection of Stable Reference Genes for Gene Expression Studies in Activated and Non-Activated PBMCs Under Normoxic and Hypoxic Conditions. International Journal of Molecular Sciences, 26(14), 6790. https://doi.org/10.3390/ijms26146790






