AlphaFold 3-Assisted Deciphering of the DNA Recognition by DREB1 Transcription Factors in Rice
Abstract
1. Introduction
2. Results and Discussion
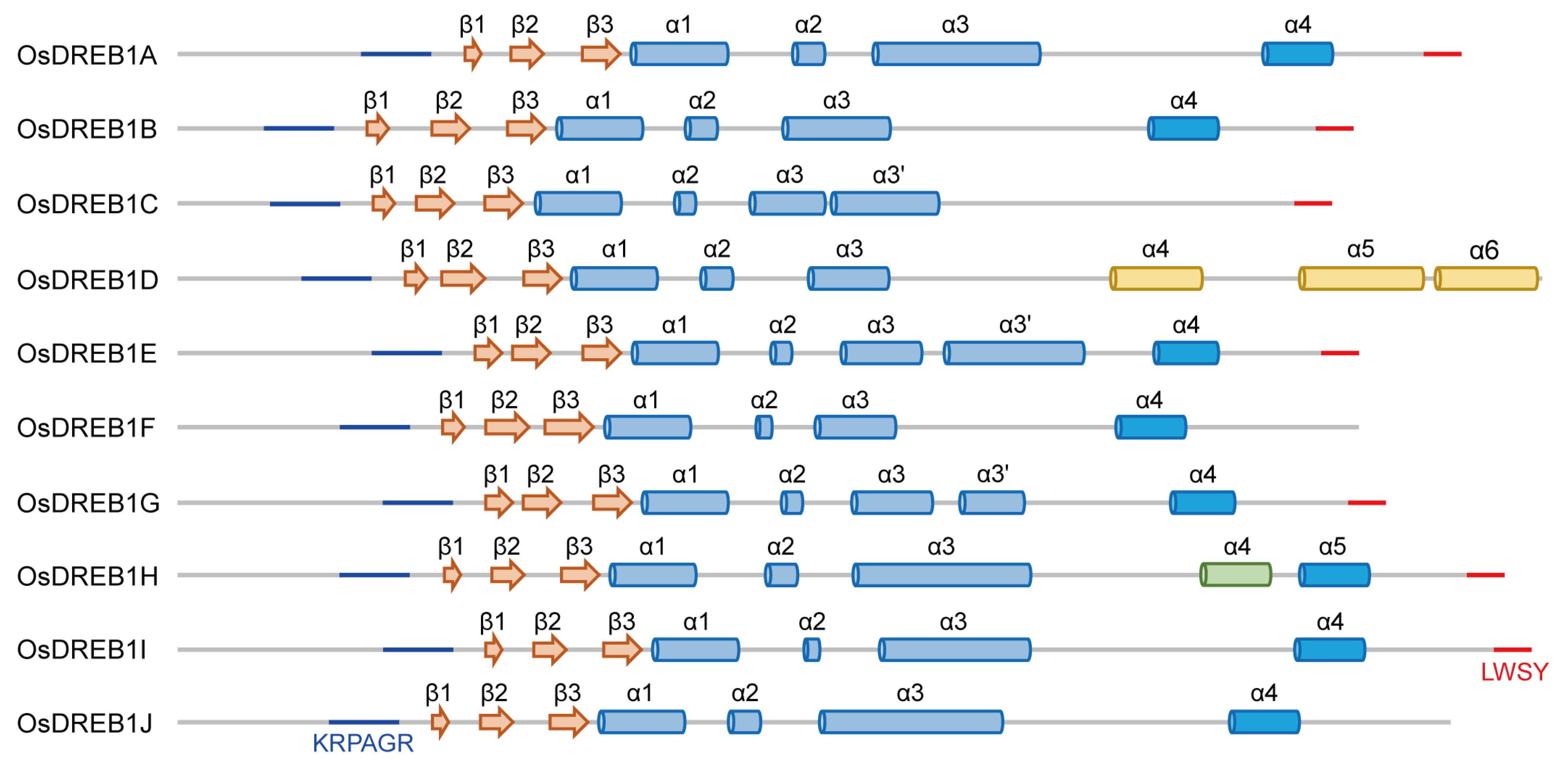
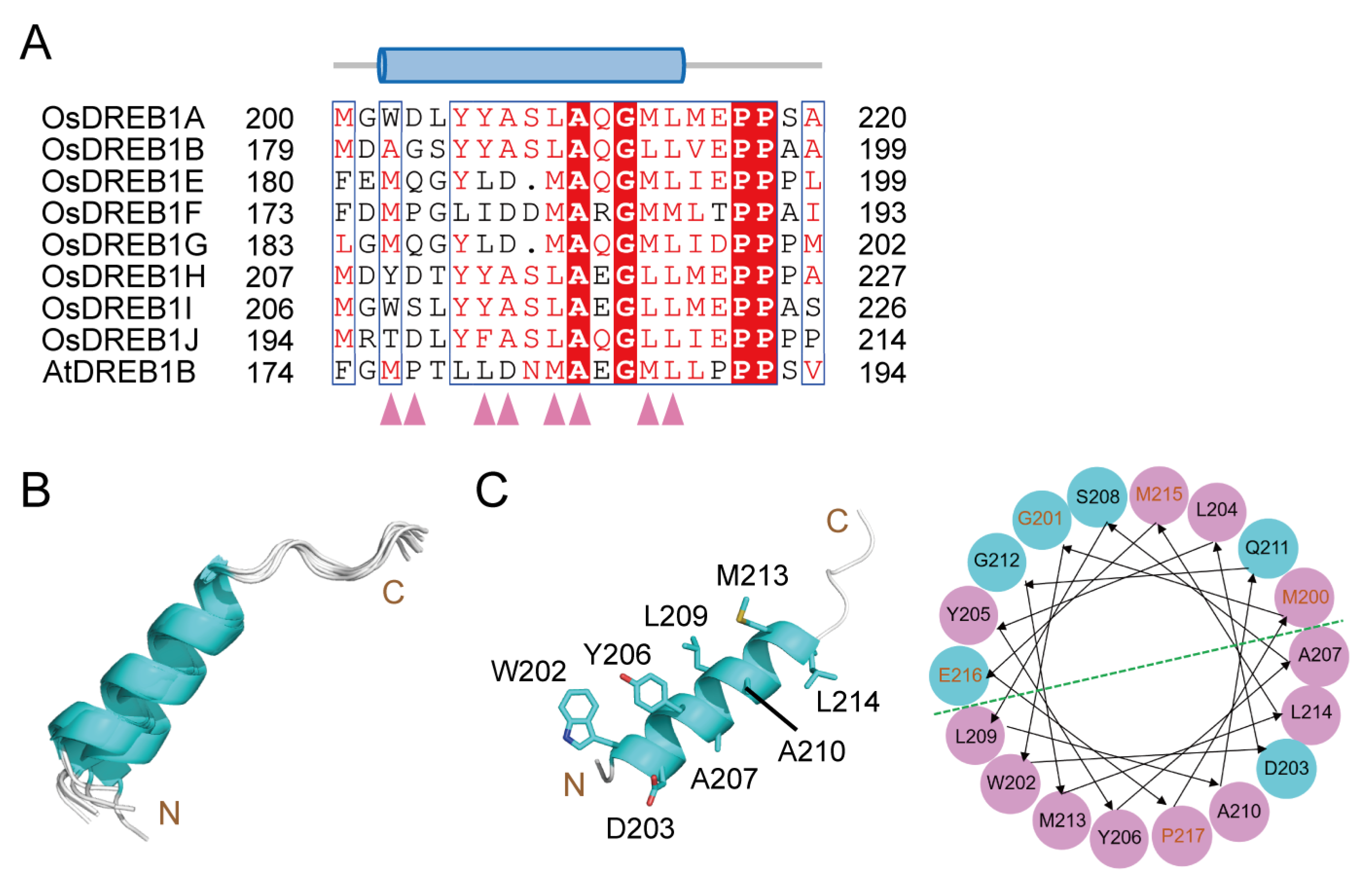
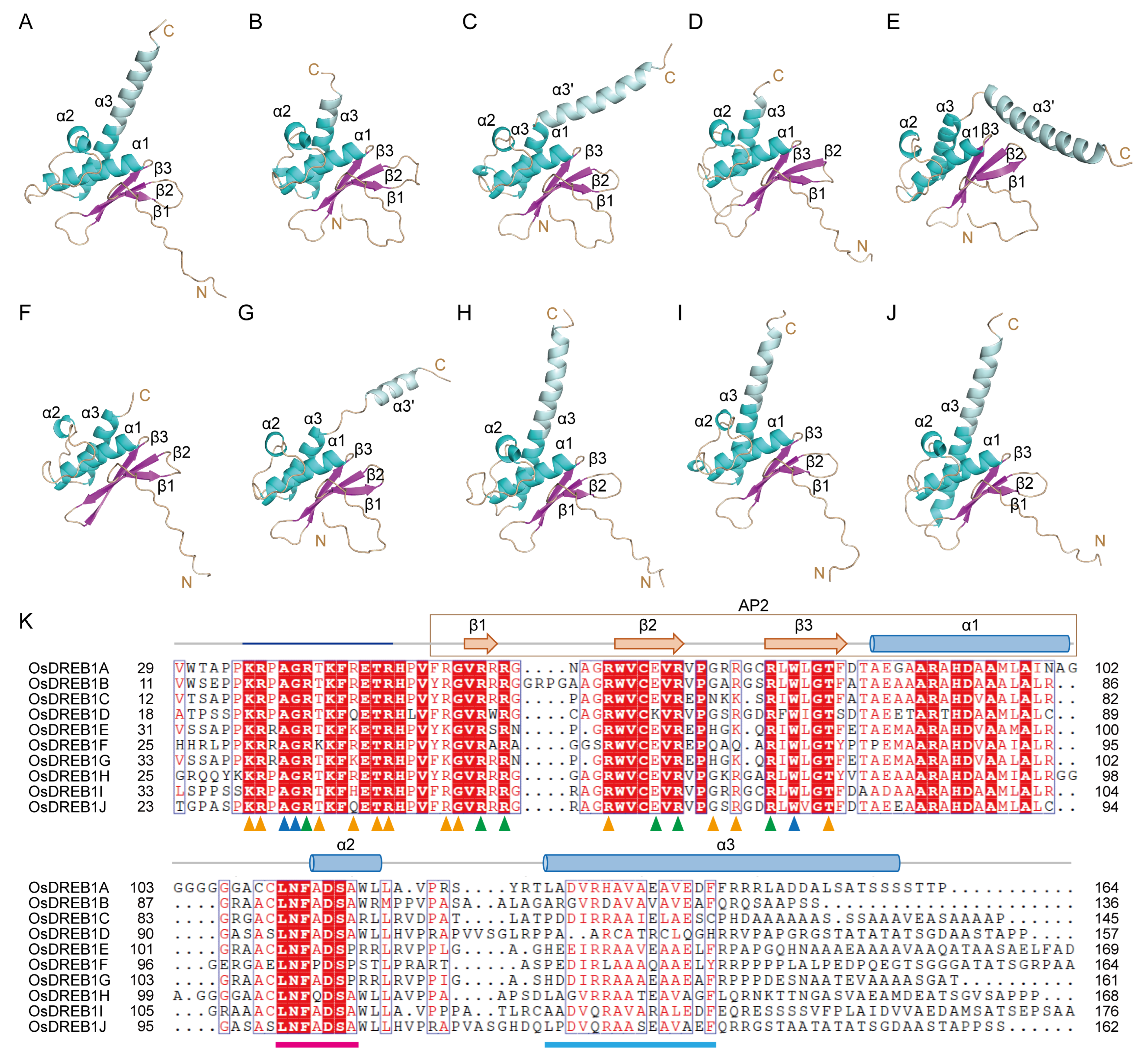
2.1. Structure Models of OsDREB1s
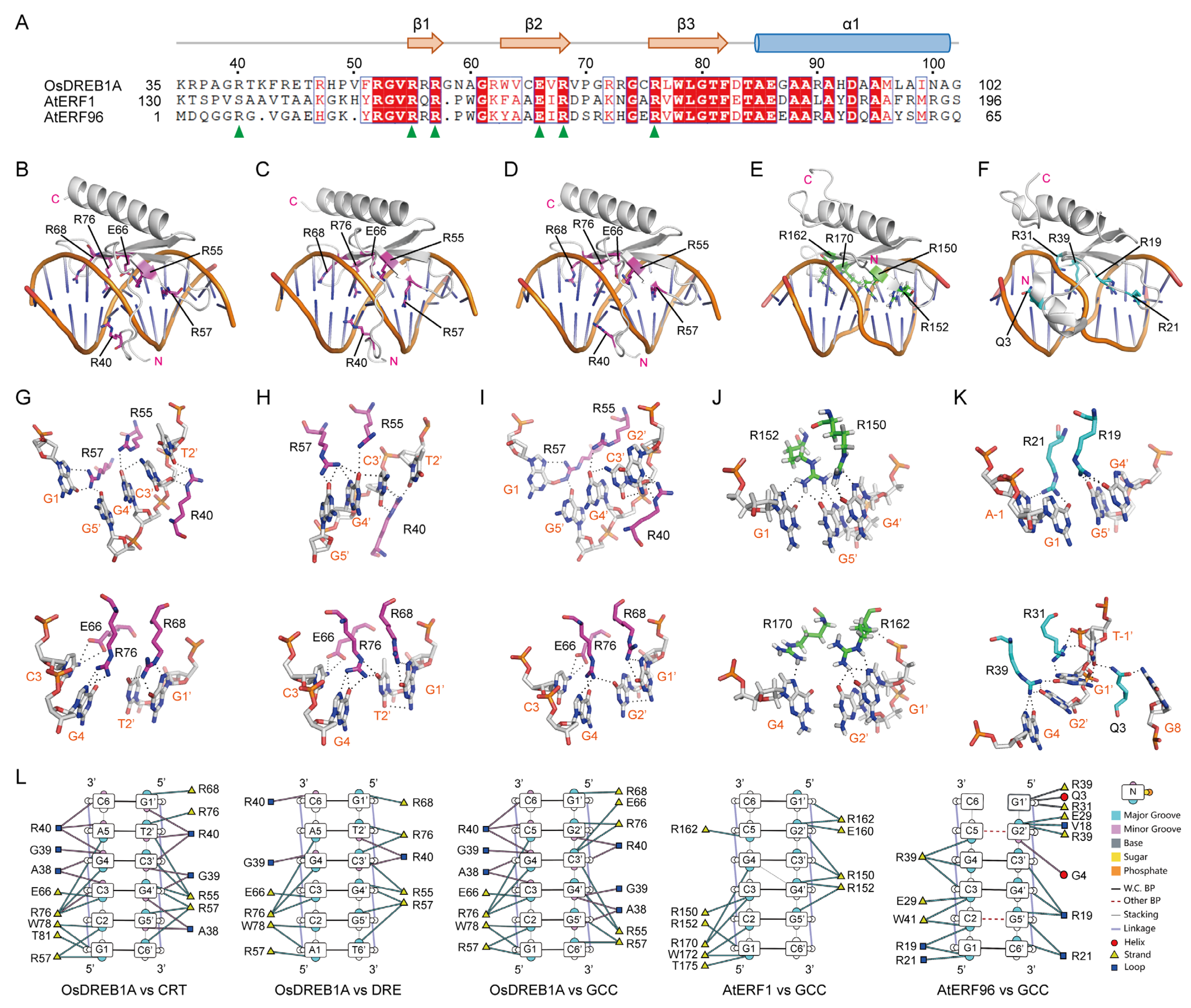
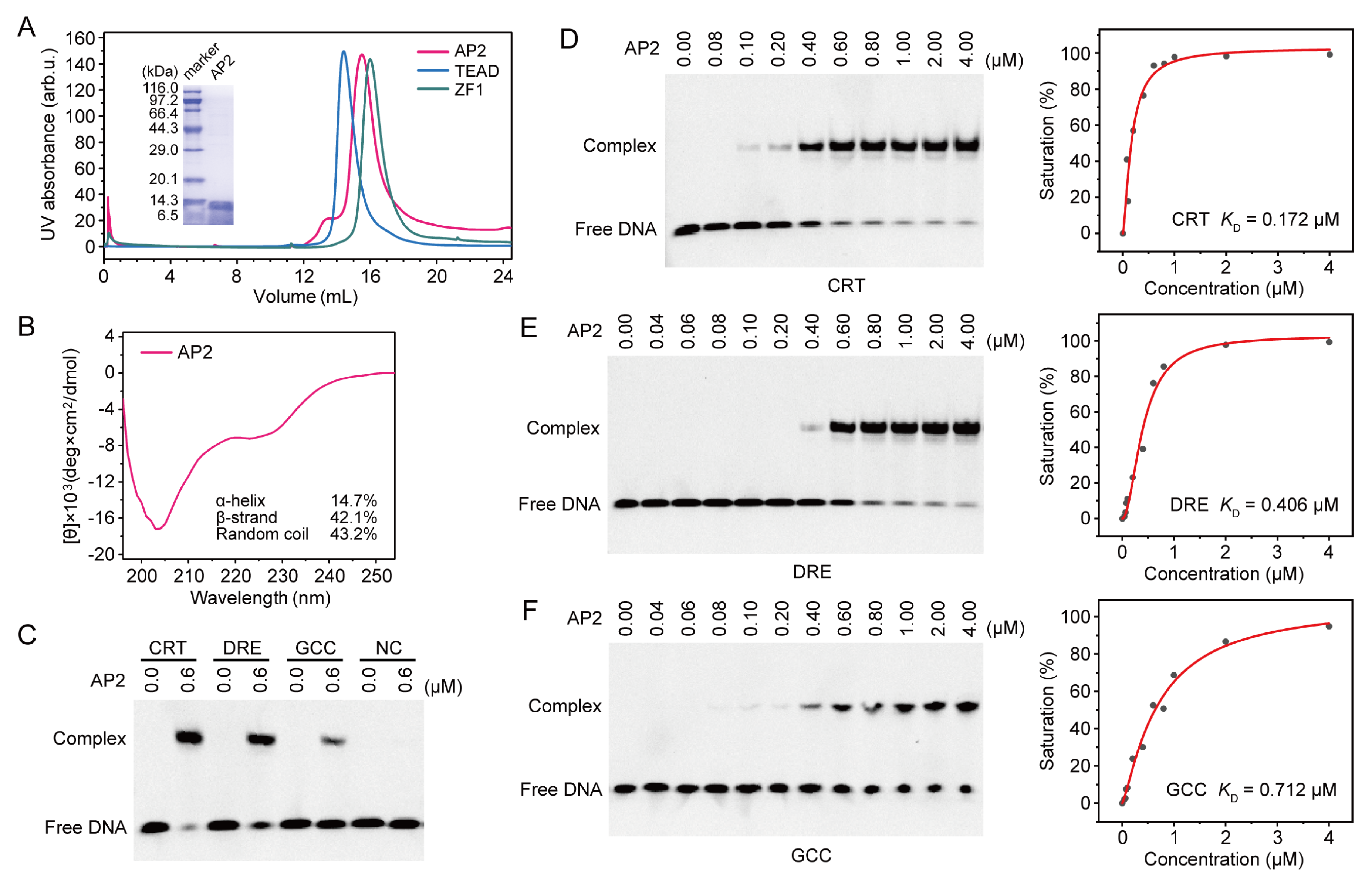
2.2. DNA Recognition of OsDREB1A DBD
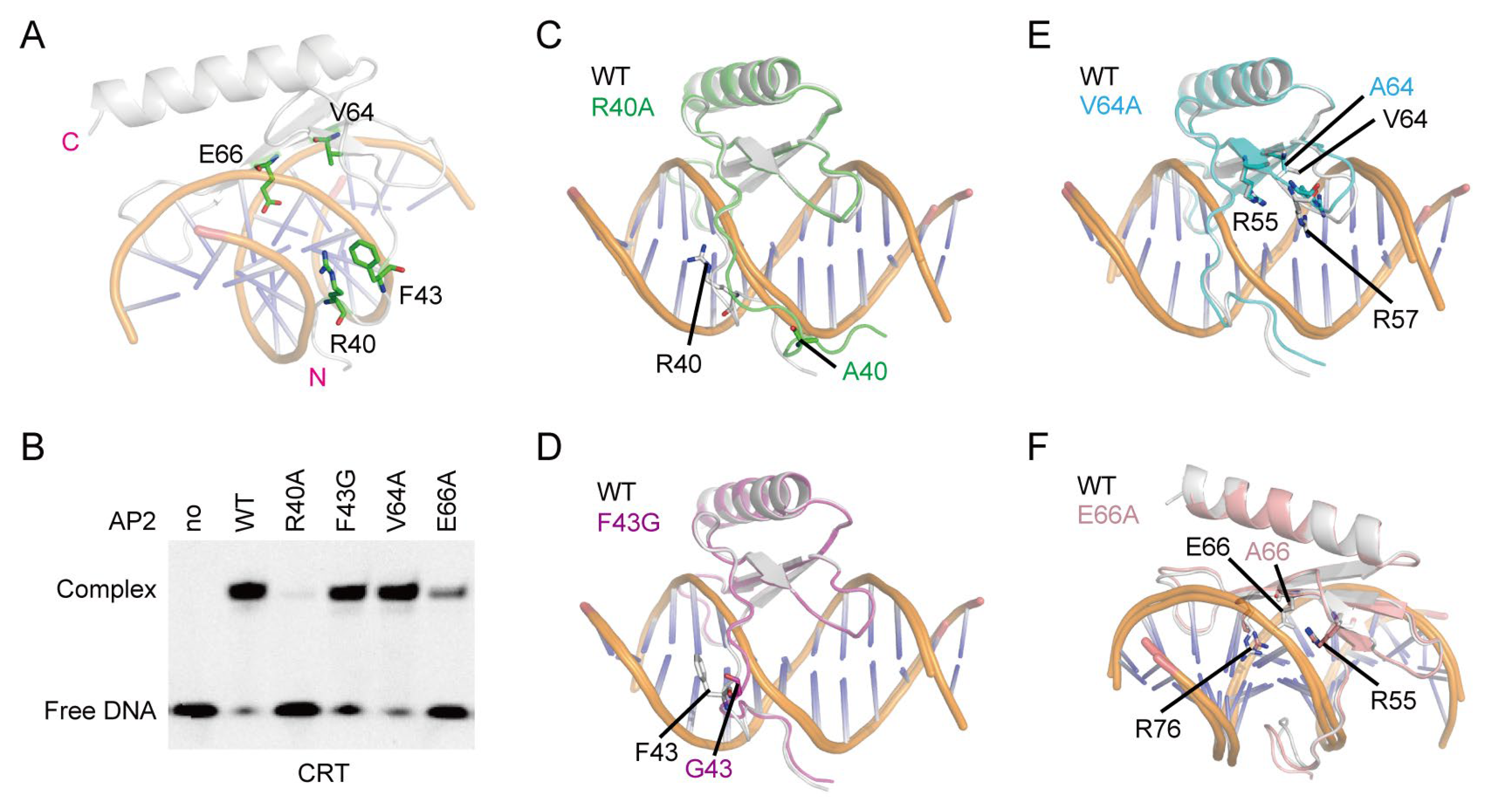
2.3. Crucial Roles of R40 and E66 of OsDREB1A in Binding to CRT DNA Element
3. Materials and Methods
3.1. Structure Modeling and Analysis
3.2. Protein Expression and Purification
3.3. Circular Dichroism (CD)
3.4. Electrophoretic Mobility Shift Assay (EMSA)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AP2 | APETALA2 |
| ERF | ethylene-responsive element binding factor |
| DREB | dehydration response element binding protein |
| CBF | C-repeat binding factor |
| RAV | related to ABI3/VP |
| CRT | C-repeat |
| DRE | dehydration response element |
| DBD | DNA-binding domain |
| PDB | Protein Data Bank |
| CD | circular dichroism |
| EMSA | electrophoretic mobility shift assay |
| FAM | 6-carboxy-fluorescein |
References
- Zhang, Y.; Xia, P. The DREB transcription factor, a biomacromolecule, responds to abiotic stress by regulating the expression of stress-related genes. Int. J. Biol. Macromol. 2023, 243, 125231. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Herath, V. Small family, big impact: In silico analysis of DREB2 transcription factor family in rice. Comput. Biol. Chem. 2016, 65, 128–139. [Google Scholar] [CrossRef]
- Liu, J.; Shi, Y.; Yang, S. Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 2018, 60, 780–795. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Nie, Y.; Guo, L.; Cui, F.; Shen, Y.; Ye, X.; Deng, D.; Wang, S.; Zhu, J.; Wu, W. Innovations and stepwise evolution of CBFs/DREB1s and their regulatory networks in angiosperms. J. Integr. Plant Biol. 2022, 64, 2111–2125. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, X.; Li, M.; Yang, H.; Fu, D.; Lv, J.; Ding, Y.; Gong, Z.; Shi, Y.; Yang, S. The direct targets of CBFs: In cold stress response and beyond. J. Integr. Plant Biol. 2021, 63, 1874–1887. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef]
- Kang, H.G.; Kim, J.; Kim, B.; Jeong, H.; Choi, S.H.; Kim, E.K.; Lee, H.Y.; Lim, P.O. Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Sci. 2011, 180, 634–641. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Hao, D.; Yamasaki, K.; Sarai, A.; Ohme-Takagi, M. Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry 2002, 41, 4202–4208. [Google Scholar] [CrossRef]
- Canella, D.; Gilmour, S.J.; Kuhn, L.A.; Thomashow, M.F. DNA binding by the Arabidopsis CBF1 transcription factor requires the PKKP/RAGRxKFxETRHP signature sequence. Biochim. Biophys. Acta 2010, 1799, 454–462. [Google Scholar] [CrossRef]
- Wang, Z.; Triezenberg, S.J.; Thomashow, M.F.; Stockinger, E.J. Multiple hydrophobic motifs in Arabidopsis CBF1 COOH-terminus provide functional redundancy in trans-activation. Plant Mol. Biol. 2005, 58, 543–559. [Google Scholar] [CrossRef]
- Mao, D.; Chen, C. Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS ONE 2012, 7, e47275. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y.; et al. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, C.; Jin, X.F.; Xiong, A.S.; Peng, R.H.; Hong, Y.H.; Yao, Q.H.; Chen, J.M. Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. BMB Rep. 2009, 42, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guan, Y.; Wu, Y.; Chen, H.; Chen, F.; Chu, C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008, 67, 589–602. [Google Scholar] [CrossRef]
- Moon, S.J.; Min, M.K.; Kim, J.A.; Kim, D.Y.; Yoon, I.S.; Kwon, T.R.; Byun, M.O.; Kim, B.G. Ectopic Expression of OsDREB1G, a Member of the OsDREB1 Subfamily, Confers Cold Stress Tolerance in Rice. Front. Plant Sci. 2019, 10, 297. [Google Scholar] [CrossRef]
- Wang, H.; Lu, S.; Guan, X.; Jiang, Y.; Wang, B.; Hua, J.; Zou, B. Dehydration-Responsive Element Binding Protein 1C, 1E, and 1G Promote Stress Tolerance to Chilling, Heat, Drought, and Salt in Rice. Front. Plant Sci. 2022, 13, 851731. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006, 47, 141–153. [Google Scholar] [CrossRef]
- Gutha, L.R.; Reddy, A.R. Rice DREB1B promoter shows distinct stress-specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol. Biol. 2008, 68, 533–555. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Q.; Meng, X.P.; Zhang, Y.; Xia, M.; Wang, X.P. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 2008, 30, 2191–2198. [Google Scholar] [CrossRef]
- Wei, S.; Li, X.; Lu, Z.; Zhang, H.; Ye, X.; Zhou, Y.; Li, J.; Yan, Y.; Pei, H.; Duan, F.; et al. A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science 2022, 377, eabi8455. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Lin, P.H.; Chen, K.H.; Cheng, Y.S. Structural insights into Arabidopsis ethylene response factor 96 with an extended N-terminal binding to GCC box. Plant Mol. Biol. 2020, 104, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Tian, S.; Xie, G.; Liu, R.; Wang, N.; Li, S.; He, Y.; Du, J. TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2103895118. [Google Scholar] [CrossRef]
- Qiao, Z.; Kong, Q.; Tee, W.T.; Lim, A.R.Q.; Teo, M.X.; Olieric, V.; Low, P.M.; Yang, Y.; Qian, G.; Ma, W.; et al. Molecular basis of the key regulator WRINKLED1 in plant oil biosynthesis. Sci. Adv. 2022, 8, eabq1211. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Zhou, H.; He, X.; Xiong, Y.; Gong, Y.; Zhang, Y.; Li, S.; Hu, R.; Li, Y.; Zhang, X.; Zhou, X.; et al. Structural insights into a highly flexible zinc finger module unravel INSM1 function in transcription regulation. Nat. Commun. 2025, 16, 2162. [Google Scholar] [CrossRef]
- Mitra, R.; Cohen, A.S.; Sagendorf, J.M.; Berman, H.M.; Rohs, R. DNAproDB: An updated database for the automated and interactive analysis of protein-DNA complexes. Nucleic Acids Res. 2025, 53, D396–D402. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Cai, W.; Zhu, J.; Zhu, Y. AlphaFold 3-Assisted Deciphering of the DNA Recognition by DREB1 Transcription Factors in Rice. Int. J. Mol. Sci. 2025, 26, 6395. https://doi.org/10.3390/ijms26136395
Wang W, Cai W, Zhu J, Zhu Y. AlphaFold 3-Assisted Deciphering of the DNA Recognition by DREB1 Transcription Factors in Rice. International Journal of Molecular Sciences. 2025; 26(13):6395. https://doi.org/10.3390/ijms26136395
Chicago/Turabian StyleWang, Wenshu, Wei Cai, Jiang Zhu, and Yongsheng Zhu. 2025. "AlphaFold 3-Assisted Deciphering of the DNA Recognition by DREB1 Transcription Factors in Rice" International Journal of Molecular Sciences 26, no. 13: 6395. https://doi.org/10.3390/ijms26136395
APA StyleWang, W., Cai, W., Zhu, J., & Zhu, Y. (2025). AlphaFold 3-Assisted Deciphering of the DNA Recognition by DREB1 Transcription Factors in Rice. International Journal of Molecular Sciences, 26(13), 6395. https://doi.org/10.3390/ijms26136395





