PD-L1 Expression and Comprehensive Genomic Profiling in Advanced NSCLC: A Single-Centre Experience
Abstract
1. Introduction
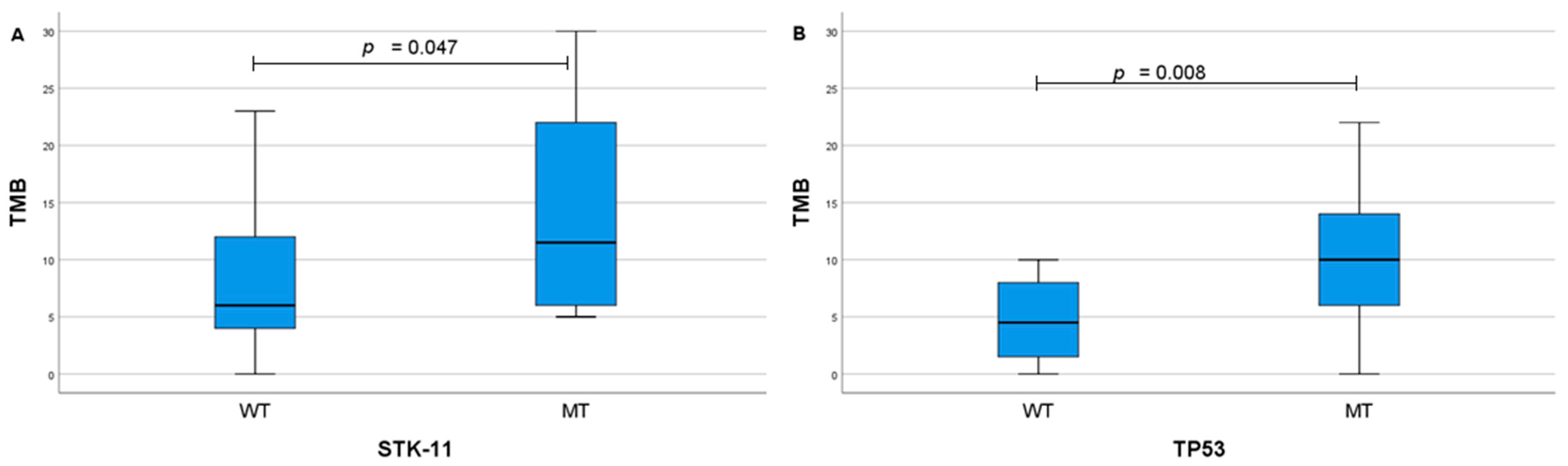
2. Results
- MT—mutation.
3. Discussion
4. Materials and Methods
4.1. Methods
4.2. Study Population
4.3. Immunohistochemistry Analysis
4.4. Targeted Tumour Next-Generation Sequencing
4.5. Clinical Outcomes
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, S.M.; Schulz, C.; Prabhash, K.; Kowalski, D.; Szczesna, A.; Han, B.; Rittmeyer, A.; Talbot, T.; Vicente, D.; Califano, R.; et al. First-line atezolizumab monotherapy versus single-agent chemotherapy in patients with non-small-cell lung cancer ineligible for treatment with a platinum-containing regimen (IPSOS): A phase 3, global, multicentre, open-label, randomised controlled study. Lancet 2023, 402, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Belaroussi, Y.; Bouteiller, F.; Bellera, C.; Pasquier, D.; Perol, M.; Debieuvre, D.; Filleron, T.; Girard, N.; Schott, R.; Mathoulin-Pélissier, S.; et al. Survival outcomes of patients with metastatic non-small cell lung cancer receiving chemotherapy or immunotherapy as first-line in a real-life setting. Sci. Rep. 2023, 13, 9584. [Google Scholar] [CrossRef]
- Spigel, D.; De Marinis, F.; Giaccone, G.; Reinmuth, N.; Vergnenegre, A.; Barrios, C.H.; Morise, M.; Felip, E.; Andric, Z.G.; Geater, S.; et al. IMpower110: Interim overall survival (OS) analysis of a phase III study of atezolizumab (atezo) vs platinum-based chemotherapy (chemo) as first-line (1L) treatment (tx) in PD-L1–selected NSCLC. Ann. Oncol. 2019, 30, v915. [Google Scholar] [CrossRef]
- Felip, E.; Altorki, N.; Zhou, C.; Vallières, E.; Martínez-Martí, A.; Rittmeyer, A.; Chella, A.; Reck, M.; Goloborodko, O.; Huang, M.; et al. Overall survival with adjuvant atezolizumab after chemotherapy in resected stage II-IIIA non-small-cell lung cancer (IMpower010): A randomised, multicentre, open-label, phase III trial. Ann. Oncol. 2023, 34, 907–919. [Google Scholar] [CrossRef]
- O’Brien, M.; Paz-Ares, L.; Marreaud, S.; Dafni, U.; Oselin, K.; Havel, L.; Esteban, E.; Isla, D.; Martinez-Marti, A.; Faehling, M.; et al. Pembrolizumab versus placebo as adjuvant therapy for completely resected stage IB–IIIA non-small-cell lung cancer (PEARLS/KEYNOTE-091): An interim analysis of a randomised, triple-blind, phase 3 trial. Lancet Oncol. 2022, 23, 1274–1286. [Google Scholar] [CrossRef] [PubMed]
- Wakelee, H.; Liberman, M.; Kato, T.; Tsuboi, M.; Lee, S.-H.; Gao, S.; Chen, K.-N.; Dooms, C.; Majem, M.; Eigendorff, E.; et al. Perioperative Pembrolizumab for Early-Stage Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2023, 389, 491–503. [Google Scholar] [CrossRef]
- Kanwal, B.; Biswas, S.; Seminara, R.S.; Jeet, C. Immunotherapy in Advanced Non-small Cell Lung Cancer Patients: Ushering Chemotherapy Through the Checkpoint Inhibitors? Cureus 2018, 10, e3254. [Google Scholar] [CrossRef] [PubMed]
- Putzu, C.; Canova, S.; Paliogiannis, P.; Lobrano, R.; Sala, L.; Cortinovis, D.L.; Colonese, F. Duration of Immunotherapy in Non-Small Cell Lung Cancer Survivors: A Lifelong Commitment? Cancers 2023, 15, 689. [Google Scholar] [CrossRef]
- Garassino, M.C.; Gadgeel, S.; Speranza, G.; Felip, E.; Esteban, E.; Dómine, M.; Hochmair, M.J.; Powell, S.F.; Bischoff, H.G.; Peled, N.; et al. Pembrolizumab Plus Pemetrexed and Platinum in Nonsquamous Non–Small-Cell Lung Cancer: 5-Year Outcomes From the Phase 3 KEYNOTE-189 Study. J. Clin. Oncol. 2023, 41, 1992–1998. [Google Scholar] [CrossRef]
- Incorvaia, L.; Fanale, D.; Badalamenti, G.; Barraco, N.; Bono, M.; Corsini, L.R.; Galvano, A.; Gristina, V.; Listì, A.; Vieni, S.; et al. Programmed Death Ligand 1 (PD-L1) as a Predictive Biomarker for Pembrolizumab Therapy in Patients with Advanced Non-Small-Cell Lung Cancer (NSCLC). Adv. Ther. 2019, 36, 2600–2617. [Google Scholar] [CrossRef]
- Catalano, M.; Iannone, L.F.; Nesi, G.; Nobili, S.; Mini, E.; Roviello, G. Immunotherapy-related biomarkers: Confirmations and uncertainties. Crit. Rev. Oncol. Hematol. 2023, 192, 104135. [Google Scholar] [CrossRef] [PubMed]
- Vryza, P.; Fischer, T.; Mistakidi, E.; Zaravinos, A. Tumor mutation burden in the prognosis and response of lung cancer patients to immune-checkpoint inhibition therapies. Transl. Oncol. 2023, 38, 101788. [Google Scholar] [CrossRef] [PubMed]
- Di Federico, A.; De Giglio, A.; Gelsomino, F.; Sperandi, F.; Melotti, B.; Ardizzoni, A. Predictors of survival to immunotherapy and chemoimmunotherapy in non-small cell lung cancer: A meta-analysis. JNCI: J. Natl. Cancer Inst. 2023, 115, 29–42. [Google Scholar] [CrossRef]
- Alessi, J.V.; Elkrief, A.; Ricciuti, B.; Wang, X.; Cortellini, A.; Vaz, V.R.; Lamberti, G.; Frias, R.L.; Venkatraman, D.; Fulgenzi, C.A.M.; et al. Clinicopathologic and Genomic Factors Impacting Efficacy of First-Line Chemoimmunotherapy in Advanced NSCLC. J. Thorac. Oncol. 2023, 18, 731–743. [Google Scholar] [CrossRef] [PubMed]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Lung Cancer—Metastatic, Patient Version [Internet]. Fort Washington (PA): NCCN; 2024. Available online: https://www.nccn.org/patients/guidelines/content/PDF/lung-metastatic-patient.pdf (accessed on 29 June 2025).
- Saller, J.J.; Boyle, T.A. Molecular Pathology of Lung Cancer. Cold Spring Harb. Perspect. Med. 2022, 12, a037812. [Google Scholar] [CrossRef]
- De Giglio, A.; De Biase, D.; Favorito, V.; Maloberti, T.; Di Federico, A.; Zacchini, F.; Venturi, G.; Parisi, C.; Gustavo Dall’Olio, F.; Ricciotti, I.; et al. STK11 mutations correlate with poor prognosis for advanced NSCLC treated with first-line immunotherapy or chemo-immunotherapy according to KRAS, TP53, KEAP1, and SMARCA4 status. Lung Cancer 2025, 199, 108058. [Google Scholar] [CrossRef]
- Shire, N.J.; Klein, A.B.; Golozar, A.; Collins, J.M.; Fraeman, K.H.; Nordstrom, B.L.; McEwen, R.; Hembrough, T.; Rizvi, N.A. STK11 (LKB1) mutations in metastatic NSCLC: Prognostic value in the real world. PLoS ONE 2020, 15, e0238358. [Google Scholar] [CrossRef] [PubMed]
- Ricciuti, B.; Arbour, K.C.; Lin, J.J.; Vajdi, A.; Vokes, N.; Hong, L.; Zhang, J.; Tolstorukov, M.Y.; Li, Y.Y.; Spurr, L.F.; et al. Diminished Efficacy of Programmed Death-(Ligand)1 Inhibition in STK11- and KEAP1-Mutant Lung Adenocarcinoma Is Affected by KRAS Mutation Status. J. Thorac. Oncol. 2022, 17, 399–410. [Google Scholar] [CrossRef]
- Sun, L.; Handorf, E.A.; Zhou, Y.; Borghaei, H.; Aggarwal, C.; Bauman, J. Outcomes in patients treated with frontline immune checkpoint inhibition (ICI) for advanced NSCLC with KRAS mutations and STK11/KEAP1 comutations across PD-L1 levels. Lung Cancer 2024, 190, 107510. [Google Scholar] [CrossRef]
- Van De Haar, J.; Mankor, J.M.; Hummelink, K.; Monkhorst, K.; Smit, E.F.; Wessels, L.F.A.; Cuppen, E.; Aerts, J.G.J.V.; Voest, E.E. Combining Genomic Biomarkers to Guide Immunotherapy in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2024, 30, 1307–1318. [Google Scholar] [CrossRef]
- Gandara, D.; Reck, M.; Moro-Sibilot, D.; Mazieres, J.; Gadgeel, S.; Morris, S.; Cardona, A.; Mendus, D.; Ballinger, M.; Rittmeyer, A.; et al. Fast progression in non–small cell lung cancer: Results from the randomized phase III OAK study evaluating second-line atezolizumab versus docetaxel. J. Immunother. Cancer 2021, 9, e001882. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, R.; Mezquita, L.; Texier, M.; Lahmar, J.; Audigier-Valette, C.; Tessonnier, L.; Mazieres, J.; Zalcman, G.; Brosseau, S.; Le Moulec, S.; et al. Comparison of Fast-Progression, Hyperprogressive Disease, and Early Deaths in Advanced Non–Small-Cell Lung Cancer Treated with PD-1/PD-L1 Inhibitors or Chemotherapy. JCO Precis. Oncol. 2020, 4, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; Novello, S.; Giannarelli, D.; Bria, E.; Galetta, D.; Gelibter, A.; Reale, M.L.; Carnio, S.; Vita, E.; Stefani, A.; et al. Early Progression in Non-Small Cell Lung Cancer (NSCLC) with High PD-L1 Treated with Pembrolizumab in First-Line Setting: A Prognostic Scoring System Based on Clinical Features. Cancers 2021, 13, 2935. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Garon, E.B.; Kim, D.-W.; Cho, B.C.; Gervais, R.; Perez-Gracia, J.L.; Han, J.-Y.; Majem, M.; Forster, M.D.; Monnet, I.; et al. Five Year Survival Update From KEYNOTE-010: Pembrolizumab Versus Docetaxel for Previously Treated, Programmed Death-Ligand 1–Positive Advanced NSCLC. J. Thorac. Oncol. 2021, 16, 1718–1732. [Google Scholar] [CrossRef]
- Hendriks, L.E.; Rouleau, E.; Besse, B. Clinical utility of tumor mutational burden in patients with non-small cell lung cancer treated with immunotherapy. Transl. Lung Cancer Res. 2018, 7, 647–660. [Google Scholar] [CrossRef]
- Willis, C.; Bauer, H.; Au, T.H.; Menon, J.; Unni, S.; Tran, D.; Rivers, Z.; Akerley, W.; Schabath, M.B.; Badin, F.; et al. Real-world survival analysis by tumor mutational burden in non-small cell lung cancer: A multisite U.S. study. Oncotarget 2022, 13, 257–270. [Google Scholar] [CrossRef]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef]
- Remon, J.; Collazo, A.; Jimenez, B. CheckMate 227 trial has not checked the immune-strategy in first-line setting in advanced non-small cell lung cancer. Transl. Cancer Res. 2020, 9, 2168–2170. [Google Scholar] [CrossRef] [PubMed]
- Cascetta, P.; Marinello, A.; Lazzari, C.; Gregorc, V.; Planchard, D.; Bianco, R.; Normanno, N.; Morabito, A. KRAS in NSCLC: State of the Art and Future Perspectives. Cancers 2022, 14, 5430. [Google Scholar] [CrossRef]
- Canale, M.; Andrikou, K.; Priano, I.; Cravero, P.; Pasini, L.; Urbini, M.; Delmonte, A.; Crinò, L.; Bronte, G.; Ulivi, P. The Role of TP53 Mutations in EGFR-Mutated Non-Small-Cell Lung Cancer: Clinical Significance and Implications for Therapy. Cancers 2022, 14, 1143. [Google Scholar] [CrossRef]
- Tseng, Y.-H.; Tran, T.T.M.; Tsai Chang, J.; Huang, Y.-T.; Nguyen, A.T.; Chang, I.Y.; Chen, Y.-T.; Hsieh, H.-W.; Juang, Y.-L.; Chang, P.M.-H.; et al. Utilizing TP53 hotspot mutations as effective predictors of gemcitabine treatment outcome in non-small-cell lung cancer. Cell Death Discov. 2025, 11, 26. [Google Scholar] [CrossRef]
- Di Federico, A.; De Giglio, A.; Parisi, C.; Gelsomino, F. STK11/LKB1 and KEAP1 mutations in non-small cell lung cancer: Prognostic rather than predictive? Eur. J. Cancer 2021, 157, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Brune, M.M.; Savic Prince, S.; Vlajnic, T.; Chijioke, O.; Roma, L.; König, D.; Bubendorf, L. MTAP as an emerging biomarker in thoracic malignancies. Lung Cancer 2024, 197, 107963. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Chen, Y.; Song, M.; Zhang, X.; Li, P.; Yu, X.; Huang, Y.; Zhang, N.; Ji, L.; Xia, L.; et al. Co-occurrence of CDKN2A/B and IFN-I homozygous deletions correlates with an immunosuppressive phenotype and poor prognosis in lung adenocarcinoma. Mol. Oncol. 2022, 16, 1746–1760. [Google Scholar] [CrossRef] [PubMed]
- Best, S.A.; Gubser, P.M.; Sethumadhavan, S.; Kersbergen, A.; Negrón Abril, Y.L.; Goldford, J.; Sellers, K.; Abeysekera, W.; Garnham, A.L.; McDonald, J.A.; et al. Glutaminase inhibition impairs CD8 T cell activation in STK11-/Lkb1-deficient lung cancer. Cell Metab. 2022, 34, 874–887.e6. [Google Scholar] [CrossRef]
- Papalexi, E.; Mimitou, E.P.; Butler, A.W.; Foster, S.; Bracken, B.; Mauck, W.M.; Wessels, H.-H.; Hao, Y.; Yeung, B.Z.; Smibert, P.; et al. Characterizing the molecular regulation of inhibitory immune checkpoints with multimodal single-cell screens. Nat. Genet. 2021, 53, 322–331. [Google Scholar] [CrossRef]
- Long, Y.; Yu, X.; Chen, R.; Tong, Y.; Gong, L. Noncanonical PD-1/PD-L1 Axis in Relation to the Efficacy of Anti-PD Therapy. Front. Immunol. 2022, 13, 910704. [Google Scholar] [CrossRef]
- Ricciuti, B.; Garassino, M.C. Precision Immunotherapy for STK11/KEAP1-Mutant NSCLC. J. Thorac. Oncol. 2024, 19, 877–882. [Google Scholar] [CrossRef]
- Jeong, Y.; Hellyer, J.A.; Stehr, H.; Hoang, N.T.; Niu, X.; Das, M.; Padda, S.K.; Ramchandran, K.; Neal, J.W.; Wakelee, H.; et al. Role of KEAP1/NFE2L2 Mutations in the Chemotherapeutic Response of Patients with Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 274–281. [Google Scholar] [CrossRef]
- Lamberti, G.; Spurr, L.F.; Li, Y.; Ricciuti, B.; Recondo, G.; Umeton, R.; Nishino, M.; Sholl, L.M.; Meyerson, M.L.; Cherniack, A.D.; et al. Clinicopathological and genomic correlates of programmed cell death ligand 1 (PD-L1) expression in nonsquamous non-small-cell lung cancer. Ann. Oncol. 2020, 31, 807–814. [Google Scholar] [CrossRef]
- Garassino, M.C.; Gadgeel, S.; Novello, S.; Halmos, B.; Felip, E.; Speranza, G.; Hui, R.; Garon, E.B.; Horinouchi, H.; Sugawara, S.; et al. Associations of Tissue Tumor Mutational Burden and Mutational Status With Clinical Outcomes With Pembrolizumab Plus Chemotherapy Versus Chemotherapy For Metastatic NSCLC. JTO Clin. Res. Rep. 2023, 4, 100431. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Goldberg, M.E.; Greenawalt, D.M.; Hellmann, M.D.; Awad, M.M.; Gainor, J.F.; Schrock, A.B.; Hartmaier, R.J.; Trabucco, S.E.; Gay, L.; et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS -Mutant Lung Adenocarcinoma. Cancer Discov. 2018, 8, 822–835. [Google Scholar] [CrossRef]
- Biton, J.; Mansuet-Lupo, A.; Pécuchet, N.; Alifano, M.; Ouakrim, H.; Arrondeau, J.; Boudou-Rouquette, P.; Goldwasser, F.; Leroy, K.; Goc, J.; et al. TP53, STK11, and EGFR Mutations Predict Tumor Immune Profile and the Response to Anti–PD-1 in Lung Adenocarcinoma. Clin. Cancer Res. 2018, 24, 5710–5723. [Google Scholar] [CrossRef]
- Lamberti, G.; Sisi, M.; Andrini, E.; Palladini, A.; Giunchi, F.; Lollini, P.-L.; Ardizzoni, A.; Gelsomino, F. The Mechanisms of PD-L1 Regulation in Non-Small-Cell Lung Cancer (NSCLC): Which Are the Involved Players? Cancers 2020, 12, 3129. [Google Scholar] [CrossRef]
- Tabernero, J.; Hyman, D.M.; Soria, J.-C. ULK1 Inhibition Restores Antigen Presentation in LKB1 -Mutant Lung Cancer. Cancer Discov. 2021, 11, OF8. [Google Scholar] [CrossRef]
- Pons-Tostivint, E.; Lugat, A.; Fontenau, J.-F.; Denis, M.G.; Bennouna, J. STK11/LKB1 Modulation of the Immune Response in Lung Cancer: From Biology to Therapeutic Impact. Cells 2021, 10, 3129. [Google Scholar] [CrossRef]
- Shackelford, D.B.; Shaw, R.J. The LKB1–AMPK pathway: Metabolism and growth control in tumour suppression. Nat. Rev. Cancer 2009, 9, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Frille, A.; Boeschen, M.; Wirtz, H.; Stiller, M.; Bläker, H.; Von Laffert, M. TP53 co-mutations in advanced lung adenocarcinoma: Comparative bioinformatic analyses suggest ambivalent character on overall survival alongside KRAS, STK11 and KEAP1 mutations. Front. Oncol. 2024, 14, 1357583. [Google Scholar] [CrossRef]
- Sun, H.; Liu, S.-Y.; Zhou, J.-Y.; Xu, J.-T.; Zhang, H.-K.; Yan, H.-H.; Huan, J.-J.; Dai, P.-P.; Xu, C.-R.; Su, J.; et al. Specific TP53 subtype as biomarker for immune checkpoint inhibitors in lung adenocarcinoma. EBioMedicine 2020, 60, 102990. [Google Scholar] [CrossRef]
- Ma, K.; Huang, F.; Wang, Y.; Kang, Y.; Wang, Q.; Tang, J.; Sun, P.; Lou, J.; Qiao, R.; Si, J.; et al. Relationship between tumor mutational burden, gene mutation status, and clinical characteristics in 340 cases of lung adenocarcinoma. Cancer Med. 2022, 11, 4389–4397. [Google Scholar] [CrossRef]
- Fu, J.; Li, Y.; Li, C.; Tong, Y.; Li, M.; Cang, S. A special prognostic indicator: Tumor mutation burden combined with immune infiltrates in lung adenocarcinoma with TP53 mutation. Transl. Cancer. Res. 2021, 10, 3963–3978. [Google Scholar] [CrossRef]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by p53 via miR-34. JNCI J. Natl. Cancer Inst. 2016, 108, djv303. [Google Scholar] [CrossRef]
- Martinkova, L.; Zatloukalova, P.; Kucerikova, M.; Friedlova, N.; Tylichova, Z.; Zavadil-Kokas, F.; Hupp, T.R.; Coates, P.J.; Vojtesek, B. Inverse correlation between TP53 gene status and PD-L1 protein levels in a melanoma cell model depends on an IRF1/SOX10 regulatory axis. Cell Mol. Biol. Lett. 2024, 29, 117. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Patel, S.; Drusbosky, L.M.; Xiong, Y.; Chen, R.; Geng, R.; Heeke, S.; Nilsson, M.; Wu, J.; Heymach, J.V.; et al. Molecular landscape of ERBB2 alterations in 3000 advanced NSCLC patients. NPJ Precis. Oncol. 2024, 8, 217. [Google Scholar] [CrossRef]
- Shih, J.-Y. ERBB2 Amplification in NSCLC: How Many Faces? J. Thorac. Oncol. 2024, 19, 668–670. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Xian, X.; Tian, P.; Li, W.; Wang, K.; Li, Y. Efficacy of Combination Chemo-Immunotherapy as a First-Line Treatment for Advanced Non-Small-Cell Lung Cancer Patients With HER2 Alterations: A Case Series. Front. Oncol. 2021, 11, 633522. [Google Scholar] [CrossRef]
- Bertero, L.; Massa, F.; Metovic, J.; Zanetti, R.; Castellano, I.; Ricardi, U.; Papotti, M.; Cassoni, P. Eighth Edition of the UICC Classification of Malignant Tumours: An overview of the changes in the pathological TNM classification criteria—What has changed and why? Virchows Arch. 2018, 472, 519–531. [Google Scholar] [CrossRef]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef] [PubMed]
- Marcus, L.; Fashoyin-Aje, L.A.; Donoghue, M.; Yuan, M.; Rodriguez, L.; Gallagher, P.S.; Philip, R.; Ghosh, S.; Theoret, M.R.; Beaver, J.A.; et al. FDA Approval Summary: Pembrolizumab for the Treatment of Tumor Mutational Burden–High Solid Tumors. Clin. Cancer Res. 2021, 27, 4685–4689. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
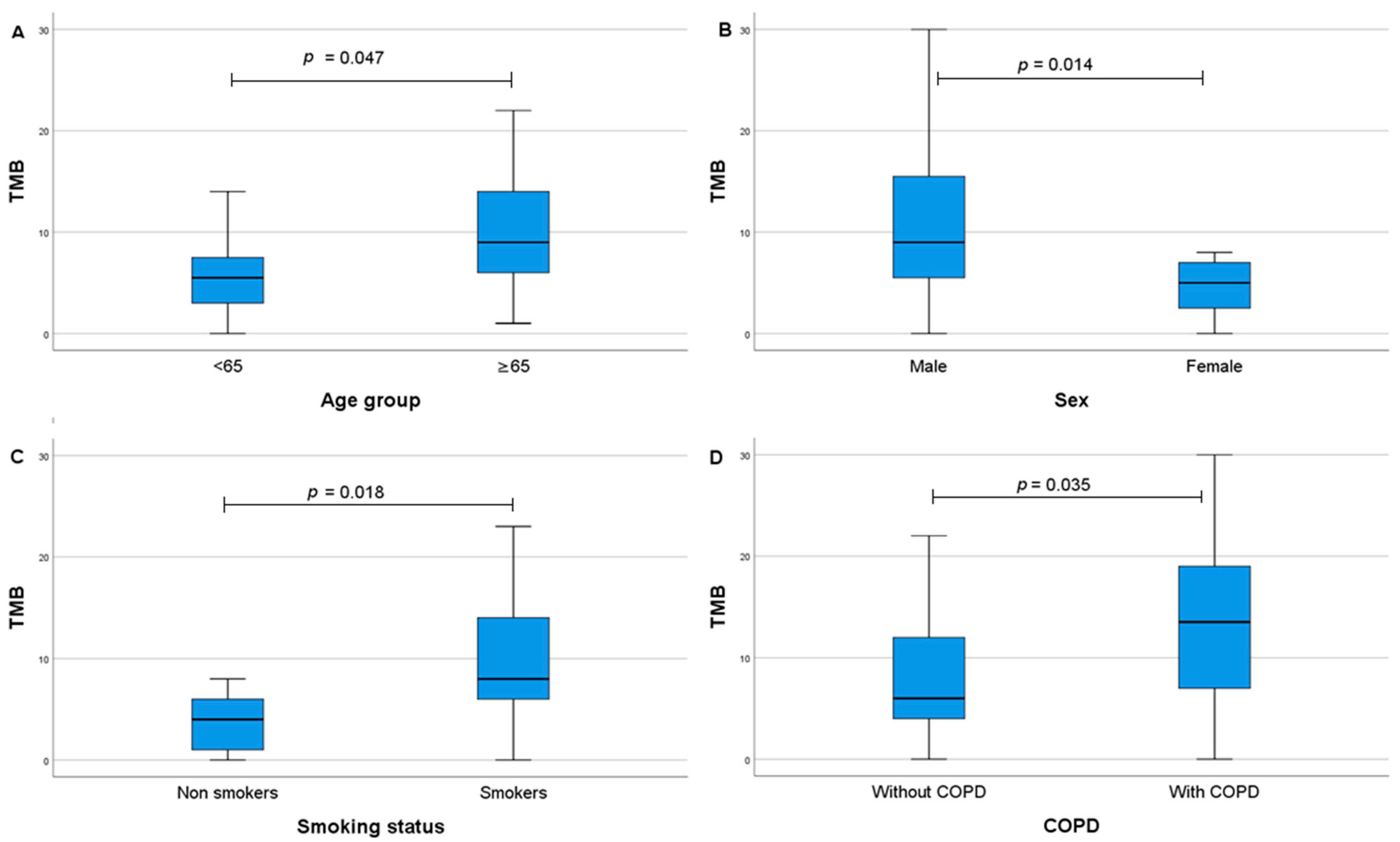
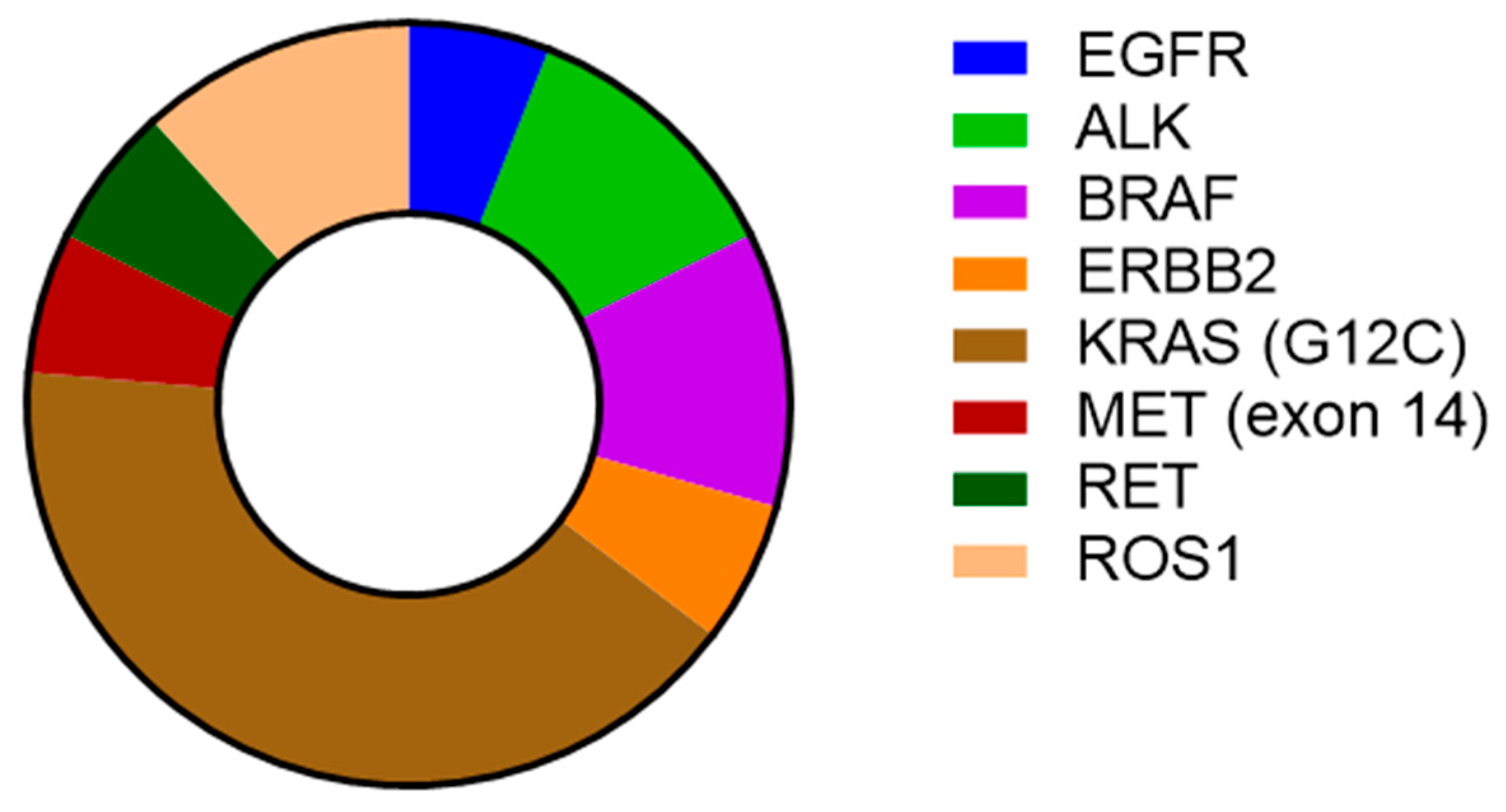
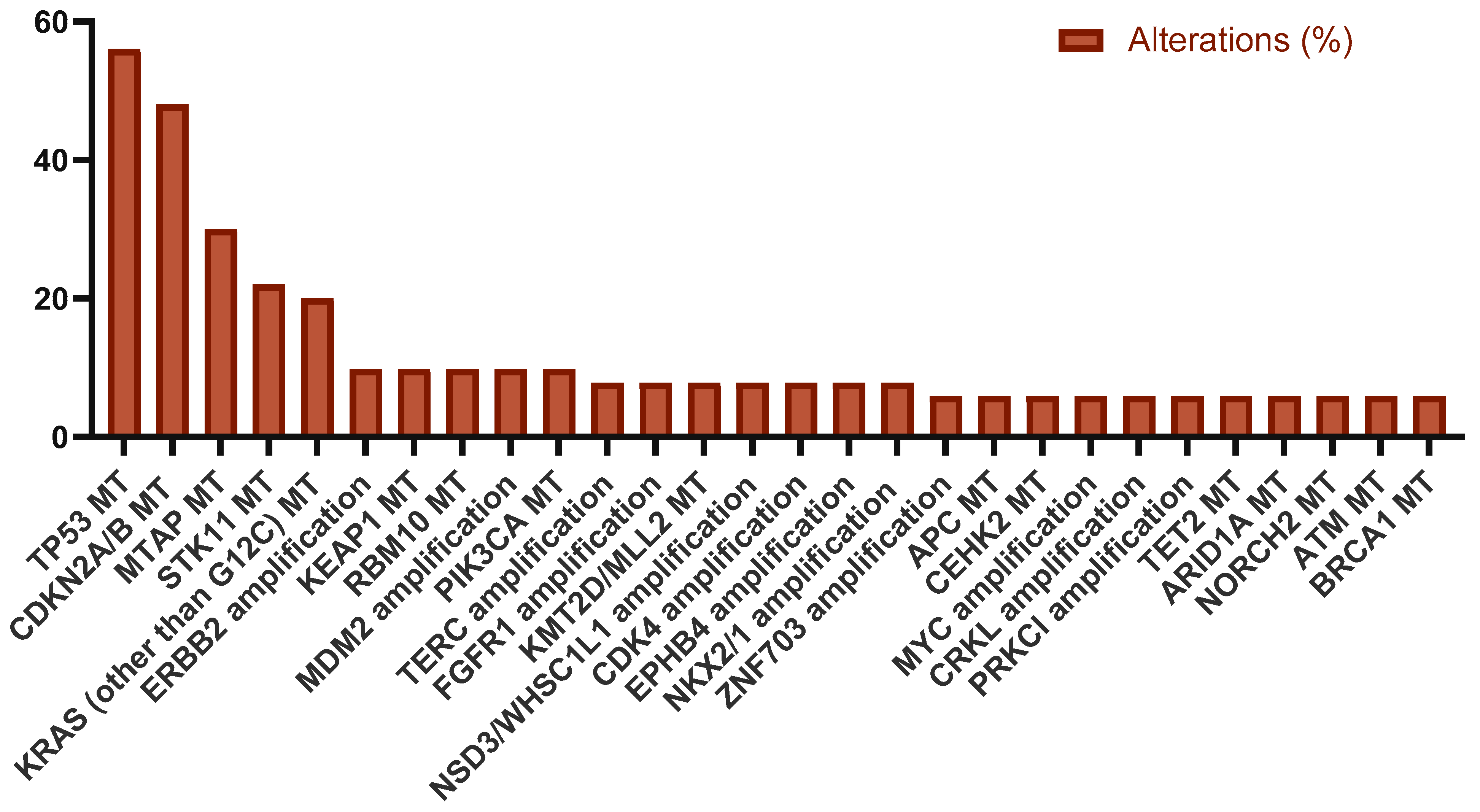
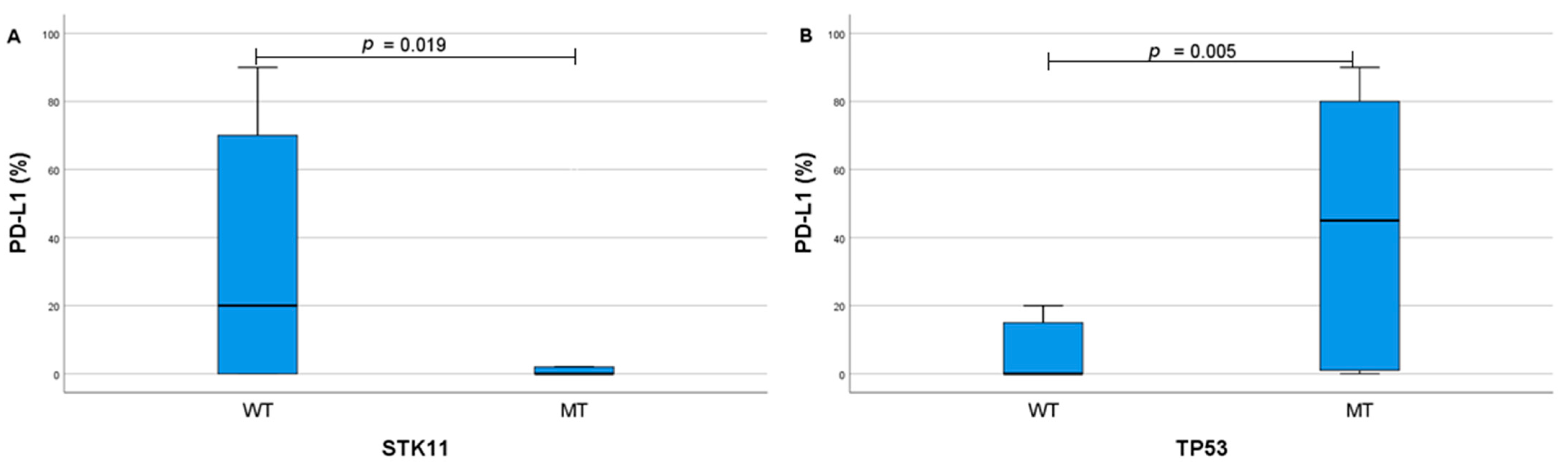
| Baseline Characteristics | n (%) |
|---|---|
| Sex | |
| Male | 38 (76) |
| Female | 12 (24) |
| Age group | |
| <65 years | 23 (46) |
| ≥65 years | 27 (54) |
| Smoking status | |
| Non-smokers | 10 (20) |
| Current or former smokers | 40 (80) |
| COPD | |
| Absent | 39 (78) |
| Present | 11 (22) |
| Histological NSCLC type | |
| Adenocarcinoma | 38 (76) |
| Squamous cell carcinoma | 10 (20) |
| Large cell carcinoma | 2 (4) |
| Differentiation | |
| Well-moderate | 22 (44) |
| Poor-undifferentiated | 18 (36) |
| NSCLC stage | |
| IVa | 21 (42) |
| IVb | 29 (58) |
| T status | |
| T1 | 8 (16) |
| T2 | 7 (14) |
| T3 | 11 (22) |
| T4 | 24 (48) |
| Lymph node status | |
| N0 | 4 (8) |
| N1 | 7 (14) |
| N2 | 14 (28) |
| N3 | 25 (50) |
| Metastases | |
| M1a | 15 (30) |
| M1b | 6 (12) |
| M1c | 29 (58) |
| Biopsy localisation | |
| Lung | 36 (72) |
| Metastasis | 14 (28) |
| Treatment | |
| Immunotherapy | 11 (22) |
| Chemotherapy + immunotherapy | 25 (50) |
| Targeted therapy | 3 (6) |
| Not given | 11 (22) |
| Alteration | Histology | p | |
|---|---|---|---|
| Adenocarcinoma | Squamous Cell Carcinoma | ||
| EGFR | |||
| WT | 35 (77.8) | 10 (22.2) | 1 |
| MT | 1 (100) | 0 (0) | |
| Amplification | 2 (100) | 0 (0) | |
| ALK | |||
| WT | 36 (78.3) | 10 (21.7) | 1 |
| MT | 2 (100) | 0 (0) | |
| BRAF | |||
| WT | 36 (78.3) | 10 (21.7) | 1 |
| MT | 2 (100) | 0 (0) | |
| ERBB2 | |||
| WT | 33 (76.7) | 10 (23.3) | 0.655 |
| MT | 1 (100) | 0 (0) | |
| Amplification | 4 (100) | 0 (0) | |
| KRAS | |||
| WT | 22 (73.3) | 8 (26.7) | 0.097 |
| MT | 15 (93.8) | 1 (6.3) | |
| Amplification | 1 (50) | 1 (50) | |
| MET | |||
| WT | 36 (78.3) | 10 (21.7) | 1 |
| MT | 1 (100) | 0 (0) | |
| Amplification | 1 (100) | 0 (0) | |
| RET | |||
| WT | 37 (78.7) | 10 (21.3) | 1 |
| MT | 1 (100) | 0 (0) | |
| ROS1 | |||
| WT | 36 (78.3) | 10 (21.7) | 1 |
| MT | 2 (100) | 0 (0) | |
| STK11 | |||
| WT | 28 (73.7) | 10 (26.3) | 0.094 |
| MT | 10 (100) | 0 (0) | |
| TP53 | |||
| WT | 21 (100) | 0 (0) | 0.003 |
| MT | 17 (63) | 10 (37) | |
| ZNF703 | |||
| WT | 36 (80) | 9 (20) | 0.512 |
| Amplification | 2 (66.7) | 1 (33.3) | |
| TERC | |||
| WT | 37 (84.1) | 7 (15.9) | 0.025 |
| Amplification | 1 (25) | 3 (75) | |
| MTAP | |||
| WT | 25 (75.8) | 8 (24.2) | 0.472 |
| MT | 13 (89.7) | 2 (13.3) | |
| CDKN2A/B | |||
| WT | 19 (79.2) | 5 (20.8) | 1 |
| MT | 19 (79.2) | 5 (20.8) | |
| FGFR1 | |||
| WT | 37 (84.1) | 7 (15.9) | 0.025 |
| Amplification | 1 (25) | 3 (75) | |
| KEAP1 | |||
| WT | 35 (79.5) | 9 (20.5) | 1 |
| MT | 3 (75) | 1 (25) | |
| KMT2D/MLL2 | |||
| WT | 37 (84.1) | 7 (15.9) | 0.025 |
| MT | 1 (25) | 3 (75) | |
| NSD3/WHSC1L1 | |||
| WT | 37 (84.1) | 7 (15.9) | 0.025 |
| Amplification | 1 (25) | 3 (75) | |
| RBM10 | |||
| WT | 33 (76.7) | 10 (23.3) | 0.569 |
| MT | 5 (100) | 0 (0) | |
| CDK4 | |||
| WT | 36 (80) | 9 (20) | 0.512 |
| Amplification | 2 (66.7) | 1 (33.3) | |
| MDM2 | |||
| WT | 33 (76.7) | 10 (23.3) | 0.569 |
| Amplification | 5 (100) | 0 (0) | |
| PIK3CA | |||
| WT | 34 (81) | 8 (19) | 0.254 |
| MT | 4 (80) | 1 (20) | |
| EPHB4 | |||
| WT | 35 (77.8) | 10 (22.2) | 1 |
| Amplification | 3 (100) | 0 (0) | |
| NKX2/1 | |||
| WT | 34 (77.3) | 10 (22.7) | 0.566 |
| Amplification | 4 (100) | 0 (0) | |
| APC | |||
| WT | 35 (77.8) | 10 (22.2) | 1 |
| MT | 3 (100) | 0 (0) | |
| CEHK2 | |||
| WT | 35 (77.8) | 10 (22.2) | 1 |
| MT | 3 (100) | 0 (0) | |
| MYC | |||
| WT | 35 (77.8) | 10 (22.2) | 1 |
| Amplification | 3 (100) | 0 (0) | |
| CRKL | |||
| WT | 37 (82.2) | 8 (17.8) | 0.106 |
| Amplification | 1 (33.3) | 2 (66.7) | |
| PRKCI | |||
| WT | 37 (82.2) | 8 (17.8) | 0.106 |
| Amplification | 1 (33.3) | 2 (66.7) | |
| TET2 | |||
| WT | 36 (80) | 9 (20) | 0.512 |
| MT | 2 (66.7) | 1 (33.3) | |
| ARID1A | |||
| WT | 35 (77.8) | 10 (22.2) | 1 |
| MT | 3 (100) | 0 (0) | |
| NORCH2 | |||
| WT | 35 (77.8) | 10 (22.2) | 1 |
| MT | 3 (100) | 0 (0) | |
| ATM | |||
| WT | 36 (80) | 9 (20) | 0.512 |
| MT | 2 (66.7) | 1 (33.3) | |
| BRCA1 | |||
| WT | 35 (77.8) | 10 (22.2) | 1 |
| MT | 3 (100) | 0 (0) | |
| Co-Occurring Mutations | PD-L1 | PD-L1 | ||||
|---|---|---|---|---|---|---|
| Negative | Positive | p | Low | High | p | |
| KRAS/STK11 | ||||||
| Not detected | 14 (31.1) | 31 (68.9) | 0.005 | 27 (60) | 18 (40) | 0.148 |
| Detected | 5 (100) | 0 (0) | 5 (100) | 0 (0) | ||
| KRAS/TP53 | ||||||
| Not detected | 17 (37) | 29 (63) | 0.629 | 30 (65.2) | 16 (34.8) | 0.612 |
| Detected | 2 (50) | 2 (50) | 2 (50) | 2 (50) | ||
| STK11/KEAP1 | ||||||
| Not detected | 16 (34) | 31 (66) | 0.049 | 29 (61.7) | 18 (38.3) | 0.544 |
| Detected | 3 (100) | 0 (0) | 3 (100) | 0 (0) | ||
| STK11/TP53 | ||||||
| Not detected | 16 (37.2) | 27 (62.8) | 1 | 26 (60.5) | 17 (39.5) | 0.398 |
| Detected | 3 (42.9) | 4 (57.1) | 6 (85.7) | 1 (14.3) | ||
| Alteration | PD-L1 | p | PD-L1 | p | TMB | p | |||
|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Low | High | Low | High | ||||
| EGFR | |||||||||
| WT | 18 (38.3) | 29 (61.7) | 0.29 | 31 (66) | 16 (34) | 0.125 | 28 (63.6) | 17 (36.4) | 1 |
| MT | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | |||
| Amplification | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 1 (50) | 1 (50) | |||
| ALK | |||||||||
| WT | 18 (37.5) | 30 (62.5) | 1 | 31 (64.6) | 17 (35.4) | 1 | 28 (62.2) | 17 (37.8) | 0.528 |
| MT | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 2 (100) | 0 (0) | |||
| BRAF | |||||||||
| WT | 18 (37.5) | 30 (62.5) | 1 | 31 (64.6) | 17 (35.4) | 1 | 29 (63) | 17 (37) | 0.528 |
| MT | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (100) | 0 (0) | |||
| ERBB2 | |||||||||
| WT | 16 (36.4) | 28 (63.6) | 0.605 | 28 (63.6) | 16 (36.4) | 1 | 27 (65.9) | 14 (34.1) | 0.581 |
| MT | 1 (5.3) | 0 (0) | 1 (102) | 0 (0) | 1 (100) | 0 (0) | |||
| Amplification | 2 (40) | 3 (60) | 3 (60) | 2 (40) | 2 (40) | 3 (60) | |||
| KRAS | |||||||||
| WT | 12 (38.7) | 19 (61.3) | 0.777 | 18 (58.1) | 13 (41.9) | 0.373 | 18 (60) | 12 (40) | 0.576 |
| MT | 7 (41.2) | 10 (58.8) | 13 (76.5) | 4 (23.5) | 11 (73.3) | 4 (26.7) | |||
| Amplification | 0 (0) | 2 (100) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | |||
| MET | |||||||||
| WT | 18 (37.5) | 30 (62.5) | 0.62 | 31 (64.6) | 17 (35.4) | 0.595 | 29 (64.4) | 16 (35.6) | 0.598 |
| MT | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | |||
| Amplification | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | |||
| RET | |||||||||
| WT | 18 (36.7) | 31 (63.6) | 0.38 | 31 (63.3) | 18 (36.7) | 1 | 29 (63) | 17 (37) | 1 |
| MT | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | |||
| ROS1 | |||||||||
| WT | 19 (36.6) | 29 (60.4) | 0.519 | 32 (66.7) | 16 (33.3) | 0.125 | 28 (62.2) | 17 (37.8) | 0.528 |
| MT | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 2 (100) | 0 (0) | |||
| STK11 | |||||||||
| WT | 12 (30.8) | 27 (69.2) | 0.078 | 22 (56.4) | 17 (43.6) | 0.072 | 25 (67.6) | 12 (32.4) | 0.46 |
| MT | 7 (63.6) | 4 (36.4) | 10 (90.9) | 1 (9.1) | 5 (50) | 5 (50) | |||
| TP53 | |||||||||
| WT | 12 (54.5) | 10 (45.5) | 0.043 | 18 (81.8) | 4 (18.2) | 0.036 | 16 (80) | 4 (20) | 0.067 |
| MT | 7 (25) | 21 (75) | 14 (50) | 14 (50) | 14 (51.9) | 13 (48.1) | |||
| ZNF703 | |||||||||
| WT | 18 (38.3) | 29 (61.7) | 1 | 29 (61.7) | 18 (38.3) | 0.544 | 30 (68.2) | 14 (31.8) | 0.042 |
| Amplification | 1 (33.3) | 2 (66.7) | 3 (100) | 0 (0) | 0 (0) | 3 (100) | |||
| TERC | |||||||||
| WT | 17 (37) | 29 (63) | 0.629 | 29 (63) | 17 (37) | 1 | 28 (65.1) | 15 (34.9) | 0.613 |
| Amplification | 2 (50) | 2 (50) | 3 (75) | 1 (25) | 2 (50) | 2 (50) | |||
| MTAP | |||||||||
| WT | 12 (34.3) | 23 (65.7) | 0.528 | 21 (60) | 14 (40) | 0.523 | 18 (56.3) | 14 (43.8) | 0.193 |
| MT | 7 (46.7) | 8 (53.3) | 11 (73.3) | 4 (26.7) | 12 (80) | 3 (20) | |||
| CDKN2A/B | |||||||||
| WT | 10 (38.5) | 16 (61.5) | 1 | 18 (69.2) | 8 (30.8) | 0.557 | 12 (52.2) | 11 (47.8) | 0.135 |
| MT | 9 (37.5) | 15 (62.5) | 14 (58.3) | 10 (41.7) | 18 (75) | 6 (25) | |||
| FGFR1 | |||||||||
| WT | 18 (39.1) | 28 (60.9) | 1 | 30 (65.2) | 16 (34.8) | 0.612 | 28 (65.1) | 15 (34.9) | 0.613 |
| Amplification | 1 (25) | 3 (75) | 2 (50) | 2 (50) | 2 (50) | 2 (50) | |||
| KEAP1 | |||||||||
| WT | 15 (33.3) | 30 (66.7) | 0.062 | 28 (62.2) | 17 (37.8) | 0.642 | 29 (69) | 13 (31) | 0.051 |
| MT | 4 (80) | 1 (20) | 4 (80) | 1 (20) | 1 (20) | 4 (80) | |||
| KMT2D/MLL2 | |||||||||
| WT | 17 (37) | 29 (63) | 0.629 | 30 (65.2) | 16 (34.8) | 0.612 | 28 (65.1) | 15 (34.9) | 0.613 |
| MT | 2 (50) | 2 (50) | 2 (50) | 2 (50) | 2 (50) | 2 (50) | |||
| NSD3/WHSC1L1 | |||||||||
| WT | 18 (39.1) | 28 (60.9) | 1 | 30 (65.2) | 16 (34.8) | 0.612 | 28 (65.1) | 15 (34.9) | 0.613 |
| Amplification | 1 (25) | 3 (75) | 2 (50) | 2 (50) | 2 (50) | 2 (50) | |||
| RBM10 | |||||||||
| WT | 17 (37.8) | 28 (62.2) | 1 | 28 (62.2) | 17 (37.8) | 0.642 | 28 (65.1) | 15 (34.9) | 0.613 |
| MT | 2 (40) | 3 (60) | 4 (80) | 1 (20) | 2 (50) | 2 (50) | |||
| CDK4 | |||||||||
| WT | 17 (37) | 29 (63) | 0.629 | 29 (63) | 17 (37) | 1 | 29 (67.4) | 14 (32.6) | 0.128 |
| Amplification | 2 (50) | 2 (50) | 3 (25) | 1 (25) | 1 (25) | 3 (75) | |||
| MDM2 | |||||||||
| WT | 15 (33.3) | 30 (66.7) | 0.062 | 28 (62.2) | 17 (37.8) | 0.642 | 27 (64.3) | 15 (35.7) | 1 |
| Amplification | 4 (80) | 1 (20) | 4 (80) | 1 (20) | 3 (60) | 2 (40) | |||
| PIK3CA | |||||||||
| WT | 16 (35.6) | 29 (64.4) | 0.355 | 28 (62.2) | 17 (37.8) | 0.642 | 25 (59.5) | 17 (40.5) | 0.143 |
| MT | 3 (60) | 2 (40) | 4 (80) | 1 (20) | 5 (100) | 0 (0) | |||
| EPHB4 | |||||||||
| WT | 17 (37) | 29 (63) | 0.629 | 28 (60.9) | 18 (39.1) | 0.283 | 29 (67.4) | 14 (32.6) | 0.128 |
| Amplification | 2 (50) | 2 (50) | 4 (100) | 0 (0) | 1 (25) | 3 (75) | |||
| NKX2/1 | |||||||||
| WT | 18 (39.1) | 28 (60.9) | 1 | 29 (63) | 17 (37) | 1 | 29 (67.4) | 14 (32.6) | 0.128 |
| Amplification | 1 (25) | 3 (75) | 3 (75) | 1 (25) | 1 (25) | 3 (75) | |||
| APC | |||||||||
| WT | 19 (40.4) | 28 (59.6) | 0.279 | 30 (63.8) | 17 (36.2) | 1 | 30 (68.2) | 14 (31.8) | 0.042 |
| MT | 0 (0) | 3 (100) | 2 (66.7) | 1 (33.3) | 0 (0) | 3 (100) | |||
| CEHK2 | |||||||||
| WT | 19 (40.4) | 28 (59.6) | 0.279 | 30 (63.8) | 17 (36.2) | 1 | 30 (68.2) | 14 (31.8) | 0.042 |
| MT | 0 (0) | 3 (100) | 2 (66.7) | 1 (33.3) | 0 (0) | 3 (100) | |||
| MYC | |||||||||
| WT | 18 (38.3) | 29 (61.7) | 1 | 29 (61.7) | 18 (38.3) | 0.544 | 28 (63.6) | 16 (36.4) | 1 |
| Amplification | 1 (33.3) | 2 (66.7) | 3 (100) | 0 (0) | 2 (66.7) | 1 (33.3) | |||
| CRKL | |||||||||
| WT | 17 (36.2) | 30 (63.8) | 0.549 | 29 (61.7) | 18 (38.3) | 0.544 | 29 (65.9) | 15 (34.1) | 0.544 |
| Amplification | 2 (66.7) | 1 (33.3) | 3 (100) | 0 (0) | 1 (33.3) | 2 (66.7) | |||
| PRKCI | |||||||||
| WT | 17 (36.2) | 30 (63.8) | 0.549 | 30 (63.8) | 17 (36.2) | 1 | 28 (63.6) | 16 (36.4) | 1 |
| Amplification | 2 (66.7) | 1 (33.3) | 2 (66.7) | 1 (33.3) | 2 (66.7) | 1 (33.3) | |||
| TET2 | |||||||||
| WT | 17 (36.2) | 30 (63.8) | 0.549 | 29 (61.7) | 18 (38.3) | 0.544 | 28 (63.6) | 16 (36.4) | 1 |
| MT | 2 (66.7) | 1 (33.3) | 3 (100) | 0 (0) | 2 (66.7) | 1 (33.3) | |||
| ARID1A | |||||||||
| WT | 18 (38.3) | 29 (61.7) | 1 | 31 (66) | 16 (34) | 0.291 | 29 (65.9) | 15 (34.1) | 0.544 |
| MT | 1 (33.3) | 2 (66.7) | 1 (33.3) | 2 (66.7) | 1 (33.3) | 2 (66.7) | |||
| NORCH2 | |||||||||
| WT | 18 (38.3) | 29 (61.7) | 1 | 29 (61.7) | 18 (38.3) | 0.544 | 30 (68.2) | 14 (31.8) | 0.042 |
| MT | 1 (33.3) | 2 (66.7) | 3 (100) | 0 (0) | 0 (0) | 3 (100) | |||
| ATM | |||||||||
| WT | 19 (40.4) | 28 (59.6) | 0.279 | 30 (63.8) | 17 (36.2) | 1 | 29 (64.4) | 16 (35.6) | 1 |
| MT | 0 (0) | 3 (100) | 2 (66.7) | 1 (33.3) | 1 (50) | 1 (50) | |||
| BRCA1 | |||||||||
| WT | 18 (38.3) | 29 (61.7) | 1 | 31 (66) | 16 (34) | 0.291 | 27 (61.4) | 17 (38.6) | 0.292 |
| MT | 1 (33.3) | 2 (66.7) | 1 (33.3) | 2 (66.7) | 3 (100) | 0 (0) | |||
| Clinical Case | Histological Type | PD-L1 Expression (%) | TMB (muts/Mb) | Treatment | STK11 MT | KEAP1 MT | Other Alterations |
|---|---|---|---|---|---|---|---|
| I | Adenocarcinoma | 60 | 10 | pembrolizumab | + | − | TP53 MT, CDKN2A/B MT |
| II | Squamous cell carcinoma | 80 | 6 | pembrolizumab | − | − | TP53 MT, KRAS amplifications, FGFR1 amplifications, NSD3/WHSC1L1 amplifications, KMT2D/MLL2 MT |
| III | Adenocarcinoma | 60 | 17 | Carboplatin, pemetrexed, pembrolizumab | − | + | TP53 MT CDKN2A/B MT, APC MT |
| IV | Adenocarcinoma | 0 | 10 | Carboplatin, pemetrexed, pembrolizumab | − | + | TERC MT EPHB4 MT NKX2/1 MT CRKL MT PRKCI MT NOTCH MT |
| V | Adenocarcinoma | 0 | 6 | Carboplatin, pemetrexed, pembrolizumab | − | − | ERBB2 amplification TP53 MT CDKN2B MT TET2 MT BRCA1 MT |
| VI | Large cell carcinoma | 0 | 22 | Carboplatin, pemetrexed, pembrolizumab | + | + | ERBB2 amplification TP53 MT CDK4 amplification EPHB4 MT |
| VII | Adenocarcinoma | 90 | 4 | Pembrolizumab | − | − | ERBB2 MT ERBB2 amplification TP53 MT MTAP MT CDKN2A/B MT |
| VIII | Adenocarcinoma | 2 | 13 | Carboplatin, pemetrexed, pembrolizumab | + | − | KRAS amplification TP53 MT |
| Non–Fast Progression | Fast Progression | OR | 95% CI | p | |
|---|---|---|---|---|---|
| n | 28 | 8 | |||
| Sex | |||||
| Male | 20 (71.4) | 8 (28.6) | 0.714 | 0.565–0.903 | 0.156 |
| Female | 8 (100) | 0 (0) | |||
| Age group | |||||
| <65 years | 12 (85.7) | 2 (14.3) | 2.25 | 0.385–13.166 | 0.441 |
| ≥65 years | 16 (72.7) | 6 (27.3) | |||
| Smoking status | |||||
| Non-smokers | 4 (80) | 1 (20) | 1.167 | 0.112–12.202 | 1 |
| Current or former smokers | 24 (77.4) | 7 (22.6) | |||
| COPD | |||||
| Absent | 20 (74.1) | 7 (25.9) | 0.357 | 0.038–3.388 | 0.648 |
| Present | 8 (88.9) | 1 (11.1) | |||
| Histological NSCLC type | |||||
| Adenocarcinoma | 18 (75) | 6 (25) | 0.333 | 0.035–3.205 | 0.644 |
| Squamous cell carcinoma | 9 (90) | 1 (10) | |||
| Differentiation | |||||
| Well-moderate | 14 (87.5) | 2 (12.5) | 4.375 | 0.684–27.983 | 0.192 |
| Poor-undifferentiated | 8 (61.5) | 5 (38.5) | |||
| NSCLC stage | |||||
| IVa | 14 (82.4) | 3 (17.6) | 1.667 | 0.333–8.352 | 0.695 |
| IVb | 14 (73.7) | 5 (26.3) | |||
| T status | |||||
| T1-3 | 14 (73.7) | 5 (26.3) | 0.6 | 0.12–3.007 | 0.695 |
| T4 | 14 (82.4) | 3 (17.6) | |||
| Lymph node status | |||||
| N0 | 3 (100) | 0 (0) | 1.32 | 1.088–1.601 | 1 |
| N1-3 | 25 (75.8) | 8 (24.2) | |||
| Metastases | |||||
| M1a | 10 (83.3) | 2 (16.7) | 1.667 | 0.282–9.856 | 0.691 |
| M1b-1c | 18 (75) | 6 (25) | |||
| TMB | |||||
| low | 17 (85) | 3 (15) | 3.148 | 0.608–16.289 | 0.228 |
| high | 9 (64.3) | 5 (35.7) | |||
| PD-L1 | |||||
| negative | 11 (78.6) | 3 (21.4) | 1.078 | 0.213–0.713 | 1 |
| positive | 17 (77.3) | 5 (22.7) | |||
| STK11 | |||||
| WT | 22 (81.5) | 5 (18.5) | 2.2 | 0.405–11.949 | 0.384 |
| MT | 6 (66.7) | 3 (33.3) | |||
| KEAP1 | |||||
| WT | 27 (84.4) | 5 (15.6) | 16.2 | 1.38–188.883 | 0.028 |
| MT | 1 (25) | 3 (75) | |||
| MDM2 | |||||
| WT | 26 (76.5) | 8 (23.5) | 0.765 | 0.635–0.921 | 1 |
| amplification | 2 (100) | 0 (0) | |||
| ERBB2 | |||||
| WT | 25 (83.3) | 5 (16.7) | 7.5 | 0.984–57.138 | 0.067 |
| amplification | 2 (40) | 3 (60) | |||
| TP53 | |||||
| WT | 11 (91.7) | 1 (8.3) | 4.529 | 0.488–42.052 | 0.224 |
| MT | 17 (70.8) | 7 (29.2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurevičienė, G.; Poškienė, L.; Miliauskas, S.; Žemaitis, M. PD-L1 Expression and Comprehensive Genomic Profiling in Advanced NSCLC: A Single-Centre Experience. Int. J. Mol. Sci. 2025, 26, 6348. https://doi.org/10.3390/ijms26136348
Gurevičienė G, Poškienė L, Miliauskas S, Žemaitis M. PD-L1 Expression and Comprehensive Genomic Profiling in Advanced NSCLC: A Single-Centre Experience. International Journal of Molecular Sciences. 2025; 26(13):6348. https://doi.org/10.3390/ijms26136348
Chicago/Turabian StyleGurevičienė, Giedrė, Lina Poškienė, Skaidrius Miliauskas, and Marius Žemaitis. 2025. "PD-L1 Expression and Comprehensive Genomic Profiling in Advanced NSCLC: A Single-Centre Experience" International Journal of Molecular Sciences 26, no. 13: 6348. https://doi.org/10.3390/ijms26136348
APA StyleGurevičienė, G., Poškienė, L., Miliauskas, S., & Žemaitis, M. (2025). PD-L1 Expression and Comprehensive Genomic Profiling in Advanced NSCLC: A Single-Centre Experience. International Journal of Molecular Sciences, 26(13), 6348. https://doi.org/10.3390/ijms26136348






