Structure–Function Analysis of the Steroid-Hydroxylating Cytochrome P450 109 (CYP109) Enzyme Family
Abstract
1. Introduction
2. Results
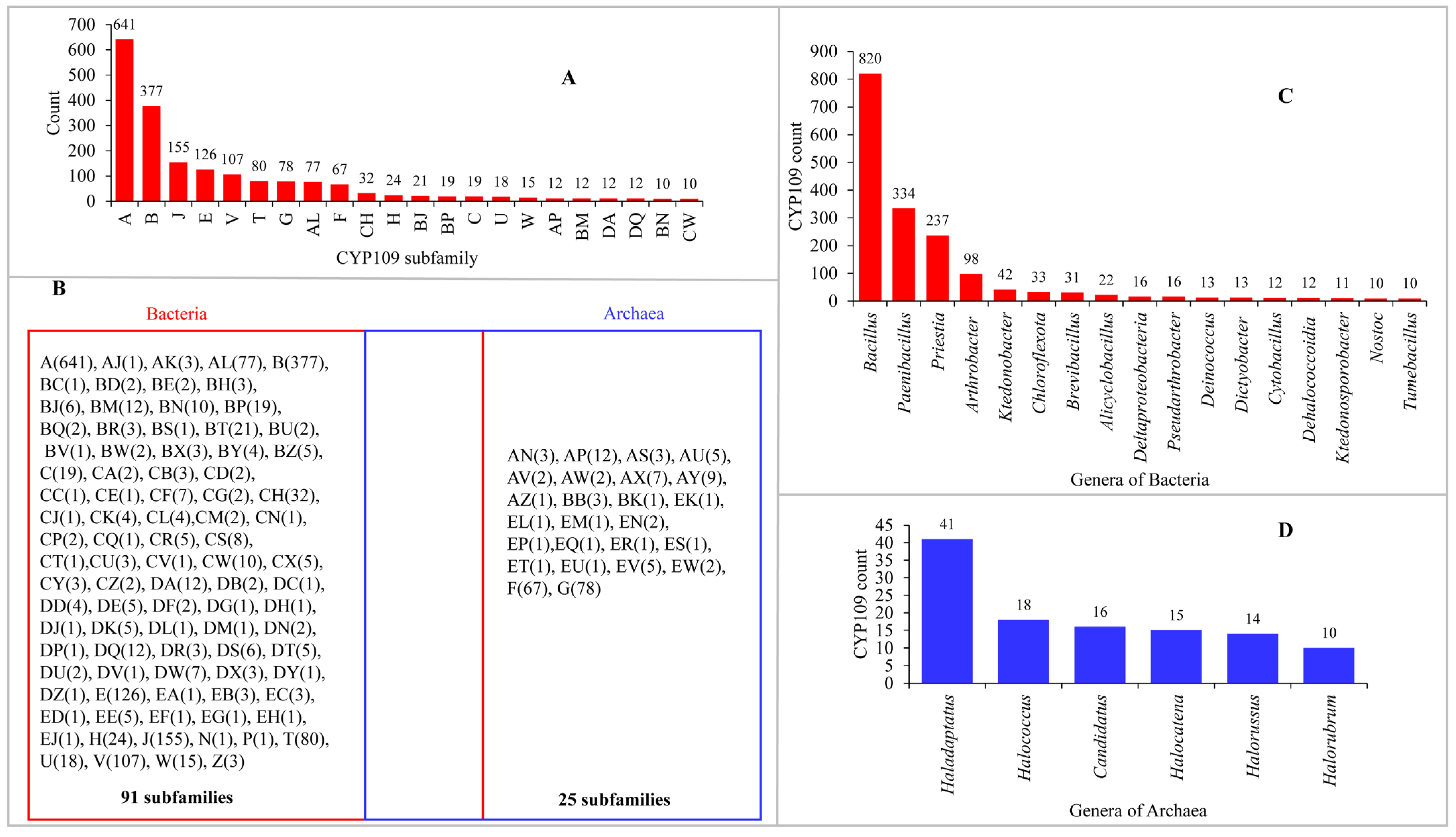
2.1. CYP109s Are Present Only in Bacteria and Archaea to Date
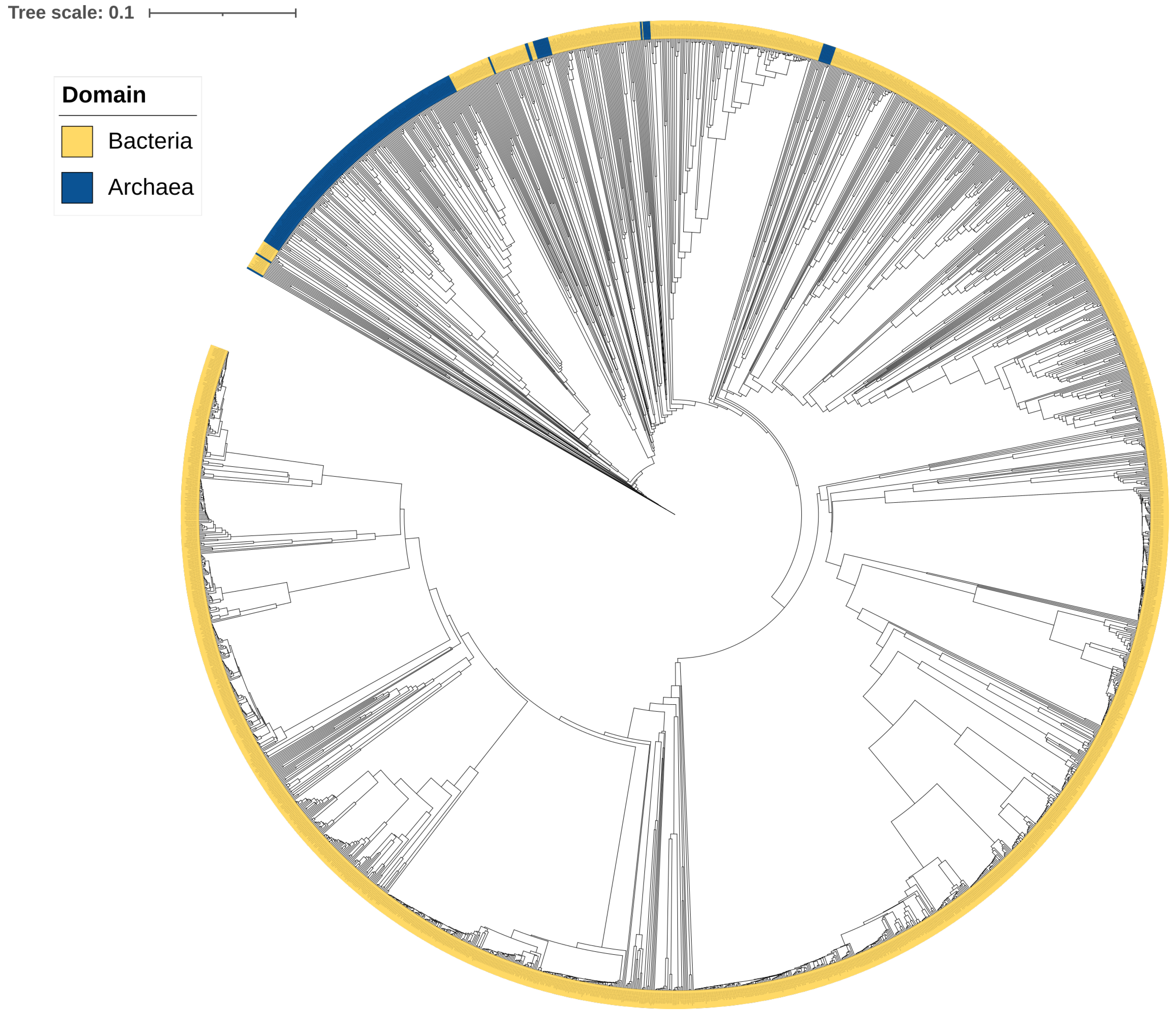
2.2. CYP109s Are of Bacterial Origin
2.3. CYP109 Members Are Structurally Highly Dynamic
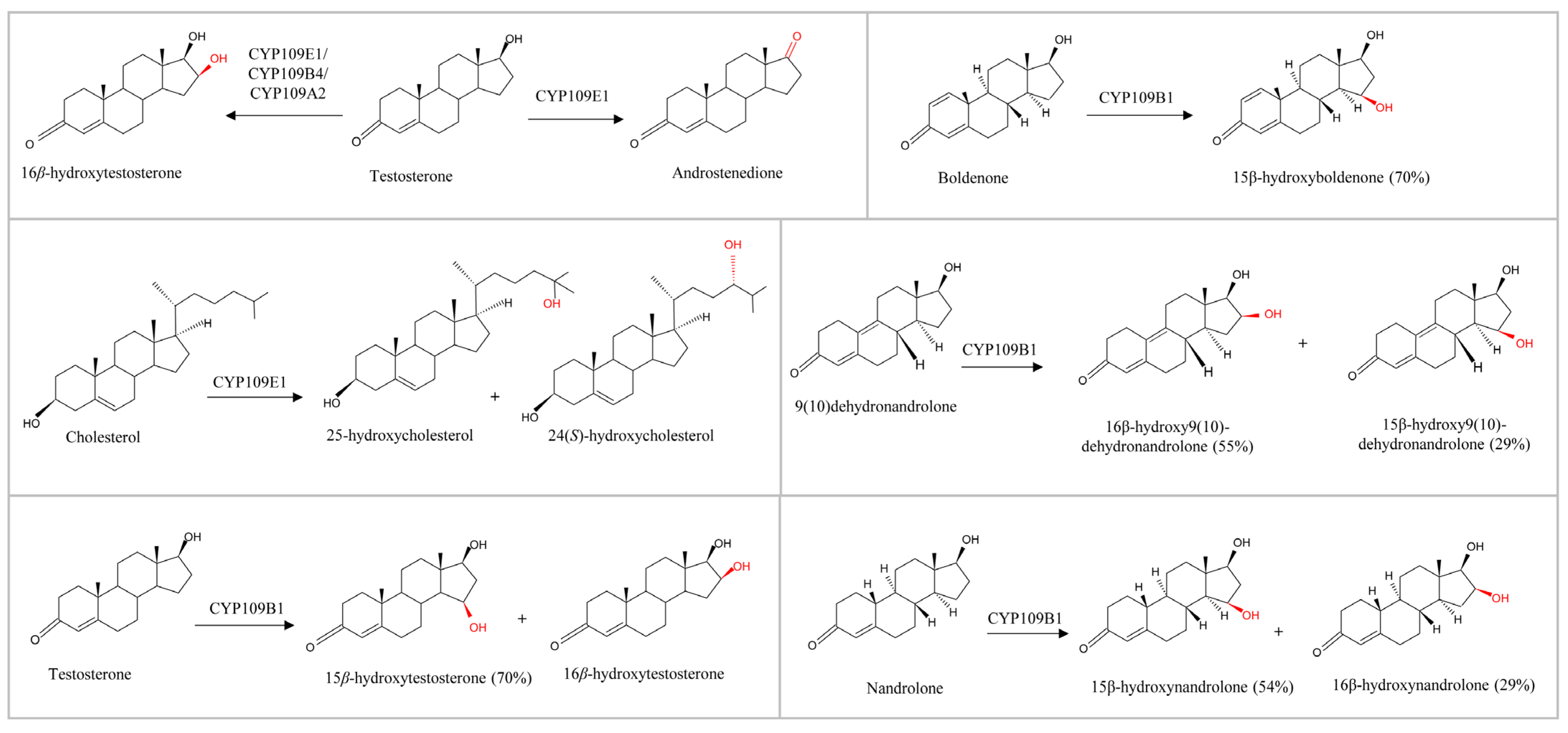
2.4. CYP109E1
2.4.1. The Type of C17 Substituent on Steroids Determines the Catalysis
2.4.2. Four Amino Acids (Val169, Lys187, Ile241, and Thr246) Are Important for Catalytic Activity
2.4.3. Thr246 Plays a Role in the Proton Shuttle
2.5. CYP109B4
2.5.1. Asn77 Hydrophilic Interaction Is Crucial in Positioning Testosterone for C16β-Hydroxylation
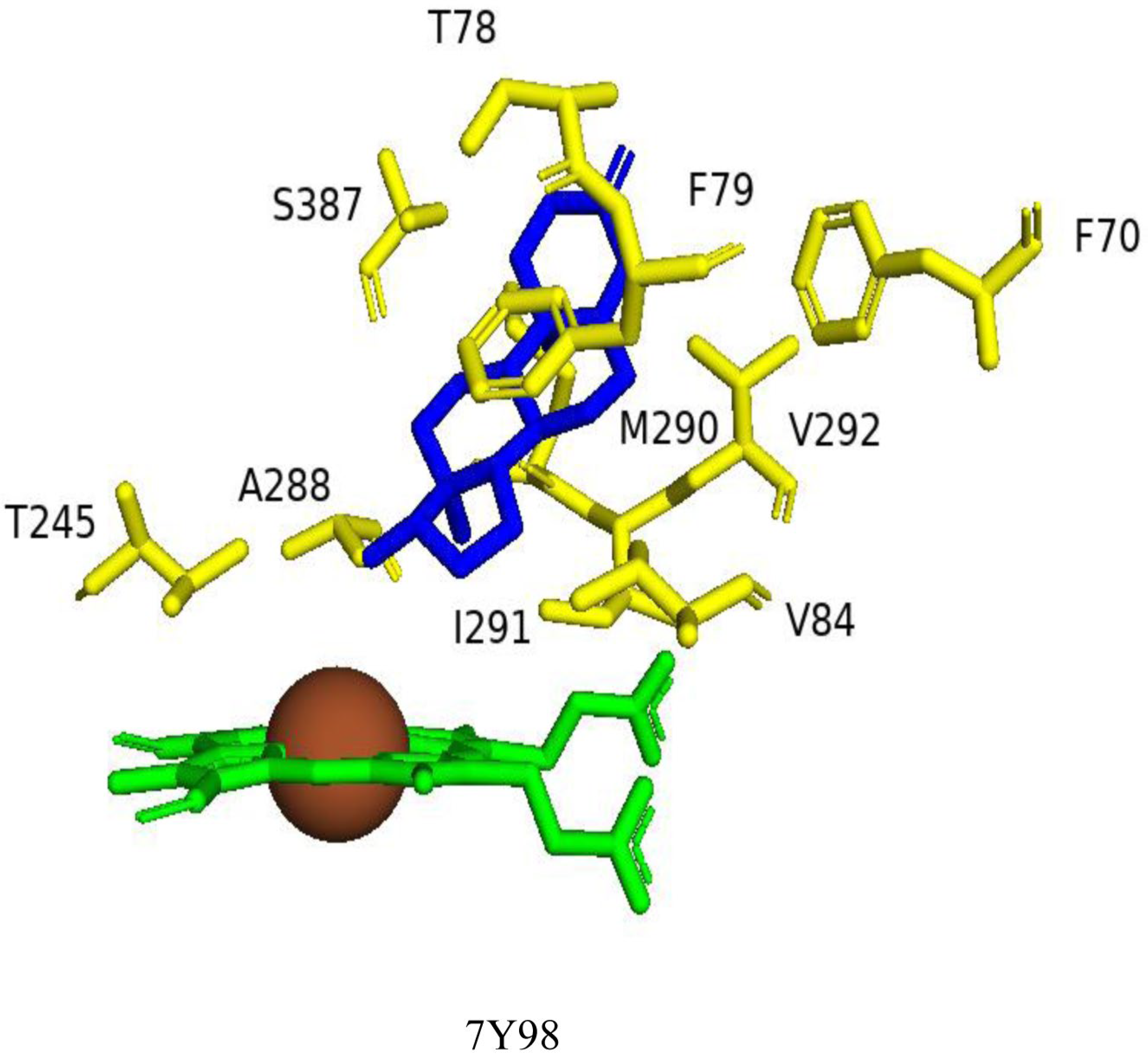
2.5.2. Val84, Val292, and Ser387 Determine the CYP109B4 Regioselectivity
2.6. CYP109A2
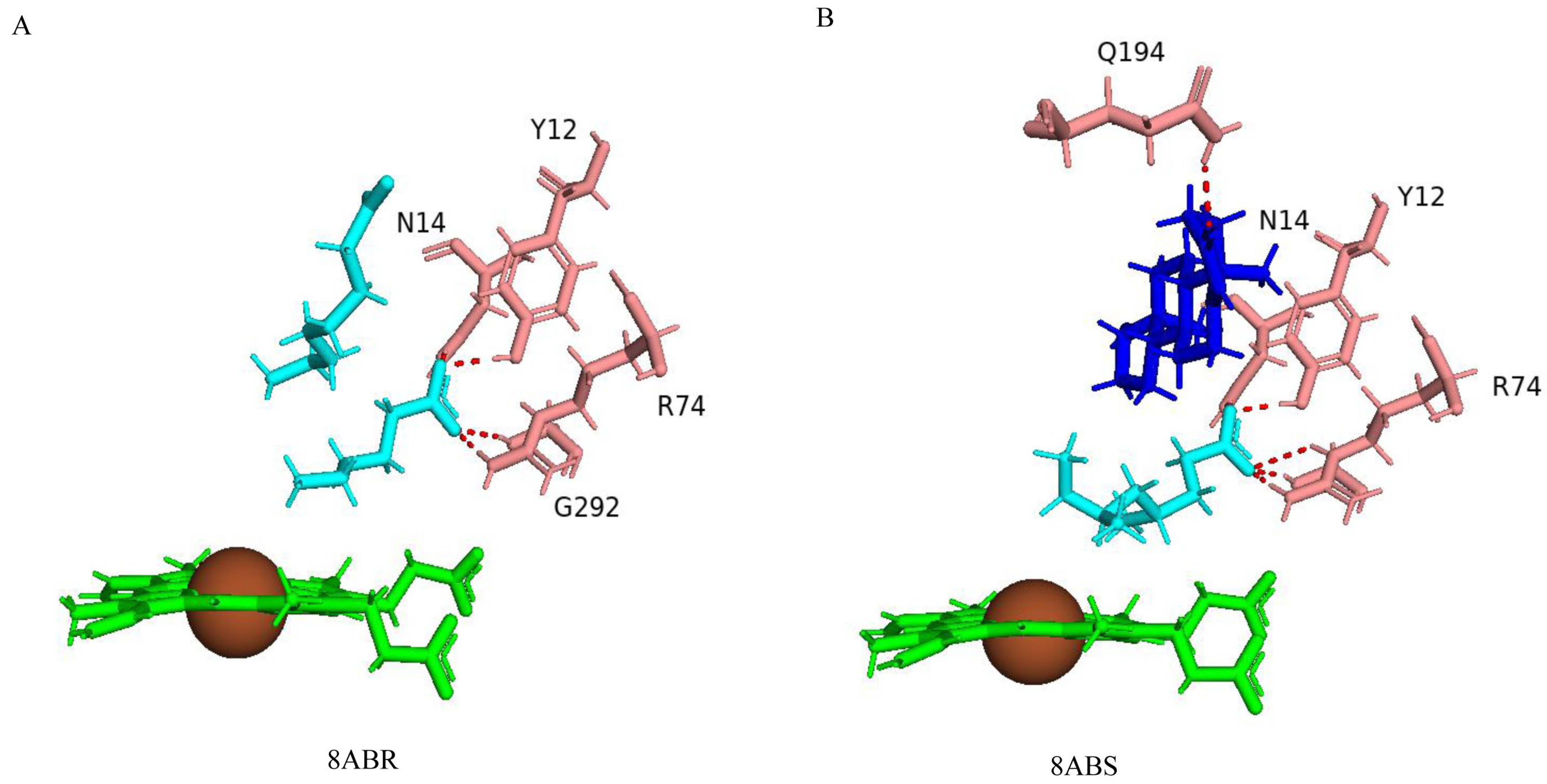
Right Positioning and Orientation of Testosterone Toward Heme Is Essential for Catalysis
3. Materials and Methods
3.1. Genome Data Mining and Annotation of CYP109s
3.2. Phylogenetic Analysis of the CYP109 Family
3.3. Retrieving of CYP109 Crystal Structures
3.4. CYP109 Active Site Analysis
3.5. Analysis of Ligand Interactions in Closed Conformation
3.6. Annotation of P450 Characteristic Secondary Structures and Identification of Substrate Recognition Sites (SRSs)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ericson-Neilsen, W.; Kaye, A.D. Steroids: Pharmacology, complications, and practice delivery issues. Ochsner J. 2014, 14, 203–207. [Google Scholar] [PubMed]
- Lewis, W. The Role of Steroids in Clinical Practice: Benefits, Risks, and Considerations for Therapeutic Use. Anna. Clin. Trai. Vacci. Res. 2023, 13, 128–131. [Google Scholar]
- Shaikh, S.; Verma, H.; Yadav, N.; Jauhari, M.; Bullangowda, J. Applications of steroid in clinical practice: A review. Int. Sch. Res. Not. 2012, 2012, 985495. [Google Scholar] [CrossRef]
- Cramer, J.; Sager, C.P.; Ernst, B. Hydroxyl Groups in Synthetic and Natural-Product-Derived Therapeutics: A Perspective on a Common Functional Group. J. Med. Chem. 2019, 62, 8915–8930. [Google Scholar] [CrossRef]
- Mao, F.; Ni, W.; Xu, X.; Wang, H.; Wang, J.; Ji, M.; Li, J. Chemical Structure-Related Drug-Like Criteria of Global Approved Drugs. Molecules 2016, 21, 75. [Google Scholar] [CrossRef]
- Guengerich, F.P. Mechanisms of Cytochrome P450-Catalyzed Oxidations. ACS Catal. 2018, 8, 10964–10976. [Google Scholar] [CrossRef]
- Abas, H.; Blencowe, P.; Brookfield, J.L.; Harwood, L.A. Selective Hydroxylation of C (sp3)−H Bonds in Steroids. Chem.–A Eur. J. 2023, 29, e202301066. [Google Scholar] [CrossRef]
- Sun, C.; Zeng, R.; Chen, T.; Yang, Y.; Song, Y.; Li, Q.; Cheng, J.; Liu, B. Recent advances and challenges in the production of hydroxylated natural products using microorganisms. Fermentation 2024, 10, 604. [Google Scholar] [CrossRef]
- Gront, D.; Syed, K.; Nelson, D.R. Exploring P450 superfamily diversity with P450Atlas-Online tool for automated subfamily assignment. Protein Sci. 2025, 34, e70057. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. 2006, 320, 1–10. [Google Scholar] [CrossRef]
- Khatri, Y.; Hannemann, F.; Girhard, M.; Kappl, R.; Même, A.; Ringle, M.; Janocha, S.; Leize-Wagner, E.; Urlacher, V.B.; Bernhardt, R. Novel family members of CYP109 from Sorangium cellulosum So ce56 exhibit characteristic biochemical and biophysical properties. Biotechnol. Appl. Biochem. 2013, 60, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Ngcobo, P.E.; Nkosi, B.V.Z.; Chen, W.; Nelson, D.R.; Syed, K. Evolution of cytochrome P450 enzymes and their redox partners in Archaea. Int. J. Mol. Sci. 2023, 24, 4161. [Google Scholar] [CrossRef] [PubMed]
- Jóźwik, I.; Kiss, F.M.; Gricman, Ł.; Abdulmughni, A.; Brill, E.; Zapp, J.; Pleiss, J.; Bernhardt, R.; Thunnissen, A.-M. Structural basis of steroid binding and oxidation by the cytochrome P450 CYP109E1 from Bacillus megaterium. FEBS J. 2016, 283, 4128–4148. [Google Scholar] [CrossRef] [PubMed]
- Putkaradze, N.; Litzenburger, M.; Hutter, M.C.; Bernhardt, R. CYP109E1 from Bacillus megaterium acts as a 24-and 25-hydroxylase for cholesterol. ChemBioChem A Eur. J. Chem. Biol. 2019, 20, 655–658. [Google Scholar] [CrossRef]
- Abdulmughni, A.; Jóźwik, I.K.; Putkaradze, N.; Brill, E.; Zapp, J.; Thunnissen, A.W.; Hannemann, F.; Bernhardt, R. Characterization of cytochrome P450 CYP109E1 from Bacillus megaterium as a novel vitamin D(3) hydroxylase. J. Biotechnol. 2017, 243, 38–47. [Google Scholar] [CrossRef]
- Putkaradze, N.; König, L.; Kattner, L.; Hutter, M.C.; Bernhardt, R. Highly regio- and stereoselective hydroxylation of vitamin D2 by CYP109E1. Biochem. Biophys. Res. Commun. 2020, 524, 295–300. [Google Scholar] [CrossRef]
- Jóźwik, I.; Bombino, E.; Abdulmughni, A.; Hartz, P.; Rozeboom, H.; Wijma, H.; Kappl, R.; Janssen, D.; Bernhardt, R.; Thunnissen, A.-M. Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling. FEBS J. 2023, 290, 5016–5035. [Google Scholar] [CrossRef]
- Abdulmughni, A.; Jóźwik, I.; Brill, E.; Hannemann, F.; Thunnissen, A.-M.; Bernhardt, R. Biochemical and structural characterization of CYP109A2, a vitamin D 3 25-hydroxylase from Bacillus megaterium. FEBS J. 2017, 284, 3881–3894. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, P.; Zhao, J.; Chen, Y.; Li, X.; Huang, J.; Zhang, L.; Li, Q.; Gao, C.; Xing, Q.; et al. Rationally Controlling Selective Steroid Hydroxylation via Scaffold Sampling of a P450 Family. ACS Catal. 2023, 13, 1280–1289. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, Y.; Peng, W.; Gao, C.; Xing, Q.; Wang, B.; Li, A. Exploring the Potential of Cytochrome P450 CYP109B1 Catalyzed Regio—And Stereoselective Steroid Hydroxylation. Front. Chem. 2021, 9, 649000. [Google Scholar] [CrossRef]
- Girhard, M.; Klaus, T.; Khatri, Y.; Bernhardt, R.; Urlacher, V. Characterization of the versatile monooxygenase CYP109B1 from Bacillus subtilis. Appl. Microbiol. Biotechnol. 2010, 87, 595–607. [Google Scholar] [CrossRef]
- Zhang, A.; Zhang, T.; Hall, E.; Hutchinson, S.; Cryle, M.; Wong, L.; Zhou, W.; Bell, S. The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis. Mol. BioSyst. 2015, 11, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Msweli, S.; Chonco, A.; Msweli, L.; Syed, P.R.; Karpoormath, R.; Chen, W.; Gront, D.; Nkosi, B.V.Z.; Nelson, D.R.; Syed, K. Lifestyles shape the cytochrome P450 repertoire of the Bacterial phylum Proteobacteria. Int. J. Mol. Sci. 2022, 23, 5821. [Google Scholar] [CrossRef] [PubMed]
- Sezutsu, H.; Le Goff, G.; Feyereisen, R. Origins of P450 diversity. Philos. Trans. R. Soc. Ser. B Biol. Sci. 2013, 368, 20120428. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Padayachee, T.; Lamb, D.C.; Nelson, D.R.; Syed, K. Structure-function analysis of the biotechnologically important cytochrome P450 107 (CYP107) enzyme family. Biomolecules 2023, 13, 1733. [Google Scholar] [CrossRef]
- Padayachee, T.; Lamb, D.C.; Nelson, D.R.; Syed, K. Structure-Function Analysis of the Essential Mycobacterium tuberculosis P450 Drug Target, CYP121A1. Int. J. Mol. Sci. 2024, 25, 4886. [Google Scholar] [CrossRef]
- Padayachee, T.; Lamb, D.C.; Nelson, D.R.; Syed, K. Structure-Function Analysis of the Self-Sufficient CYP102 Family Provides New Insights into Their Biochemistry. Int. J. Mol. Sci. 2025, 26, 2161. [Google Scholar] [CrossRef]
- Herzog, K.; Bracco, P.; Onoda, A.; Hayashi, T.; Hoffmann, K.; Schallmey, A. Enzyme-substrate complex structures of CYP154C5 shed light on its mode of highly selective steroid hydroxylation. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 2875–2889. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature. Methods Mol. Biol. 1998, 107, 15–24. [Google Scholar] [CrossRef]
- Msweli, S.; Pakala, S.B.; Syed, K. NF-κB Transcription Factors: Their Distribution, Family Expansion, Structural Conservation, and Evolution in Animals. Int. J. Mol. Sci. 2024, 25, 9793. [Google Scholar] [CrossRef] [PubMed]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Berman, H.; Battistuz, T.; Bhat, T.; Bluhm, W.; Bourne, P.; Burkhardt, K.; Feng, Z.; Gilliland, G.; Iype, L.; Jain, S.; et al. The Protein data Bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Binkowski, T.A.; Naghibzadeh, S.; Liang, J. CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 2003, 31, 3352–3355. [Google Scholar] [CrossRef]
- Schrödinger, L.D. The PyMOL Molecular Graphics System; Version 2.0; Schrödinger, LLC.: New York, NY, USA, 2020. [Google Scholar]
- Graham, S.E.; Peterson, J.A. How similar are P450s and what can their differences teach us? Arch. Biochem. Biophys. 1999, 369, 24–29. [Google Scholar] [CrossRef]
- Midlik, A.; Navrátilová, V.; Moturu, T.R.; Koča, J.; Svobodová, R.; Berka, K. Uncovering of cytochrome P450 anatomy by SecStrAnnotator. Sci. Rep. 2021, 11, 12345. [Google Scholar] [CrossRef]
- Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992, 267, 83–90. [Google Scholar] [CrossRef]
- Size, E.R.M. Share Analysis—Growth Trends & Forecasts (2025–2030); Mordor Intelligence: Hyderabad, India, 2024; Available online: https://www.mordorintelligence.com/industry-reports/androgens-and-anabolic-steroids-market (accessed on 25 April 2025).
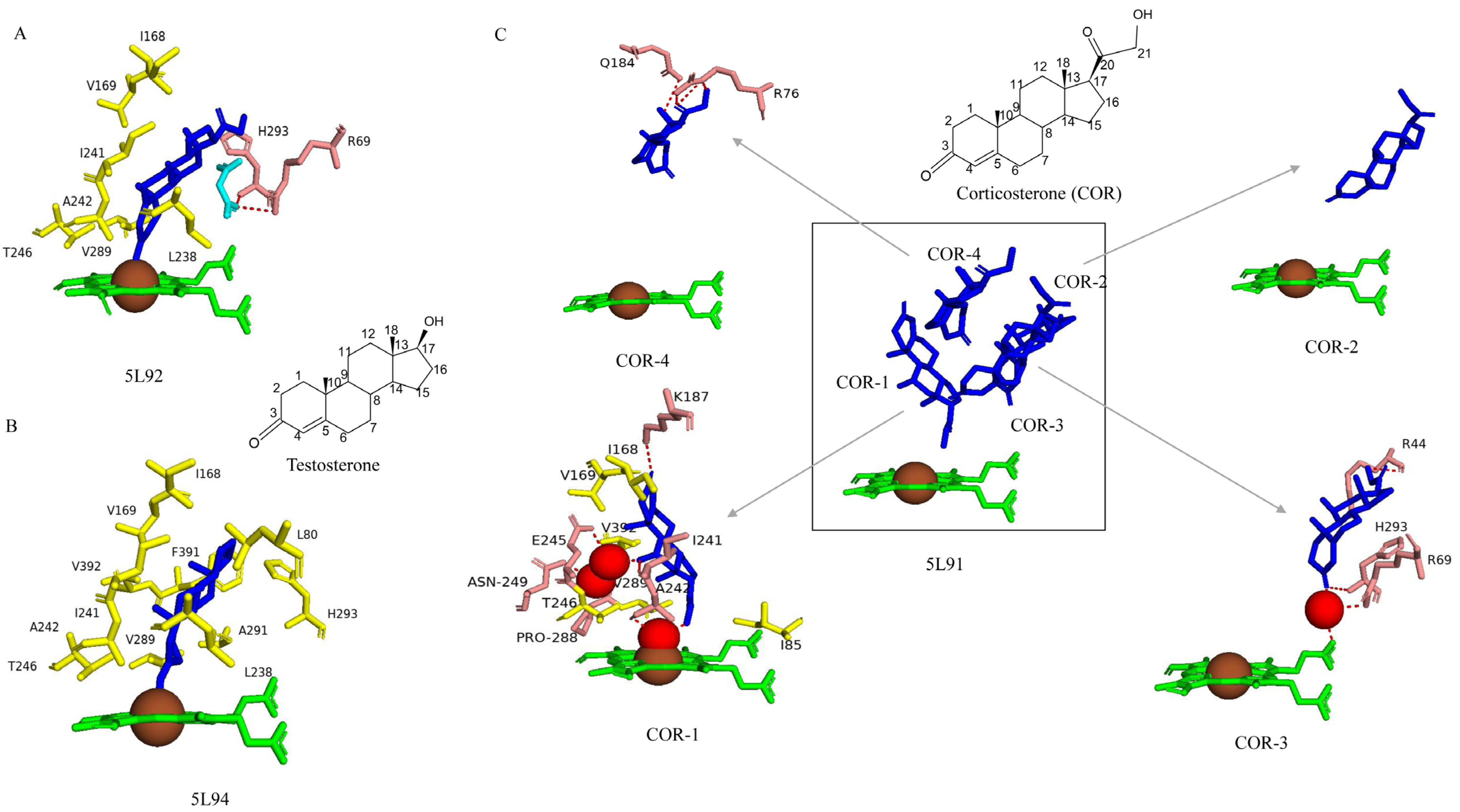



| P450 Name | Organism | PDB CODE | Surface Area (Å2) | Volume (Å3) | Conformation | Change in Surface Area (Å2) | Change in Volume (Å3) |
|---|---|---|---|---|---|---|---|
| CYP109B1 | Bacillus subtilis subsp. subtilis | 4RM4 | 1682 | 1652 | Open | na | na |
| CYP109Q5 | Chondromyces apiculatus DSM 436 | 6GMF | 1161 | 769 | Open | na | na |
| CYP109E1 | Priestia megaterium DSM 319 (formerly Bacillus megaterium) | 5L90 | 1633 | 2699 | Open | - | - |
| CYP109E1 | Priestia megaterium DSM 319 | 5L91 | 1762 | 2809 | Closed | −129 | −110 |
| CYP109E1 | Priestia megaterium DSM 319 | 5L92 | 2021 | 2260 | Closed | −388 | 439 |
| CYP109E1 | Priestia megaterium DSM 319 | 5L94 | 1569 | 1391 | Closed | 64 | 1308 |
| CYP109B4 | Bacillus sonorensis L12 | 7Y97 | 1515 | 1793 | Open | - | - |
| CYP109B4 | Bacillus sonorensis L12 | 7Y9O | 875 | 646 | Closed | 640 | 1147 |
| CYP109B4 | Bacillus sonorensis L12 | 7Y98 | 1773 | 1965 | Closed | −258 | −172 |
| CYP109A2 | Priestia megaterium DSM 319 | 5OFQ | 1561 | 1337 | Open | - | - |
| CYP109A2 | Priestia megaterium DSM 319 | 8ABS | 961 | 647 | Closed | 600 | 690 |
| CYP109A2 | Priestia megaterium DSM 319 | 8ABR | 806 | 446 | Closed | 755 | 891 |
| P450 Name | PDB Code | Conformation | Number of Amino Acids in the Active Site Cavity | Common Amino Acids | Unique Amino Acids (Found Only in Either Open or Closed Confirmation) | Amino Acids Interacting With the Ligand | RMSD (Å) |
|---|---|---|---|---|---|---|---|
| CYP109B4 | 7Y97 | Open | 34 | 33 | Ala354 | - | 0.7 |
| 7Y98 | Closed | 33 | - | Phe70, Thr78, Phe79, Val84 *, Thr245 *, Ala288 *, Met290, Ile291 *, Ser387 | |||
| 7Y97 | Open | 34 | 30 | Val84, Ile291, Val92, Phe234 | - | 2.4 | |
| 7Y9O | Closed | 37 | Ile60, Leu84, Ala286, Thr291, Ser292, Ile342, Glu360 | - | |||
| CYP109E1 | 5L90 | Open | 34 | 33 | Gly243 | - | 2.7 |
| 5L92 | Closed | 36 | Ser67, Ala355, Glu361 | Arg69 *, Leu80, Ile168, Val169, Leu238 *, Ile241, Ala242 *, Thr246 *, Val289 *, His293 | |||
| 5L90 | Open | 34 | 33 | Leu239 | - | 0.5 | |
| 5L91 | Closed | 35 | Ser67, Glu361 | Arg44, Ile85 *, Ile168, Val169, Lys187, Ile241, Ala242 *, Glu245, Asn249, Pro288, Thr246 *, Val289 *, His293, Val392 | |||
| 5L90 | Open | 34 | 33 | Gly243 | - | 2.9 | |
| 5L94 | Closed | 35 | Ala355, Glu361 | Leu80, Ile168, Val169, Leu238 *, Ile241, Ala242 *, Thr246 *, Val289 *, Ala291, His293, Phe391, Val392 | |||
| CYP109A2 | 5OFQ | Open | 33 | 33 | - | - | 2.5 |
| 8ABR | Closed | 40 | Ser72, Arg74, Leu149, Asn243, Phe346, Ala354, Glu360 | Tyr12, Asn14, Arg74 *, Gln80, Leu84 *, Leu167, Leu168, Val240, Ile291 *, Gly292 *, Phe389 | |||
| CYP109A2 | 5OFQ | Open | 33 | 33 | - | - | 2.5 |
| 8ABS | Closed | 40 | Ser72, Arg74, Leu149, Asn243, Phe346, Ala354, Glu360 | Tys12, Asn14, Arg74 *, Gln80, Leu84 *, Leu167, Val168, Arg186, Gln194, Val240 |
| P450 Name | Change in Average Area (Å2) | Change in Average Volume (Å3) | RMSD (Å) | Reference |
|---|---|---|---|---|
| CYP107FH5 | 276 # | 494 # | 3.0 | [26] |
| CYP121A1 | 37 | 8 | 0.2 | [27] |
| CYP102A1 | 179 # | 23 # | 4.4 | [28] |
| CYP109E1 | 151 # | 545 | 2.9 | Current work |
| PDB CODE | Amino Acid Residues |
|---|---|
| 5L91 COR-1 | Ile85, Ile168, Val169, Ala170, Lys187, Ile241, Ala242, Glu245, Asn249, Pro288, Thr246, Val289, Ala291, Leu292, Ser389, Phe391, Val392 |
| 5L91 COR-2 | Arg69, Pro71, Gln75, Leu80, Gly81, Ser83, Ile85, Asn86, Leu238, Ile241, Ala242 |
| 5L91 COR-3 | Arg44, Val46, Arg69, Ala291, Leu292, His293, Arg294, Phe391, His312 |
| 5L91 COR-4 | Arg76, Thr78, Leu80, Ile168, Gln184, Lys187, Met188, Asn191, Ile241 |
| 5L92 | Arg69, Leu80, Ile85, Asn86, Ile168, Val169, Leu238, Ile241, Ala242, Thr246, Val289, His293, Cys352, Phe391, Val392 |
| 5L94 | Leu80, Ile85, Ile168, Val169, Leu238, Ile241, Ala242, Glu245, Thr246, Val289, Ala291, Leu292, His293, Cys352, Ser389, Phe391, Val392 |
| 7Y98 | Phe70, N77, Thr78, Phe79, Val84, Ala241, Thr245, Ala288, Met290, Ile291, Val292, Ser387 |
| 7Y9O | Leu84, Leu237, Ala241, Thr245, Ala288, Cys351 |
| 8ABS | Tys12, Asn14, Arg74, Glu77, Gln80, Glu81, Ser82, Leu84, Met85, Val168, Leu167, Arg186, Val190, Gln194, Gly233, Phe234, Ile236, Leu237, Val240, Ala241, Glu244, Thr245, Ile288, Ala290, Ile291, Gly292, Arg293, Phe389 |
| 8ABR | Tyr12, Asn14, Arg74, Glu78, Gln80, Ser82, Leu84, Met85, Leu167, Leu168, Arg186, Val190, Ile236, Leu237, Val240, Ala241, Glu244, Thr245, Ile288, Ala290, Ile291, Gly292, Arg293, Phe389 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Msweli, S.M.; Padayachee, T.; Khumalo, T.; Nelson, D.R.; Lamb, D.C.; Syed, K. Structure–Function Analysis of the Steroid-Hydroxylating Cytochrome P450 109 (CYP109) Enzyme Family. Int. J. Mol. Sci. 2025, 26, 6219. https://doi.org/10.3390/ijms26136219
Msweli SM, Padayachee T, Khumalo T, Nelson DR, Lamb DC, Syed K. Structure–Function Analysis of the Steroid-Hydroxylating Cytochrome P450 109 (CYP109) Enzyme Family. International Journal of Molecular Sciences. 2025; 26(13):6219. https://doi.org/10.3390/ijms26136219
Chicago/Turabian StyleMsweli, Siphesihle M., Tiara Padayachee, Thembeka Khumalo, David R. Nelson, David C. Lamb, and Khajamohiddin Syed. 2025. "Structure–Function Analysis of the Steroid-Hydroxylating Cytochrome P450 109 (CYP109) Enzyme Family" International Journal of Molecular Sciences 26, no. 13: 6219. https://doi.org/10.3390/ijms26136219
APA StyleMsweli, S. M., Padayachee, T., Khumalo, T., Nelson, D. R., Lamb, D. C., & Syed, K. (2025). Structure–Function Analysis of the Steroid-Hydroxylating Cytochrome P450 109 (CYP109) Enzyme Family. International Journal of Molecular Sciences, 26(13), 6219. https://doi.org/10.3390/ijms26136219







