Post-Transcriptional Regulation of the MiaA Prenyl Transferase by CsrA and the Small RNA CsrB in Escherichia coli
Abstract
1. Introduction
2. Results
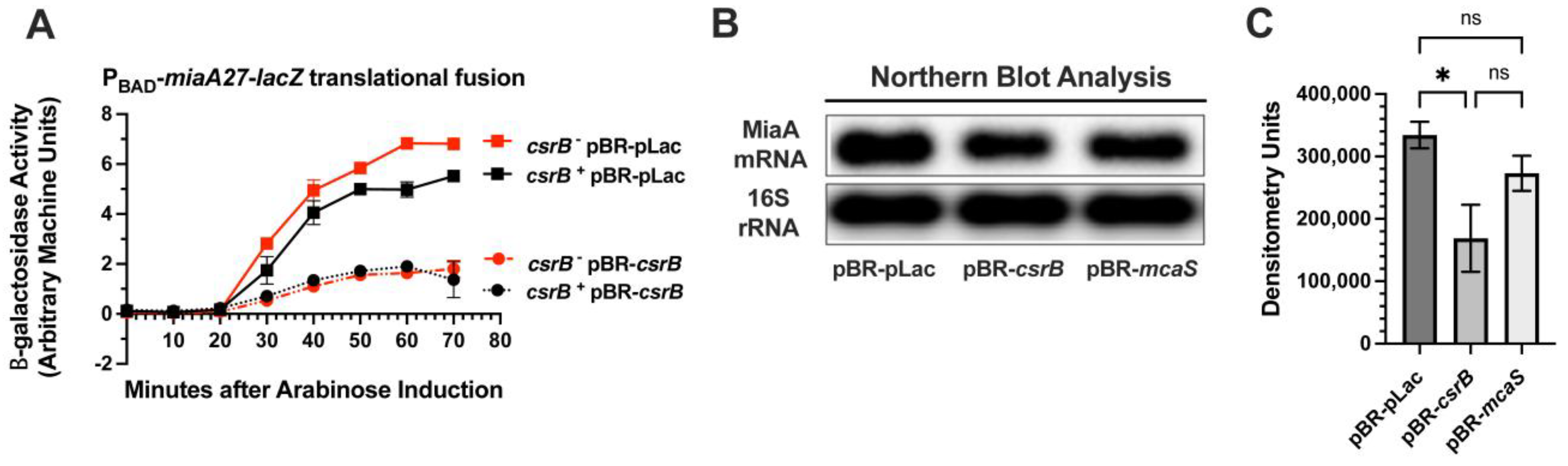
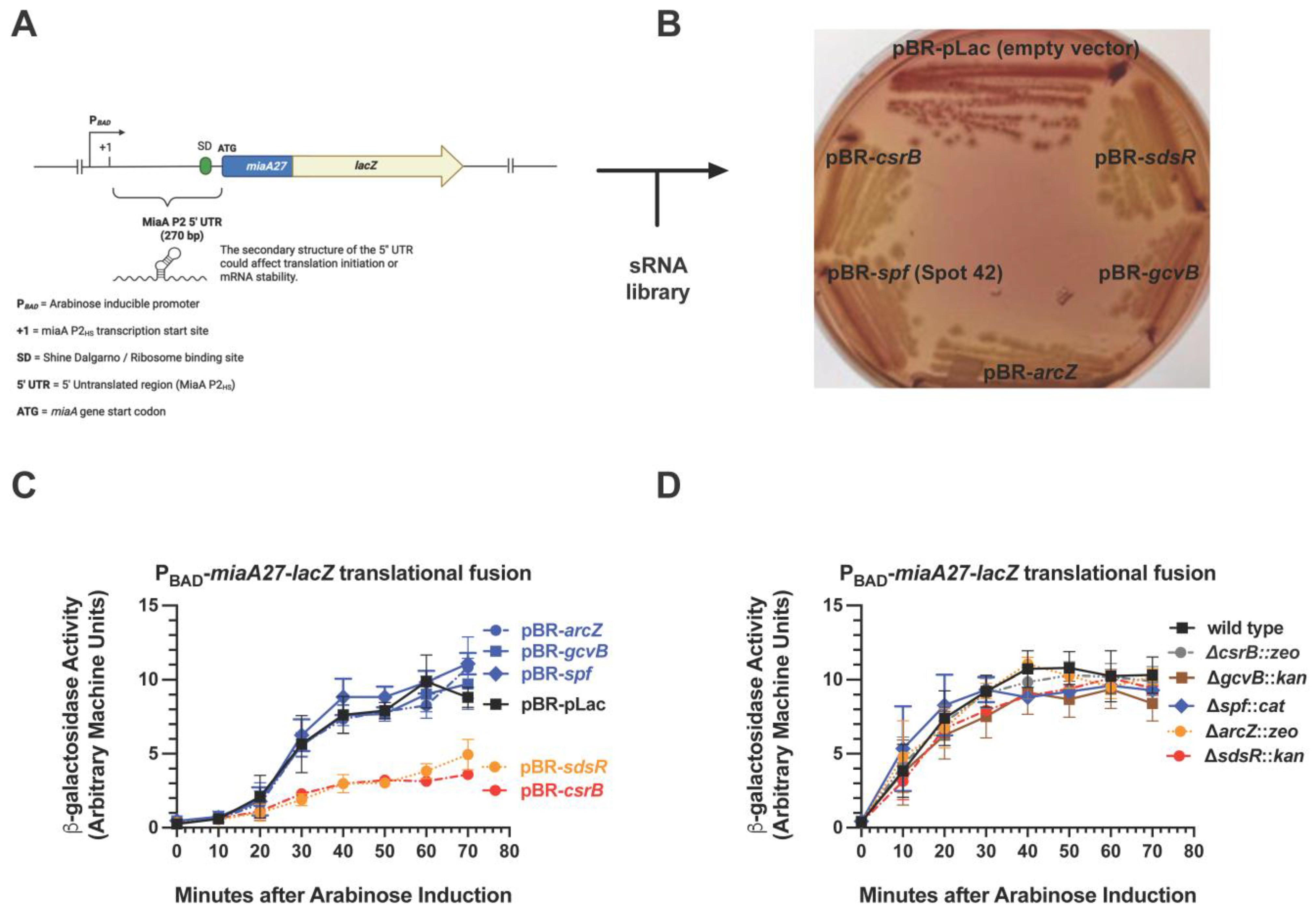
2.1. CsrB Was Selected as a Multi-Copy Repressor of MiaA Translation in a Targeted Screen of sRNA Regulators
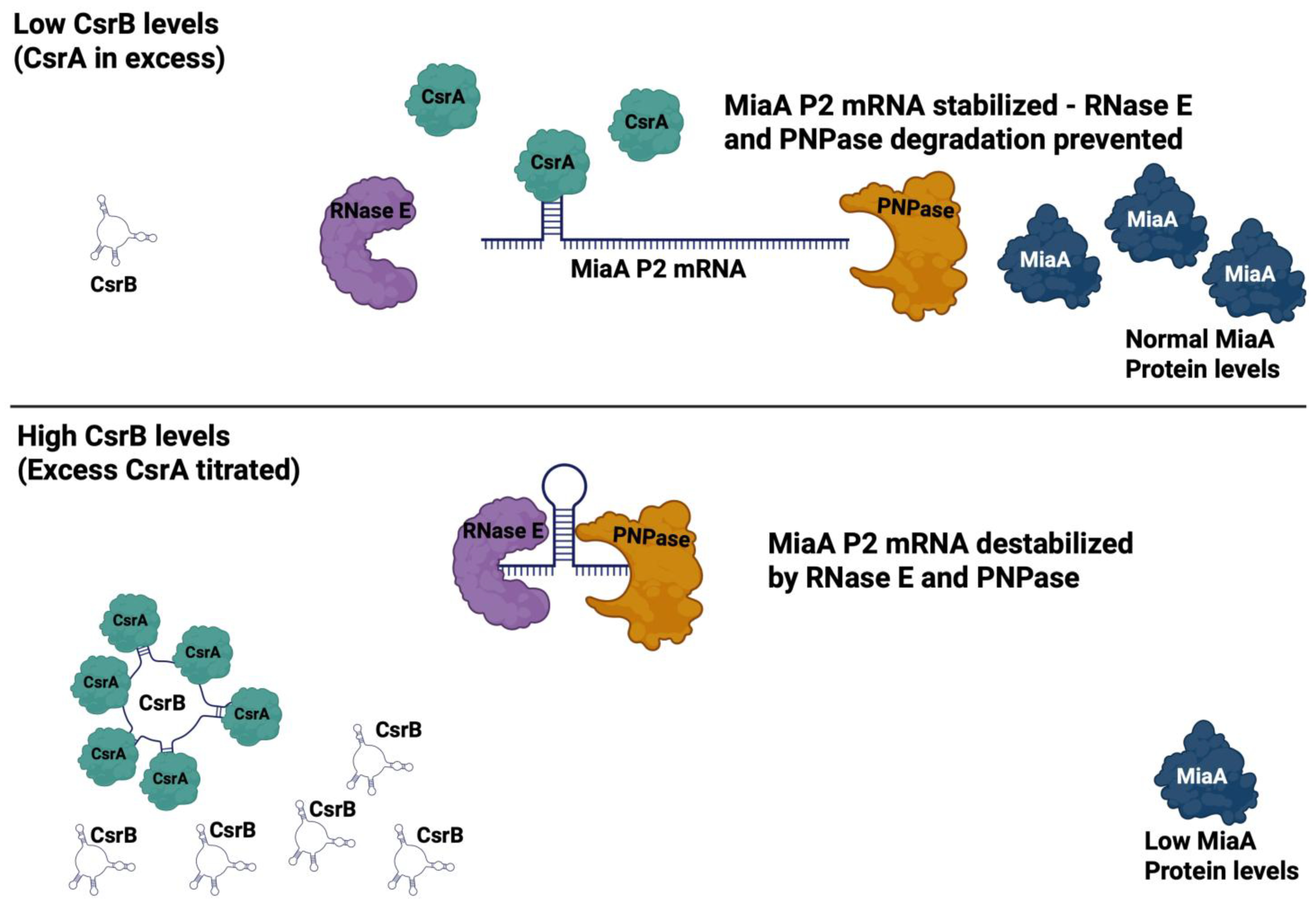
2.2. CsrB Affects MiaA mRNA Levels and Translation
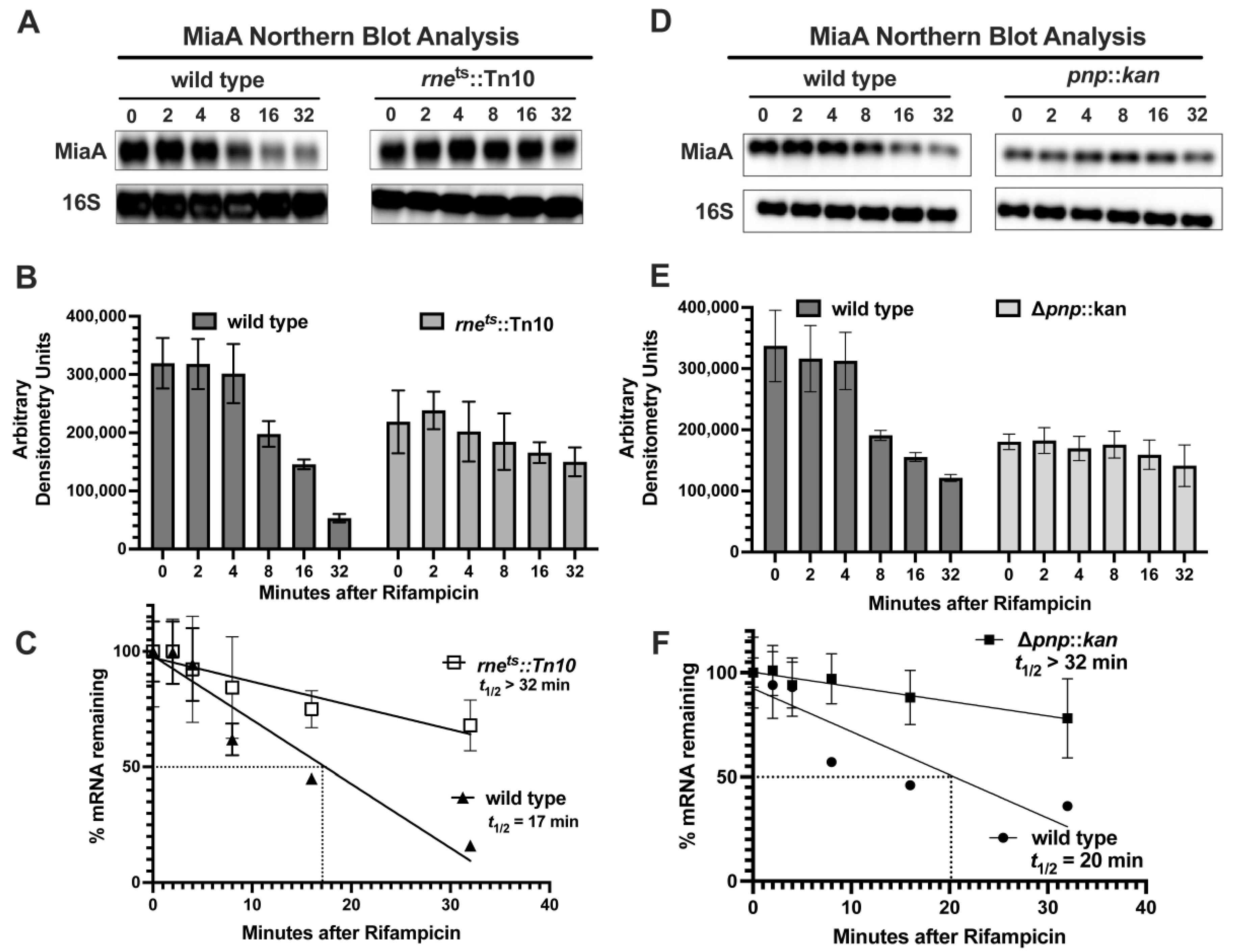
2.3. MiaA Is Regulated at the Level of mRNA Stability by PNPase and RNase E
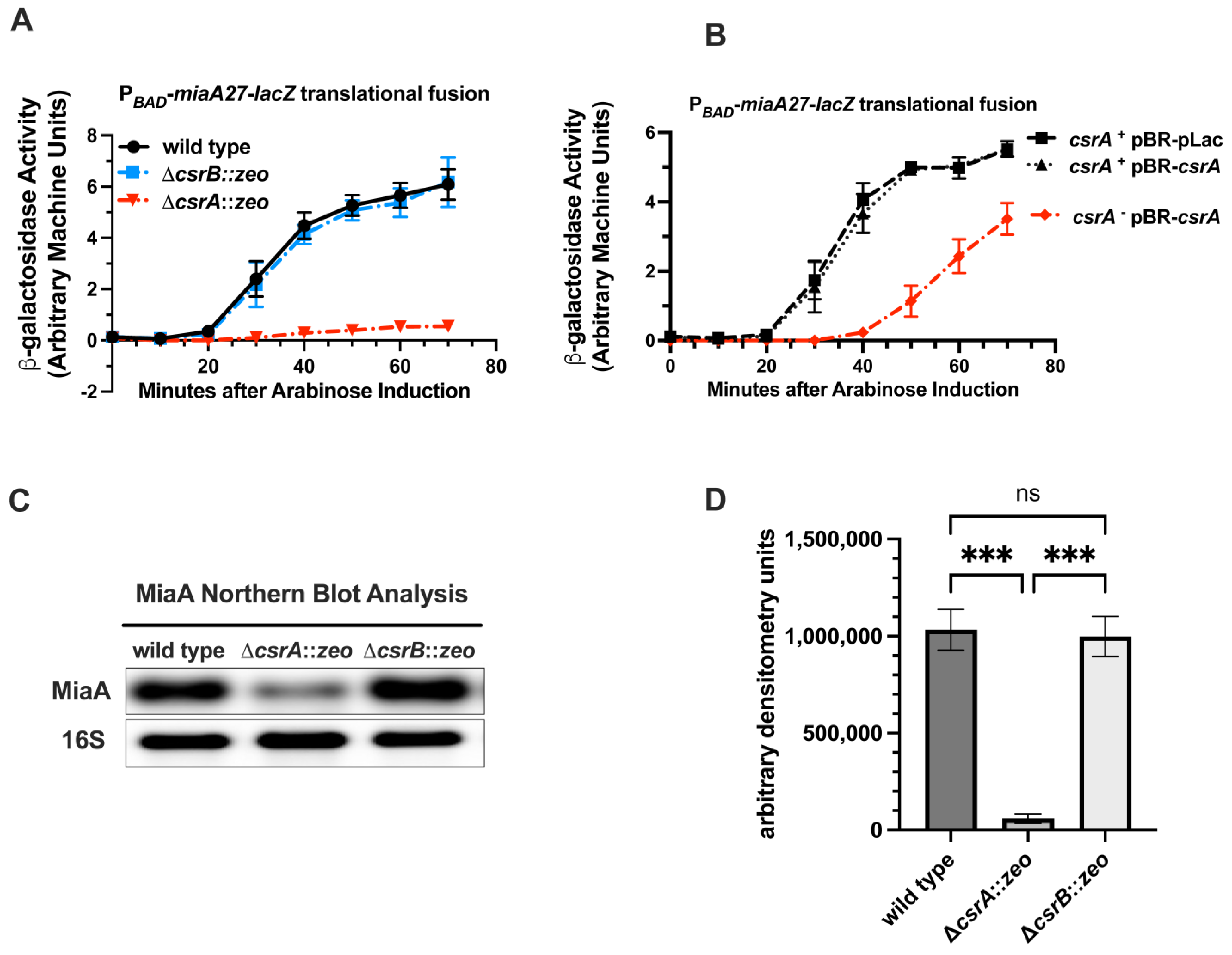
2.4. CsrA Is Necessary for the Full Expression of MiaA
3. Discussion
3.1. RNA Modifications and the Bacterial Epitranscriptome
3.2. New Regulators of MiaA Expression
3.3. MiaA Is an Additional Potential Stimulatory Target of the CsrA-CsrB System and Interactions with Other Post-Transriptional Regulators of the MiaA Operon
4. Materials and Methods
4.1. Strains and Plasmids
4.2. Media and Growth Conditions
4.3. General Molecular Biology Techniques
4.4. Genetic Engineering and Strain Construction
4.4.1. Insertional Inactivation Mutagenesis Using Recombineering
4.4.2. P1 Transduction to Move Mutants Between Strains
4.4.3. Heat Shock Transformation of Chemically Competent Cells for Cloning
4.5. RNA Isolation
4.6. Agarose Northern Blot
4.7. β-Galactosidase Assays (Kinetic Microtiter Assays)
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caillet, J.; Droogmans, L. Molecular cloning of the Escherichia coli miaA gene involved in the formation of delta 2-isopentyl adenosine in tRNA. J. Bacteriol 1988, 170, 4147–4152. [Google Scholar] [CrossRef]
- Connolly, D.M.; Winkler, M.E. Genetic and physiological relationships among the miaA gene, 2-methylthio-N6-(delta 2-isopentenyl)-adenosine tRNA modification, and spontaneous mutagenesis in Escherichia coli K-12. J. Bacteriol. 1989, 171, 3233–3246. [Google Scholar] [CrossRef]
- Connolly, D.M.; Winkler, M.E. Structure of Escherichia coli K-12 miaA and characterization of the mutator phenotype caused by miaA insertion mutations. J. Bacteriol 1991, 173, 1711–1721. [Google Scholar] [CrossRef]
- Blum, P.H. Reduced leu operon expression in a miaA mutant of Salmonella typhimurium. J. Bacteriol 1988, 170, 5125–5133. [Google Scholar] [CrossRef]
- Diaz, I.; Ehrenberg, M.; Kurland, C.G. How do combinations of rpsL- and miaA- generate streptomycin dependence? Mol. Gen. Genet. MGG 1986, 202, 207–211. [Google Scholar] [CrossRef]
- Pierrel, F.; Bjork, G.R.; Fontecave, M.; Atta, M. Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 2002, 277, 13367–13370. [Google Scholar] [CrossRef]
- Esberg, B.; Leung, H.C.; Tsui, H.C.; Bjork, G.R.; Winkler, M.E. Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1999, 181, 7256–7265. [Google Scholar] [CrossRef]
- Tukenmez, H.; Xu, H.; Esberg, A.; Bystrom, A.S. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2015, 43, 9489–9499. [Google Scholar] [CrossRef]
- Bjork, G.R.; Durand, J.M.; Hagervall, T.G.; Leipuviene, R.; Lundgren, H.K.; Nilsson, K.; Chen, P.; Qian, Q.; Urbonavicius, J. Transfer RNA modification: Influence on translational frameshifting and metabolism. FEBS Lett. 1999, 452, 47–51. [Google Scholar] [CrossRef]
- Bjork, G.R.; Wikstrom, P.M.; Bystrom, A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 1989, 244, 986–989. [Google Scholar] [CrossRef]
- Urbonavicius, J.; Qian, Q.; Durand, J.M.; Hagervall, T.G.; Bjork, G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001, 20, 4863–4873. [Google Scholar] [CrossRef] [PubMed]
- Urbonavicius, J.; Stahl, G.; Durand, J.M.; Ben Salem, S.N.; Qian, Q.; Farabaugh, P.; Bjork, G.R. Transfer RNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting. RNA 2003, 9, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; Bjork, G.R. Structural requirements for the formation of 1-methylguanosine in vivo in tRNA(Pro)GGG of Salmonella typhimurium. J. Mol. Biol. 1997, 266, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; Bjork, G.R. Structural alterations far from the anticodon of the tRNAProGGG of Salmonella typhimurium induce +1 frameshifting at the peptidyl-site. J. Mol. Biol. 1997, 273, 978–992. [Google Scholar] [CrossRef]
- Qian, Q.; Curran, J.F.; Bjork, G.R. The methyl group of the N6-methyl-N6-threonylcarbamoyladenosine in tRNA of Escherichia coli modestly improves the efficiency of the tRNA. J. Bacteriol. 1998, 180, 1808–1813. [Google Scholar] [CrossRef]
- Zhao, J.; Leung, H.E.; Winkler, M.E. The miaA mutator phenotype of Escherichia coli K-12 requires recombination functions. J. Bacteriol. 2001, 183, 1796–1800. [Google Scholar] [CrossRef]
- Schweizer, U.; Bohleber, S.; Fradejas-Villar, N. The modified base isopentenyladenosine and its derivatives in tRNA. RNA Biol. 2017, 14, 1197–1208. [Google Scholar] [CrossRef]
- Nishii, K.; Wright, F.; Chen, Y.Y.; Moller, M. Tangled history of a multigene family: The evolution of ISOPENTENYLTRANSFERASE genes. PLoS ONE 2018, 13, e0201198. [Google Scholar] [CrossRef]
- Soman, S.; Ram, S. MiaA (Rv2727c) mediated tRNA isopentenylation of Mycobacterium tuberculosis H37Rv. Mol. Biol. Res. Commun. 2022, 11, 97–104. [Google Scholar] [CrossRef]
- Fleming, B.A.; Blango, M.G.; Rousek, A.A.; Kincannon, W.M.; Tran, A.; Lewis, A.J.; Russell, C.W.; Zhou, Q.; Baird, L.M.; Barber, A.E.; et al. A tRNA modifying enzyme as a tunable regulatory nexus for bacterial stress responses and virulence. Nucleic. Acids Res. 2022, 50, 7570–7590. [Google Scholar] [CrossRef]
- Durand, J.M.; Bjork, G.R.; Kuwae, A.; Yoshikawa, M.; Sasakawa, C. The modified nucleoside 2-methylthio-N6-isopentenyladenosine in tRNA of Shigella flexneri is required for expression of virulence genes. J. Bacteriol. 1997, 179, 5777–5782. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Liu, H.; Jiang, Y.; Shao, L.; Yang, S.; Chen, D. New Mutations Involved in Colistin Resistance in Acinetobacter baumannii. mSphere 2020, 5, e00895-19. [Google Scholar] [CrossRef] [PubMed]
- Koshla, O.; Yushchuk, O.; Ostash, I.; Dacyuk, Y.; Myronovskyi, M.; Jager, G.; Sussmuth, R.D.; Luzhetskyy, A.; Bystrom, A.; Kirsebom, L.A.; et al. Gene miaA for post-transcriptional modification of tRNA(XXA) is important for morphological and metabolic differentiation in Streptomyces. Mol. Microbiol. 2019, 112, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Koshla, O.; Kravets, V.; Dacyuk, Y.; Ostash, I.; Sussmuth, R.; Ostash, B. Genetic analysis of Streptomyces albus J1074 mia mutants suggests complex relationships between post-transcriptional tRNA(XXA) modifications and physiological traits. Folia Microbiol. 2020, 65, 1009–1015. [Google Scholar] [CrossRef]
- Thompson, K.M.; Gottesman, S. The MiaA tRNA modification enzyme is necessary for robust RpoS expression in Escherichia coli. J. Bacteriol. 2014, 196, 754–761. [Google Scholar] [CrossRef]
- Aubee, J.I.; Olu, M.; Thompson, K.M. TrmL and TusA are necessary for rpoS and MiaA is required for hfq expression in Escherichia coli. Biomolecules 2017, 7, 39. [Google Scholar] [CrossRef]
- Aubee, J.I.; Olu, M.; Thompson, K.M. The i6A37 tRNA modification is essential for proper decoding of UUX-Leucine codons during rpoS and iraP translation. RNA 2016, 22, 729–742. [Google Scholar] [CrossRef]
- Moller, T.; Franch, T.; Hojrup, P.; Keene, D.R.; Bachinger, H.P.; Brennan, R.G.; Valentin-Hansen, P. Hfq: A bacterial Sm-like proteins that mediates RNA-RNA interaction. Mol. Cell. 2002, 9, 23–30. [Google Scholar] [CrossRef]
- Kajitani, M.; Kato, A.; Wada, A.; Inokuchi, H.; Ishihama, A. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J. Bacteriol. 1994, 176, 531–534. [Google Scholar] [CrossRef]
- Tsui, H.C.; Feng, G.; Winkler, M.E. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Esigma32-specific promoters during heat shock. J. Bacteriol. 1996, 178, 5719–5731. [Google Scholar] [CrossRef]
- Tsui, H.C.; Winkler, M.E. Transcriptional patterns of the mutL-miaA superoperon of Escherichia coli K-12 suggest a model for posttranscriptional regulation. Biochimie 1994, 76, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Tsui, H.C.; Feng, G.; Winkler, M.E. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 1997, 179, 7476–7487. [Google Scholar] [CrossRef] [PubMed]
- Py, B.; Causton, H.; Mudd, E.A.; Higgins, C.F. A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 1994, 14, 717–729. [Google Scholar] [CrossRef]
- Py, B.; Higgins, C.F.; Krisch, H.M.; Carpousis, A.J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 1996, 381, 169–172. [Google Scholar] [CrossRef]
- Mudd, E.A.; Higgins, C.F. Escherichia coli endoribonuclease RNase E: Autoregulation of expression and site-specific cleavage of mRNA. Mol. Microbiol. 1993, 9, 557–568. [Google Scholar] [CrossRef]
- Masse, E.; Escorcia, F.E.; Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003, 17, 2374–2383. [Google Scholar] [CrossRef]
- Guillier, M.; Gottesman, S.; Storz, G. Modulating the outer membrane with small RNAs. Genes Dev. 2006, 20, 2338–2348. [Google Scholar] [CrossRef]
- Mandin, P.; Gottesman, S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010, 29, 3094–3107. [Google Scholar] [CrossRef]
- Luo, X.; Majdalani, N. Directed Screening for sRNA Targets in E. coli Using a Plasmid Library. Methods Mol. Biol. 2024, 2741, 291–306. [Google Scholar] [CrossRef]
- Liu, M.Y.; Gui, G.; Wei, B.; Preston, J.F., III; Oakford, L.; Yuksel, U.; Giedroc, D.P.; Romeo, T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997, 272, 17502–17510. [Google Scholar] [CrossRef]
- Liu, M.Y.; Romeo, T. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 1997, 179, 4639–4642. [Google Scholar] [CrossRef] [PubMed]
- Sabnis, N.A.; Yang, H.; Romeo, T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 1995, 270, 29096–29104. [Google Scholar] [CrossRef] [PubMed]
- Pannuri, A.; Vakulskas, C.A.; Zere, T.; McGibbon, L.C.; Edwards, A.N.; Georgellis, D.; Babitzke, P.; Romeo, T. Circuitry Linking the Catabolite Repression and Csr Global Regulatory Systems of Escherichia coli. J. Bacteriol. 2016, 198, 3000–3015. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dubey, A.K.; Suzuki, K.; Baker, C.S.; Babitzke, P.; Romeo, T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 2005, 56, 1648–1663. [Google Scholar] [CrossRef]
- Wei, B.L.; Brun-Zinkernagel, A.M.; Simecka, J.W.; Pruss, B.M.; Babitzke, P.; Romeo, T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 2001, 40, 245–256. [Google Scholar] [CrossRef]
- Wang, D.; McAteer, S.P.; Wawszczyk, A.B.; Russell, C.D.; Tahoun, A.; Elmi, A.; Cockroft, S.L.; Tollervey, D.; Granneman, S.; Tree, J.J.; et al. An RNA-dependent mechanism for transient expression of bacterial translocation filaments. Nucleic. Acids Res. 2018, 46, 3366–3381. [Google Scholar] [CrossRef]
- Liaw, S.J.; Lai, H.C.; Ho, S.W.; Luh, K.T.; Wang, W.B. Role of RsmA in the regulation of swarming motility and virulence factor expression in Proteus mirabilis. J. Med. Microbiol. 2003, 52, 19–28. [Google Scholar] [CrossRef]
- Nava-Galeana, J.; Yakhnin, H.; Babitzke, P.; Bustamante, V.H. CsrA Positively and Directly Regulates the Expression of the pdu, pocR, and eut Genes Required for the Luminal Replication of Salmonella Typhimurium. Microbiol. Spectr. 2023, 11, e0151623. [Google Scholar] [CrossRef]
- Zhu, D.; Wang, S.; Sun, X. FliW and CsrA Govern Flagellin (FliC) Synthesis and Play Pleiotropic Roles in Virulence and Physiology of Clostridioides difficile R20291. Front Microbiol 2021, 12, 735616. [Google Scholar] [CrossRef]
- Hubloher, J.J.; Schabacker, K.; Muller, V.; Averhoff, B. CsrA Coordinates Compatible Solute Synthesis in Acinetobacter baumannii and Facilitates Growth in Human Urine. Microbiol Spectr 2021, 9, e0129621. [Google Scholar] [CrossRef]
- Butz, H.A.; Mey, A.R.; Ciosek, A.L.; Crofts, A.A.; Davies, B.W.; Payne, S.M. Regulatory Effects of CsrA in Vibrio cholerae. mBio 2021, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Xu, L.; Xiao, L.; Zhu, K.; Song, Y.; Li, C.; Zhu, L.; Shen, X.; Wang, Y. RovM and CsrA Negatively Regulate Urease Expression in Yersinia pseudotuberculosis. Front. Microbiol. 2018, 9, 348. [Google Scholar] [CrossRef] [PubMed]
- Potts, A.H.; Leng, Y.; Babitzke, P.; Romeo, T. Examination of Csr regulatory circuitry using epistasis analysis with RNA-seq (Epi-seq) confirms that CsrD affects gene expression via CsrA, CsrB and CsrC. Sci. Rep. 2018, 8, 5373. [Google Scholar] [CrossRef]
- Heroven, A.K.; Bohme, K.; Dersch, P. The Csr/Rsm system of Yersinia and related pathogens: A post-transcriptional strategy for managing virulence. RNA Biol. 2012, 9, 379–391. [Google Scholar] [CrossRef]
- Abbott, Z.D.; Yakhnin, H.; Babitzke, P.; Swanson, M.S. csrR, a Paralog and Direct Target of CsrA, Promotes Legionella pneumophila Resilience in Water. mBio 2015, 6, e00595. [Google Scholar] [CrossRef]
- Ozturk, G.; LeGrand, K.; Zheng, Y.; Young, G.M. Yersinia enterocolitica CsrA regulates expression of the Ysa and Ysc type 3 secretion system in unique ways. FEMS Microbiol. Lett. 2017, 364, fnx204. [Google Scholar] [CrossRef]
- Muller, P.; Gimpel, M.; Wildenhain, T.; Brantl, S. A new role for CsrA: Promotion of complex formation between an sRNA and its mRNA target in Bacillus subtilis. RNA Biol. 2019, 16, 972–987. [Google Scholar] [CrossRef]
- Heroven, A.K.; Nuss, A.M.; Dersch, P. RNA-based mechanisms of virulence control in Enterobacteriaceae. RNA Biol. 2017, 14, 471–487. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Potts, A.H.; Babitzke, P.; Ahmer, B.M.; Romeo, T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. MMBR 2015, 79, 193–224. [Google Scholar] [CrossRef]
- Cerca, N.; Jefferson, K.K. Effect of growth conditions on poly-N-acetylglucosamine expression and biofilm formation in Escherichia coli. FEMS Microbiol. Lett. 2008, 283, 36–41. [Google Scholar] [CrossRef]
- Lee, J.H.; Ancona, V.; Chatnaparat, T.; Yang, H.; Zhao, Y. The RNA-binding protein CsrA controls virulence in Erwinia amylovora by regulating RelA, RcsB, and FlhD at the posttranscriptional level. Mol. Plant Microbe Interact. 2019, 32, 1448–1459. [Google Scholar] [CrossRef] [PubMed]
- Rojano-Nisimura, A.M.; Simmons, T.R.; Leistra, A.N.; Mihailovic, M.K.; Buchser, R.; Ekdahl, A.M.; Joseph, I.; Curtis, N.C.; Contreras, L.M. CsrA selectively modulates sRNA-mRNA regulator outcomes. Front. Mol. Biosci. 2023, 10, 1249528. [Google Scholar] [CrossRef] [PubMed]
- Stenum, T.S.; Holmqvist, E. CsrA enters Hfq’s territory: Regulation of a base-pairing small RNA. Mol. Microbiol. 2022, 117, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.J.; Yakhnin, H.; Pannuri, A.; Pourciau, C.; Babitzke, P.; Romeo, T. CsrA regulation via binding to the base-pairing small RNA Spot 42. Mol. Microbiol. 2022, 117, 32–53. [Google Scholar] [CrossRef]
- London, L.Y.; Aubee, J.I.; Nurse, J.; Thompson, K.M. Post-transcriptional regulation of rseA by small RNAs ryhB and fnrS in Escherichia coli. Front. Mol. Biosci. 2021, 8, 668613. [Google Scholar] [CrossRef]
- Jorgensen, M.G.; Thomason, M.K.; Havelund, J.; Valentin-Hansen, P.; Storz, G. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev. 2013, 27, 1132–1145. [Google Scholar] [CrossRef]
- Rojano-Nisimura, A.M.; Simmons, T.R.; Lukasiewicz, A.J.; Buchser, R.; Ruzek, J.S.; Avila, J.L.; Contreras, L.M. Concentration-Dependent CsrA Regulation of the uxuB Transcript Leads to Development of a Post-Transcriptional Bandpass Filter. ACS Synth. Biol. 2025, 14, 1084–1098. [Google Scholar] [CrossRef]
- Rojano-Nisimura, A.M.; Grismore, K.B.; Ruzek, J.S.; Avila, J.L.; Contreras, L.M. The Post-Transcriptional Regulatory Protein CsrA Amplifies Its Targetome through Direct Interactions with Stress-Response Regulatory Hubs: The EvgA and AcnA Cases. Microorganisms 2024, 12, 636. [Google Scholar] [CrossRef]
- Leistra, A.N.; Gelderman, G.; Sowa, S.W.; Moon-Walker, A.; Salis, H.M.; Contreras, L.M. A Canonical Biophysical Model of the CsrA Global Regulator Suggests Flexible Regulator-Target Interactions. Sci. Rep. 2018, 8, 9892. [Google Scholar] [CrossRef]
- Romeo, T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 1998, 29, 1321–1330. [Google Scholar] [CrossRef]
- Romeo, T.; Babitzke, P. Global Regulation by CsrA and Its RNA Antagonists. Microbiol. Spectr. 2018, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Kudla, G.; Murray, A.W.; Tollervey, D.; Plotkin, J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science 2009, 324, 255–258. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bailly, M.; de Crecy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef]
- de Crecy-Lagard, V.; Jaroch, M. Functions of Bacterial tRNA Modifications: From Ubiquity to Diversity. Trends Microbiol. 2021, 29, 41–53. [Google Scholar] [CrossRef]
- de Crecy-Lagard, V.; Ross, R.L.; Jaroch, M.; Marchand, V.; Eisenhart, C.; Bregeon, D.; Motorin, Y.; Limbach, P.A. Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus subtilis sp. Subtilis Strain 168. Biomolecules 2020, 10, 977. [Google Scholar] [CrossRef]
- Quaiyum, S.; Sun, J.; Marchand, V.; Sun, G.; Reed, C.J.; Motorin, Y.; Dedon, P.C.; Minnick, M.F.; de Crecy-Lagard, V. Mapping the tRNA modification landscape of Bartonella henselae Houston I and Bartonella quintana Toulouse. Front. Microbiol. 2024, 15, 1369018. [Google Scholar] [CrossRef]
- McGuffey, J.C.; Jackson-Litteken, C.D.; Di Venanzio, G.; Zimmer, A.A.; Lewis, J.M.; Distel, J.S.; Kim, K.Q.; Zaher, H.S.; Alfonzo, J.; Scott, N.E.; et al. The tRNA methyltransferase TrmB is critical for Acinetobacter baumannii stress responses and pulmonary infection. mBio 2023, 14, e0141623. [Google Scholar] [CrossRef]
- Diaz-Rullo, J.; Gonzalez-Pastor, J.E. tRNA queuosine modification is involved in biofilm formation and virulence in bacteria. Nucleic Acids Res. 2023, 51, 9821–9837. [Google Scholar] [CrossRef]
- Shippy, D.C.; Eakley, N.M.; Lauhon, C.T.; Bochsler, P.N.; Fadl, A.A. Virulence characteristics of Salmonella following deletion of genes encoding the tRNA modification enzymes GidA and MnmE. Microb. Pathog. 2013, 57, 1–9. [Google Scholar] [CrossRef]
- Krueger, J.; Preusse, M.; Oswaldo Gomez, N.; Frommeyer, Y.N.; Doberenz, S.; Lorenz, A.; Kordes, A.; Grobe, S.; Musken, M.; Depledge, D.P.; et al. tRNA epitranscriptome determines pathogenicity of the opportunistic pathogen Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2024, 121, e2312874121. [Google Scholar] [CrossRef]
- Zhong, W.; Koay, A.; Ngo, A.; Li, Y.; Nah, Q.; Wong, Y.H.; Chionh, Y.H.; Ng, H.Q.; Koh-Stenta, X.; Poulsen, A.; et al. Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel tRNA-(N(1)G37) Methyltransferase (TrmD) Inhibitors. ACS Infect. Dis. 2019, 5, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Pasunooti, K.K.; Balamkundu, S.; Wong, Y.H.; Nah, Q.; Gadi, V.; Gnanakalai, S.; Chionh, Y.H.; McBee, M.E.; Gopal, P.; et al. Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N(1))-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism. J. Med. Chem. 2019, 62, 7788–7805. [Google Scholar] [CrossRef] [PubMed]
- Hofer, K.; Jaschke, A. Epitranscriptomics: RNA Modifications in Bacteria and Archaea. Microbiol. Spectr. 2018, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Luo, H.; Wei, J.; Wu, S.; Zheng, Q.; Zhang, N.; Chen, P. Exploring CircRNA N6-methyladenosine in human rheumatoid arthritis: Hyper-methylated hsa_circ_0007259 as a potential biomarker and its involvement in the hsa_circ_0007259/hsa_miR-21-5p/STAT3 axis. Int. Immunopharmacol. 2023, 124, 110938. [Google Scholar] [CrossRef]
- He, T.; Hia, H.; Chen, B.; Duan, Z.; Huang, C. m6A Writer mettl3-mediated lncRNA linc01125 prevents the malignancy of papillary thyroid cancer. Crit. Rev. Immunol. 2023, 43, 43–52. [Google Scholar] [CrossRef]
- Ma, L.; Ma, Q.; Deng, Q.; Zhou, J.; Zhou, Y.; Wei, Q.; Huang, Z.; Lao, X.; Du, P. N7-methylguanosine-related miRNAs predict hepatocellular carcinoma prognosis and immune therapy. Aging 2023, 3, 12192–12208. [Google Scholar] [CrossRef]
- Chan, C.T.; Deng, W.; Li, F.; DeMott, M.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Highly Predictive Reprogramming of tRNA Modifications Is Linked to Selective Expression of Codon-Biased Genes. Chem. Res. Toxicol. 2015, 28, 978–988. [Google Scholar] [CrossRef]
- Deng, W.; Babu, I.R.; Su, D.; Yin, S.; Begley, T.J.; Dedon, P.C. Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet. 2015, 11, e1005706. [Google Scholar] [CrossRef]
- de Crecy-Lagard, V.; Boccaletto, P.; Mangleburg, C.G.; Sharma, P.; Lowe, T.M.; Leidel, S.A.; Bujnicki, J.M. Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic. Acids. Res. 2019, 47, 2143–2159. [Google Scholar] [CrossRef]
- Helm, M.; Alfonzo, J.D. Posttranscriptional RNA Modifications: Playing Metabolic Games in a Cell’s Chemical Legoland. Chem. Biol. 2014, 21, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Begley, T.J.; Dedon, P.C. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014, 588, 4287–4296. [Google Scholar] [CrossRef] [PubMed]
- Dedon, P.C.; Begley, T.J. A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem. Res. Toxicol. 2014, 27, 330–337. [Google Scholar] [CrossRef]
- Pollo-Oliveira, L.; Davis, N.K.; Hossain, I.; Ho, P.; Yuan, Y.; Salguero Garcia, P.; Pereira, C.; Byrne, S.R.; Leng, J.; Sze, M.; et al. The absence of the queuosine tRNA modification leads to pleiotropic phenotypes revealing perturbations of metal and oxidative stress homeostasis in Escherichia coli K12. Metallomics 2022, 14, mfac065. [Google Scholar] [CrossRef]
- Chionh, Y.H.; McBee, M.; Babu, I.R.; Hia, F.; Lin, W.; Zhao, W.; Cao, J.; Dziergowska, A.; Malkiewicz, A.; Begley, T.J.; et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 2016, 7, 13302. [Google Scholar] [CrossRef]
- Lampi, M.; Gregorova, P.; Qasim, M.S.; AAhlblad, N.C.V.; Sarin, L.P. Bacteriophage infection of the marine bacterium shewanella glacialimarina induces dynamic change in tRNA modifications. Microorganisms 2023, 11, 355. [Google Scholar] [CrossRef]
- Nakayashiki, T.; Inokuchi, H. Novel temperature-sensitive mutants of Escherichia coli that are unable to grow in the absence of wild type tRNA6Leu. J. Bacteriol. 1998, 180, 2931–2935. [Google Scholar] [CrossRef]
- Renda, A.; Poly, S.; Lai, Y.J.; Pannuri, A.; Yakhnin, H.; Potts, A.H.; Bevilacqua, P.C.; Romeo, T.; Babitzke, P. CsrA-Mediated Translational Activation of ymdA Expression in Escherichia coli. mBio 2020, 11, 5. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Leng, Y.; Abe, H.; Amaki, T.; Okayama, A.; Babitzke, P.; Suzuki, K.; Romeo, T. Antagonistic control of the turnover pathway for the global regulatory sRNA CsrB by the CsrA and CsrD proteins. Nucleic. Acids Res. 2016, 44, 7896–7910. [Google Scholar] [CrossRef]
- Kitagawa, R.; Mitsuki, H.; Okazaki, T.; Ogawa, T. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 1996, 19, 1137–1147. [Google Scholar] [CrossRef]
- Maciag, A.; Peano, C.; Pietrelli, A.; Egli, T.; De Bellis, G.; Landini, P. In vitro transcription profiling of the sigmaS subunit of bacterial RNA polymerase: Re-definition of the sigmaS regulon and identification of sigmaS-specific promoter sequence elements. Nucleic. Acids Res. 2011, 39, 5338–5355. [Google Scholar] [CrossRef] [PubMed]
- Tsui, H.C.; Leung, H.C.; Winkler, M.E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994, 13, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; Elliot, T. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 1996, 178, 3763–3770. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; Elliot, T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J. Bacteriol. 1997, 179, 656–662. [Google Scholar] [CrossRef]
- Zhang, A.; Altuvia, S.; Tiwari, A.; Argaman, L.; Hengge-Aronis, R.; Storz, G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998, 17, 6061–6068. [Google Scholar] [CrossRef]
- Sledjeski, D.D.; Whitman, C.; Zhang, A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001, 183, 1997–2005. [Google Scholar] [CrossRef]
- Majdalani, N.; Chen, S.; Murrow, J.; St John, K.; Gottesman, S. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol. Microbiol. 2001, 39, 1382–1394. [Google Scholar] [CrossRef]
- Baker, C.S.; Eory, L.A.; Yakhnin, H.; Mercante, J.; Romeo, T.; Babitzke, P. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the shine-dalgarno aequence. J. Bacteriol. 2007, 189, 5472–5481. [Google Scholar] [CrossRef]
- Yu, D.; Ellis, H.M.; Lee, E.C.; Jenkins, N.A.; Copeland, N.G.; Court, D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 5978–5983. [Google Scholar] [CrossRef]
- Court, D.L.; Swaminathan, S.; Yu, D.; Wilson, H.; Baker, T.; Bubunenko, M.; Sawitzke, J.; Sharan, S.K. Mini-lambda: A tractable system for chromosome and BAC engineering. Gene 2003, 315, 63–69. [Google Scholar] [CrossRef]
- Chung, C.T.; Niemela, S.L.; Miller, R.H. One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solutions. Proc. Natl. Acad. Sci. USA 1989, 86, 2172–2175. [Google Scholar] [CrossRef] [PubMed]
- Sharan, S.K.; Thomason, L.C.; Kuznetsov, S.G.; Court, D.L. Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 2009, 4, 206–223. [Google Scholar] [CrossRef] [PubMed]
- Thomason, L.C.; Costantino, N.; Court, D.L. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 2007, 79, 1–8. [Google Scholar] [CrossRef]
- Karlsson, J.; Eichner, H.; Loh, E. Total Bacterial RNA Isolation and Northern Blotting Analysis. Methods Mol. Biol. 2023, 2674, 73–85. [Google Scholar] [CrossRef]
- Ares, M. Bacterial RNA isolation. Cold Spring Harb. Protoc. 2012, 2012, 1024–1027. [Google Scholar] [CrossRef]
- Masse, E.; Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 4620–4625. [Google Scholar] [CrossRef]
- Rio, D.C. Northern blots: Capillary transfer of RNA from agarose gels and filter hybridization using standard stringency conditions. Cold Spring Harb. Protoc. 2015, 2015, 306–313. [Google Scholar] [CrossRef]
- Thibodeau, S.A.; Fang, R.; Joung, J.K. High-throughput beta-galactosidase assay for bacterial cell-based reporter systems. BioTechniques 2004, 36, 410–415. [Google Scholar] [CrossRef]
| Repressed | Activated |
|---|---|
| glgC | moaABC operon |
| glgS | ymdAB-clsC |
| sdiA | |
| dgcZ | |
| dgcT | |
| pgaA | |
| pdeI | |
| nhaR | |
| rpoE | |
| iraD |
| Strain Number | Genotype | Construction, Source, or Comment |
|---|---|---|
| DY330 | W3110 λlacU169 gal490 pgl l8 [λ cI857 λ(cro-bioA)] | 62 |
| JIA4000 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion | KMT590 × PBAD-miaA27(P2HS)-lacZ gBlock |
| JIA4001 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion pBR-pLac | JIA4000 + pBR-pLac |
| JIA4010 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion pBR-sdsR | JIA4000 + pBR-sdsR |
| JIA4015 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion pBR-gcvB | JIA4000 + pBR-gcvB |
| JIA4018 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion pBR-arcZ | JIA4000 + pBR-arcZ |
| JIA4024 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion pBR-spf | JIA4000 + pBR-spf |
| JIA4029 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion pBR-csrB | JIA4000 + pBR-csrB |
| JIA4040 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion ∆arcZ::zeo | JIA4000 × P1 (KMT657) |
| JIA4041 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion ∆gcvB::kan | JIA4000 × P1 (KMT660) |
| JIA4042 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion ∆csrB::zeo | JIA4000 × P1 (DJ480 ∆csrB::zeo) |
| JIA4043 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion ∆spf::cat | JIA4000 × P1 (KMT658) |
| JIA4044 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion ∆csrA::zeo | JIA4000 × P1 (DJ480 ∆csrA::zeo) |
| JIA4045 | MG1655 ΔaraBAD, araC+, mal::lacIq Φ80− lacI::PBAD-miaA27P2-lacZ translational fusion ∆sdsR::kan | JIA4000 × P1 (KMT662) |
| KMT195 | DJ480 mini- λ::tet | NM300 obtained from Susan Gottesman Lab, NCI-NIH |
| KMT414 | MG1655 ΔlacZYAfrtfrt lacIq ∆ara714 ∆ParaE::frtfrtPCP18araE | Thompson Lab collection |
| KMT621 | C600 rne3071::Tn10-TetR | C600 × P1 (rne3071::Tn10-Tet), temperature-sensitive |
| KMT624 | MG1655 Δpnp::kan | NRD465, cold-sensitive, obtained from Gottesman Lab, NCI-NIH |
| KMT657 | MG1655 ΔarcZ::zeo | NM665 (NM1100 ∆arcZ::zeo) gift from Nadim Majdalani-Gottesman Lab, NCI-NIH |
| KMT658 | W3110 λ lacU169 gal490 pgl l8 [λ cI857 λ (cro-bioA)] ∆spf::cat | NM18 (DY330 ∆spf::cat) gift from Nadim Majdalani-Gottesman Lab, NCI-NIH |
| KMT660 | MG1655 mal::IacIq ΔaraBAD leu+ araC+ ∆ParaE::frtfrtPCP18araE | KM357 (PM101 ∆gcvB::kan) obtained from Gottesman Lab, NCI-NIH |
| KMT662 | MG1655 ΔsdsR::kan | ASP7023 (∆sdsR::kan) obtained from Gottesman Lab, NCI-NIH |
| KMT665 | MG1655 lacIq | NM525 obtained from Gottesman Lab, NCI-NIH |
| KMT719 | MG1655 lacIq ∆csrB::zeo | KMT665 × P1 (DJ480 ∆csrB::zeo) |
| KMT720 | MG1655 lacIq ∆csrA::zeo | KMT665 × P1 (DJ480 ∆csrA::zeo) |
| KMT791 | MG1655 lacIq pBR-csrA | KMT665 + pBR-csrA (TSS Transformation) |
| KMT792 | MG1655 lacIq pBR-csrB | KMT665 + pBR-csrB (TSS Transformation) |
| KMT796 | MG1655 lacIq ∆csrB::zeo pBR-pLac-csrB | KMT719 + pBR-csrB (TSS Transformation) |
| KMT798 | MG1655 lacIq ∆csrA::zeo pBR-pLac-csrA | KMT720 + pBR-csrA (TSS Transformation) |
| KMT799 | MG1655 lacIq pBR-pLac-mcaS | KMT665 + pBR-mcaS (TSS Transformation) |
| KMT800 | MG1655 lacIq ∆pnpA::kan | KMT665 × P1 (KMT624-∆pnp::kan) |
| KMT801 | MG1655 lacIq rne3071::Tn10 (TetR) | KMT665 × P1 (C600 rne3071::Tn10-TetR) |
| KMT590 | MG1655 lacI::PBAD::cat-sacB::lacZ, ΔaraBAD, araC+, mal::lacIq, mini- λ::tet Φ80− | PM1805 obtained from Nadim Majdalani-Gottesman Lab, NCI-NIH |
| Oligo Primer | Sequence (5′ to 3′ Orientation) | Purpose Description |
|---|---|---|
| KT1205 | 5′-tcgacgtcCTTTCAAGGAGCAAAGAatgCTGAT-3′ | csrA into pBR-pLac AatII |
| KT1206 | 5′-tcagaattcttaGTAACTGGACTGCTGGG-3′ | csrA into pBR-pLac EcoRI |
| KT1207 | 5′-CAGAGAGACCCGACTCTTTTAATCTTTCAAGGAGCAAAGA-3′ | ΔcsrA::zeo forward screening |
| KT1208 | 5′-TGAGGGTGCGTCTCACCGATAAAGATGAGACGCGGAAAGA-3′ | ΔcsrA::zeo reverse screening |
| KT1209 | 5′-CAGAGAGACCCGACTCTTTTAATCTTTCAAGGAGCAAAGACACGTGTTGACAATTAATCA-3′ | ΔcsrA::zeo forward mutagenesis |
| KT1210 | 5′-TGAGGGTGCGTCTCACCGATAAAGATGAGACGCGGAAAGATCAGTCCTGCTCCTCGGCCA-3′ | ΔcsrA::zeo reverse mutagenesis |
| KT1211 | 5′-GCGCCTTGTAAGACTTCGCGAAAAAGACGATTCTATCTTC-3′ | ΔcsrB::zeo forward screening |
| KT1212 | 5′-AGCAACCTCAATAAGAAAAACTGCCGCGAAGGATAGCAGG-3′ | ΔcsrB::zeo reverse screening |
| KT1213 | 5′-GCGCCTTGTAAGACTTCGCGAAAAAGACGATTCTATCTTCCACGTGTTGACAATTAATCA-3′ | ΔcsrB::zeo forward mutagenesis |
| KT1214 | 5′-AGCAACCTCAATAAGAAAAACTGCCGCGAAGGATAGCAGGTCAGTCCTGCTCCTCGGCCA-3′ | ΔcsrB::zeo reverse mutagenesis |
| MiaA probe | 5′-Biosg/CGCGGCGAGTAACTCTTCAGCGTTCGGCTTCGCCG-3′ | Biotinylated oligo antisense to miaA |
| 16S probe | 5′-Biosg/CACAACACGAGCTGACGACAGCCATGCAGCACCTG-3′ | Biotinylated oligo antisense to 16S rrnA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aubee, J.I.; Williams, K.; Adigun, A.; Olusanya, O.; Nurse, J.; Thompson, K.M. Post-Transcriptional Regulation of the MiaA Prenyl Transferase by CsrA and the Small RNA CsrB in Escherichia coli. Int. J. Mol. Sci. 2025, 26, 6068. https://doi.org/10.3390/ijms26136068
Aubee JI, Williams K, Adigun A, Olusanya O, Nurse J, Thompson KM. Post-Transcriptional Regulation of the MiaA Prenyl Transferase by CsrA and the Small RNA CsrB in Escherichia coli. International Journal of Molecular Sciences. 2025; 26(13):6068. https://doi.org/10.3390/ijms26136068
Chicago/Turabian StyleAubee, Joseph I., Kinlyn Williams, Alexandria Adigun, Olufolakemi Olusanya, Jalisa Nurse, and Karl M. Thompson. 2025. "Post-Transcriptional Regulation of the MiaA Prenyl Transferase by CsrA and the Small RNA CsrB in Escherichia coli" International Journal of Molecular Sciences 26, no. 13: 6068. https://doi.org/10.3390/ijms26136068
APA StyleAubee, J. I., Williams, K., Adigun, A., Olusanya, O., Nurse, J., & Thompson, K. M. (2025). Post-Transcriptional Regulation of the MiaA Prenyl Transferase by CsrA and the Small RNA CsrB in Escherichia coli. International Journal of Molecular Sciences, 26(13), 6068. https://doi.org/10.3390/ijms26136068






