Comparative Analysis of Chloroplast Genomes of 19 Saxifraga Species, Mostly from the European Alps
Abstract
1. Introduction
2. Results
2.1. Comparative Chloroplast Genomes of Saxifraga Species
2.1.1. General Feature Comparison
2.1.2. IR Boundary Variation Analysis
2.1.3. Polymorphic Analysis Among Chloroplast Genome Sequences
2.1.4. Repeat Sequences Analysis
2.1.5. Codon Usage Analysis
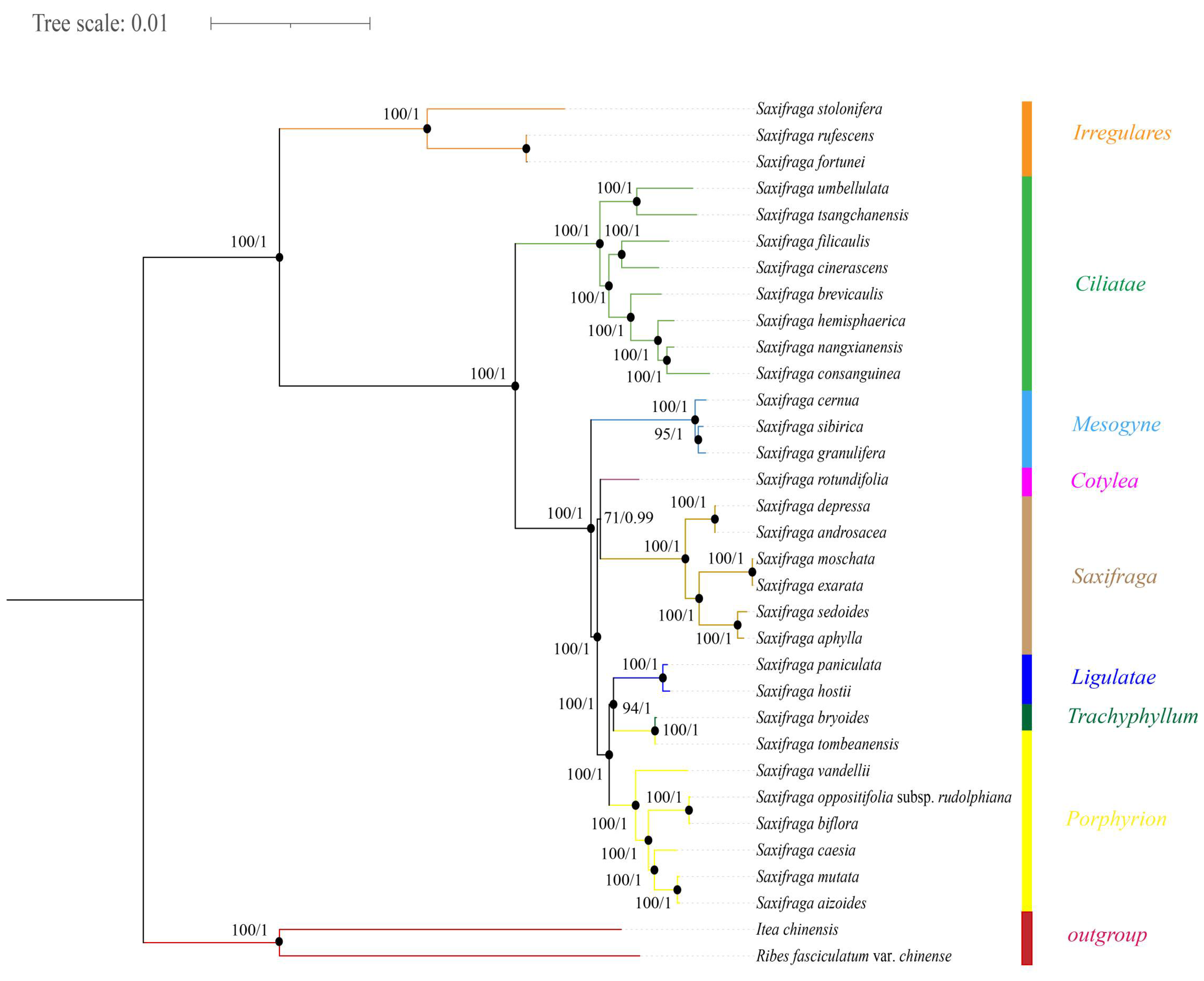
2.2. Phylogenetic Analysis
3. Discussion
3.1. Chloroplast Genome Structure Variation Within the 19 Saxifraga Species
3.2. Phylogeny of Saxifraga Species
4. Materials and Methods
4.1. Sample Collection, DNA Extraction, and Sequencing
4.2. Chloroplast Genome Assembly and Annotation
4.3. Comparative Analysis of Chloroplast Genomes
4.4. Phylogenetic Analysis
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ebersbach, J.; Schnitzler, J.; Favre, A.; Muellner-Riehl, A.N. Evolutionary radiations in the species-rich mountain genus Saxifraga L. BMC Evol. Biol. 2017, 17, 119. [Google Scholar]
- Webb, D.A.; Gornall, R.J. Saxifrages of Europe; Helm: London, UK, 1989. [Google Scholar]
- Pan, J.T.; Gornall, R.J.; Ohba, H. Saxifraga. In Flora of China; Wu, C.Y., Raven, P.H., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2001; Volume 8. [Google Scholar]
- Gao, Q.B.; Li, Y.H.; Gornall, R.J.; Zhang, Z.X.; Zhang, F.Q.; Xing, R.; Fu, P.C.; Wang, J.L.; Liu, H.R.; Tian, Z.Z.; et al. Phylogeny and speciation in Saxifraga sect. Ciliatae (Saxifragaceae): Evidence from psbA-trnH, trnL-F and ITS sequences. Taxon 2015, 64, 703–713. [Google Scholar] [CrossRef]
- Ebersbach, J.; Muellner-Riehl, A.N.; Favre, A.; Paule, J.; Winterfeld, G.; Schnitzler, J. Driving forces behind evolutionary radiations: Saxifraga section Ciliatae (Saxifragaceae) in the region of the Qinghai-Tibet Plateau. Bot. J. Linn. Soc. 2018, 186, 304–320. [Google Scholar] [CrossRef]
- Ebersbach, J.; Tkach, N.; Röser, M.; Favre, A. The role of hybridisation in the making of the species-rich arctic-alpine genus Saxifraga (Saxifragaceae). Diversity 2020, 12, 440. [Google Scholar] [CrossRef]
- Liu, L.; Xu, X.; Zhang, L.; Li, Y.; Shrestha, N.; Neves, D.M.; Wang, Q.; Chang, H.; Su, X.; Liu, Y.; et al. Global patterns of species richness of the Holarctic alpine herb Saxifraga: The role of temperature and habitat heterogeneity. J. Plant. Ecol. 2022, 15, 237–252. [Google Scholar] [CrossRef]
- Soltis, D.; Soltis, P.; Endress, P.; Chase, M.; Manchester, S.; Judd, W.; Majure, L.; Mavrodiev, E. Phylogeny and Evolution of the Angiosperm: Revised and Updated Edition; The University of Chicago Press: Chicago, IL, USA, 2018. [Google Scholar]
- Li, Y.; Jia, L.; Wang, Z.; Xing, R.; Chi, X.; Chen, S.; Gao, Q. The complete chloroplast genome of Saxifraga sinomontana (Saxifragaceae) and comparative analysis with other Saxifragaceae species. Braz. J. Bot. 2019, 42, 601–611. [Google Scholar] [CrossRef]
- Tkach, N.; Röser, M.; Miehe, G.; Muellner-Riehl, A.N.; Ebersbach, J.; Favre, A.; Hoffmann, M.H. Molecular phylogenetics morphology and a revised classification of the complex genus Saxifraga (Saxifragaceae). Taxon 2015, 64, 1159–1187. [Google Scholar] [CrossRef]
- Burke, S.V.; Lin, C.S.; Wysocki, W.P.; Clark, L.G.; Duvall, M.R. Phylogenomics and plastome evolution of tropical forest grasses (Leptaspis, Streptochaeta: Poaceae). Front. Plant. Sci. 2016, 7, 1993. [Google Scholar] [CrossRef]
- Simmons, M.P. Mutually exclusive phylogenomic inferences at the root of the angiosperms: Amborella is supported as sister and observed variability is biased. Cladistics 2017, 33, 488–512. [Google Scholar] [CrossRef]
- Dong, W.P.; Xu, C.; Wu, P.; Cheng, T.; Yu, J.; Zhou, S.L. Resolving the systematic positions of enigmatic taxa: Manipulating the chloroplast genome data of Saxifragales. Mol. Phylogenet. Evol. 2018, 126, 321–330. [Google Scholar] [CrossRef]
- Yuan, R.; Ma, X.; Zhang, Z.; Gornall, R.J.; Wang, Y.; Chen, S.; Gao, Q. Chloroplast phylogenomics and the taxonomy of Saxifraga section Ciliatae (Saxifragaceae). Ecol. Evol. 2023, 13, e9694. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.C.; Zhang, Y.Z.; Geng, H.M.; Chen, S.L. The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ 2016, 4, e2540. [Google Scholar] [PubMed]
- Sun, S.S.; Fu, P.C.; Zhou, X.J.; Cheng, Y.W.; Zhang, F.Q.; Chen, S.L.; Gao, Q.B. The complete plastome sequences of seven species in Gentiana sect. Kudoa (Gentianaceae): Insights into plastid gene loss and molecular evolution. Front. Plant. Sci. 2018, 9, 493. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huo, N.; Dong, L.; Wang, Y.; Zhang, S.; Young, H.A.; Feng, X.; Gu, Y.Q. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS ONE 2013, 8, e57533. [Google Scholar] [CrossRef]
- Walker, J.F.; Zanis, M.J.; Emery, N.C. Comparative analysis of complete chloroplast genome sequence and inversion variation in Lasthenia burkei (Madieae Asteraceae). Am. J. Bot. 2014, 101, 722–729. [Google Scholar] [CrossRef]
- Doyle, J.J.; Davis, J.I.; Soreng, R.J.; Garvin, D.; Anderson, M.J. Chloroplast DNA inversions and the origin of the grass family (Poaceae). Proc. Natl. Acad. Sci. USA 1992, 89, 7722–7726. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L.; Ballenger, J.A.; Palmer, J.D. The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol. Phylogenet. Evol. 1996, 5, 429–438. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; de Pamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Song, T.; Liu, Z.B.; Li, J.J.; Zhu, Q.K.; Tan, R.; Chen, J.S.; Zhou, J.Y.; Liao, H. Comparative transcriptome of rhizome and leaf in Ligusticum Chuanxiong. Plant Syst. Evol. 2015, 301, 2073–2085. [Google Scholar] [CrossRef]
- Duan, L.; Li, S.J.; Su, C.; Sirichamorn, Y.; Han, L.N.; Ye, W.; Lôc, P.K.; Wen, J.; Compton, J.A.; Schrire, B.; et al. Phylogenomic framework of the IRLC legumes (Leguminosae subfamily Papilionoideae) and intercontinental biogeography of tribe Wisterieae. Mol. Phylogenet. Evol. 2021, 163, 107235. [Google Scholar] [CrossRef]
- Palmer, J.D. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Y.; Yang, J.X.; Li, H.K.; Zhao, H.S. Chloroplast genomes of two species of Cypripedium: Expanded genome size and proliferation of AT-biased repeat sequences. Front. Plant Sci. 2021, 12, 609729. [Google Scholar] [CrossRef]
- Tsuji, S.; Ueda, K.; Nishiyama, T.; Hasebe, M.; Yoshikawa, S.; Konagaya, A.; Nishiuchi, T.; Yamaguchi, K. The chloroplast genome from a lycophyte (microphyllophyte), Selaginella uncinata, has a unique inversion, transpositions and many gene losses. J. Plant Res. 2007, 120, 281–290. [Google Scholar] [CrossRef]
- Plunkett, G.M.; Downie, S.R. Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Syst. Bot. 2000, 25, 648–667. [Google Scholar] [CrossRef]
- Wu, C.S.; Chaw, S.M. Large-scale comparative analysis reveals the mechanisms driving plastomic compaction, reduction, and inversions in conifers II (Cupressophytes). Genome Biol. Evol. 2016, 8, 3740–3750. [Google Scholar] [CrossRef]
- Zhu, A.; Guo, W.; Gupta, S.; Fan, W.; Mower, J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef]
- Guo, Y.Y.; Yang, J.X.; Bai, M.Z.; Zhang, G.Q.; Liu, Z.J. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 2021, 21, 248. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Thompson, W.F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 1982, 29, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Chen, S.; Xu, W.; Zhao, Y.; Zhang, D. Origination and function of plant pseudogenes. Plant Signal. Behav. 2019, 14, e1625698. [Google Scholar] [CrossRef]
- Xia, C.; Wang, M.; Guan, Y.; Li, J. Comparative analysis of the chloroplast genome for Aconitum species: Genome structure and phylogenetic relationships. Front. Genet. 2022, 13, 878182. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, X.; Yang, Y.; Wei, P.; Zhang, W.; Li, X.; Liu, C.; Zhao, S.; Li, X.; Liu, X. Comparative Analysis of Chloroplast Genomes within Saxifraga (Saxifragaceae) Takes Insights into Their Genomic Evolution and Adaption to the High-Elevation Environment. Genes 2022, 13, 1673. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Morgante, M.; McDevitt, R.; Vendramin, G.G.; Rafalski, J.A. Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. USA 1995, 92, 7759–7763. [Google Scholar] [CrossRef] [PubMed]
- Provan, J.; Russell, J.R.; Booth, A.; Powell, W. Polymorphic chloroplast simple sequence repeat primers for systematic and population studies in the genus Hordeum. Mol. Ecol. 1999, 8, 505–511. [Google Scholar] [CrossRef]
- Provan, J.; Powell, W.; Hollingsworth, P.M. Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends Ecol. Evol. 2001, 16, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Flannery, M.L.; Mitchell, F.J.G.; Coyne, S.; Kavanagh, T.A.; Burke, J.I.; Salamin, N.; Dowding, P.; Hodkinson, T.R. Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theor. Appl. Genet. 2006, 113, 1221–1231. [Google Scholar] [CrossRef]
- Li, L.; Hu, Y.; He, M.; Zhang, B.; Wu, W.; Cai, P.; Huo, D.; Hong, Y. Comparative chloroplast genomes: Insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genom. 2021, 22, 138. [Google Scholar] [CrossRef]
- Yan, C.; Du, J.; Gao, L.; Li, Y.; Hou, X. The complete chloroplast genome sequence of watercress (Nasturtium officinale R. Br.): Genome organization, adaptive evolution and phylogenetic relationships in Cardamineae. Gene 2019, 699, 24–36. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Liao, M.; Cheng, Y.H.; Feng, Y.; Ju, W.B.; Deng, H.N.; Li, X.; Plenković-Moraj, A.; Xu, B. Comparative chloroplast genomics of seven endangered Cypripedium species and phylogenetic relationships of Orchidaceae. Front. Plant Sci. 2022, 13, 911702. [Google Scholar] [CrossRef]
- Kuang, D.Y.; Wu, H.; Wang, Y.L.; Gao, L.M.; Zhang, S.Z.; Lu, L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome 2011, 54, 663–673. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, H.T.; Kim, J.H. The largest plastid genome of monocots: A novel genome type containing AT residue repeats in the slipper orchid Cypripedium japonicum. Plant Mol. Biol. Rep. 2015, 33, 1210–1220. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Waqas, M.; Kang, S.M.; Yun, B.W.; Lee, I.J. Chloroplast genomes of Arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: Structures and comparative analysis. Sci. Rep. 2017, 7, 7556. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Cowe, E.; Higgins, D.G.; Shields, D.C.; Wolff, K.H.; Wright, F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988, 16, 8207–8211. [Google Scholar] [CrossRef]
- Zhang, X.J.; Huang, X.H.; Landis, J.B.; Fu, Q.S.; Chen, J.T.; Luo, P.R.; Li, L.J.; Lu, H.Y.; Sun, H.; Deng, T. Shifts in reproductive strategies in the evolutionary trajectory of plant lineages. Sci. China Life Sci. 2024, 67, 2499–2510. [Google Scholar] [CrossRef] [PubMed]
- Gornall, R.J. An outline of a revised classification of Saxifraga L. Bot. J. Linn. Soc. 1987, 95, 273–292. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; de Pamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Liu, S.; Ni, Y.; Li, J.; Zhang, X.; Yang, H.; Chen, H.; Liu, C. CPGView: A package for visualizing detailed chloroplast genome structures. Mol. Ecol. Resour. 2023, 23, 694–704. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Huang, L.; Yu, H.; Wang, Z.; Xu, W. CPStools: A package for analyzing chloroplast genome sequences. iMetaOmics 2024, 1, e25. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczail, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Yang, T.; Aishan, S.; Zhu, J.; Qin, Y.; Liu, J.; Liu, H.; Tie, J.; Wang, J.; Qin, R. Chloroplast genomes and phylogenetic analysis of three Carthamus (Asteraceae) Species. Int. J. Mol. Sci. 2023, 24, 15634. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Effective Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
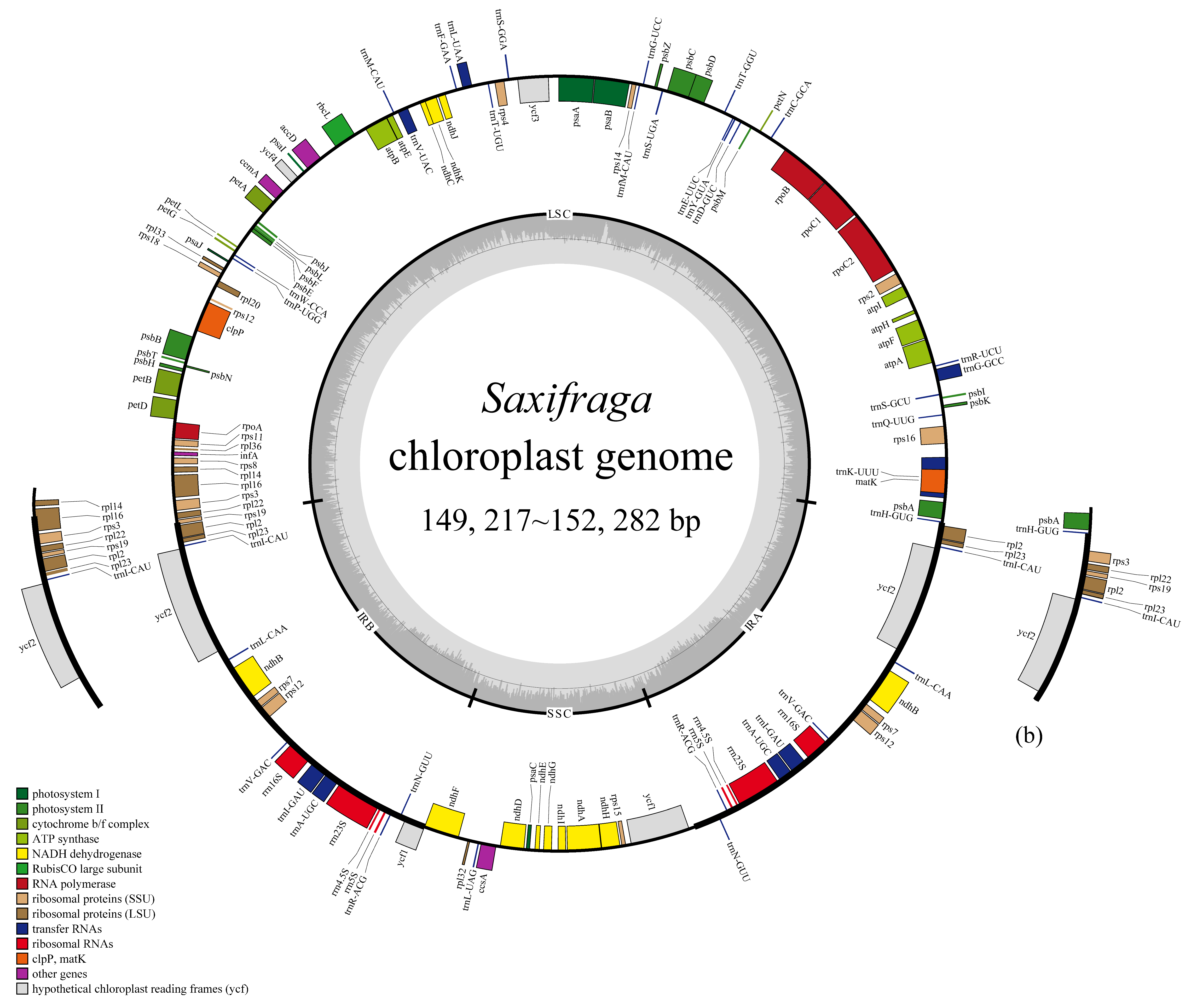

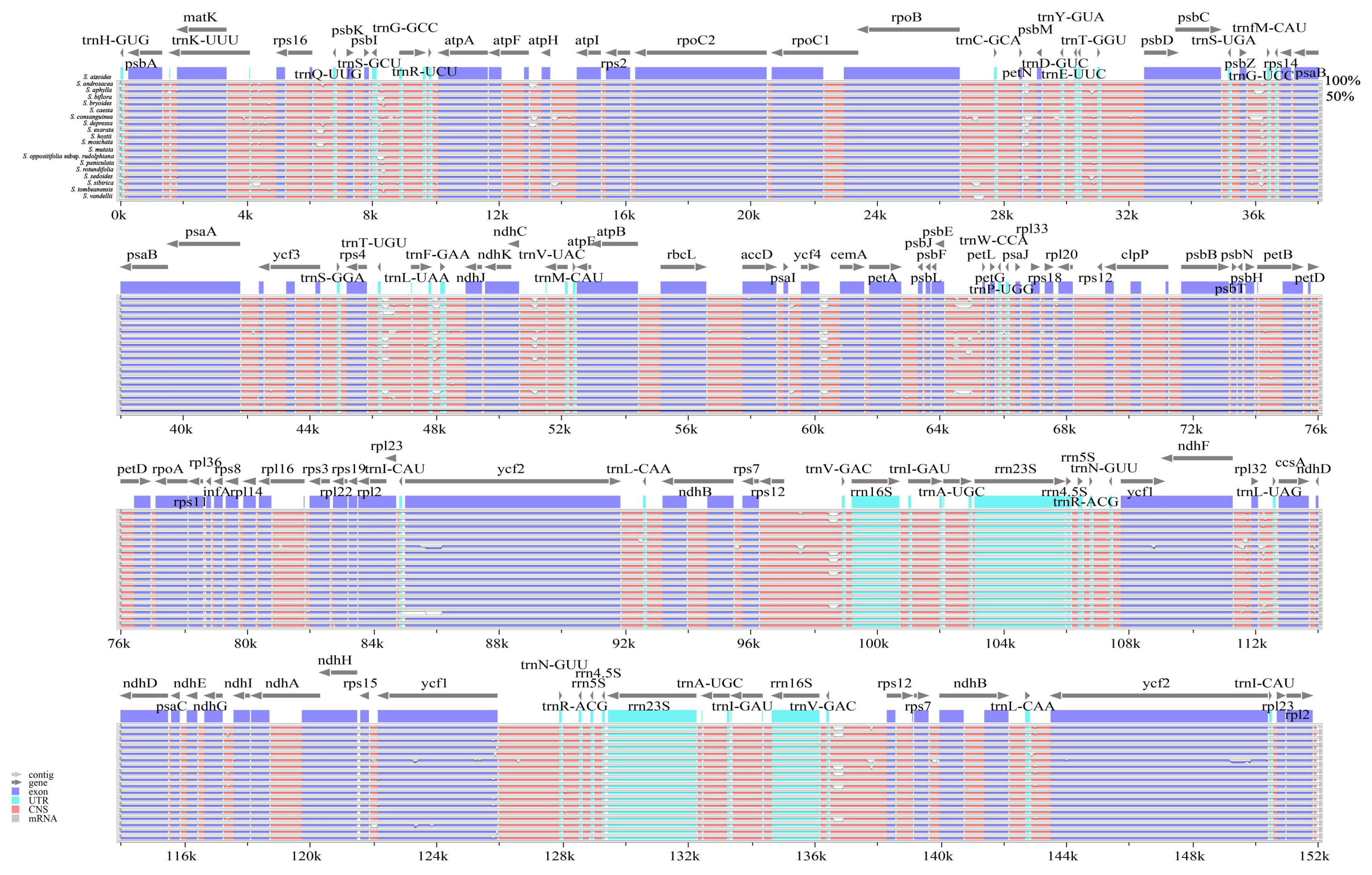
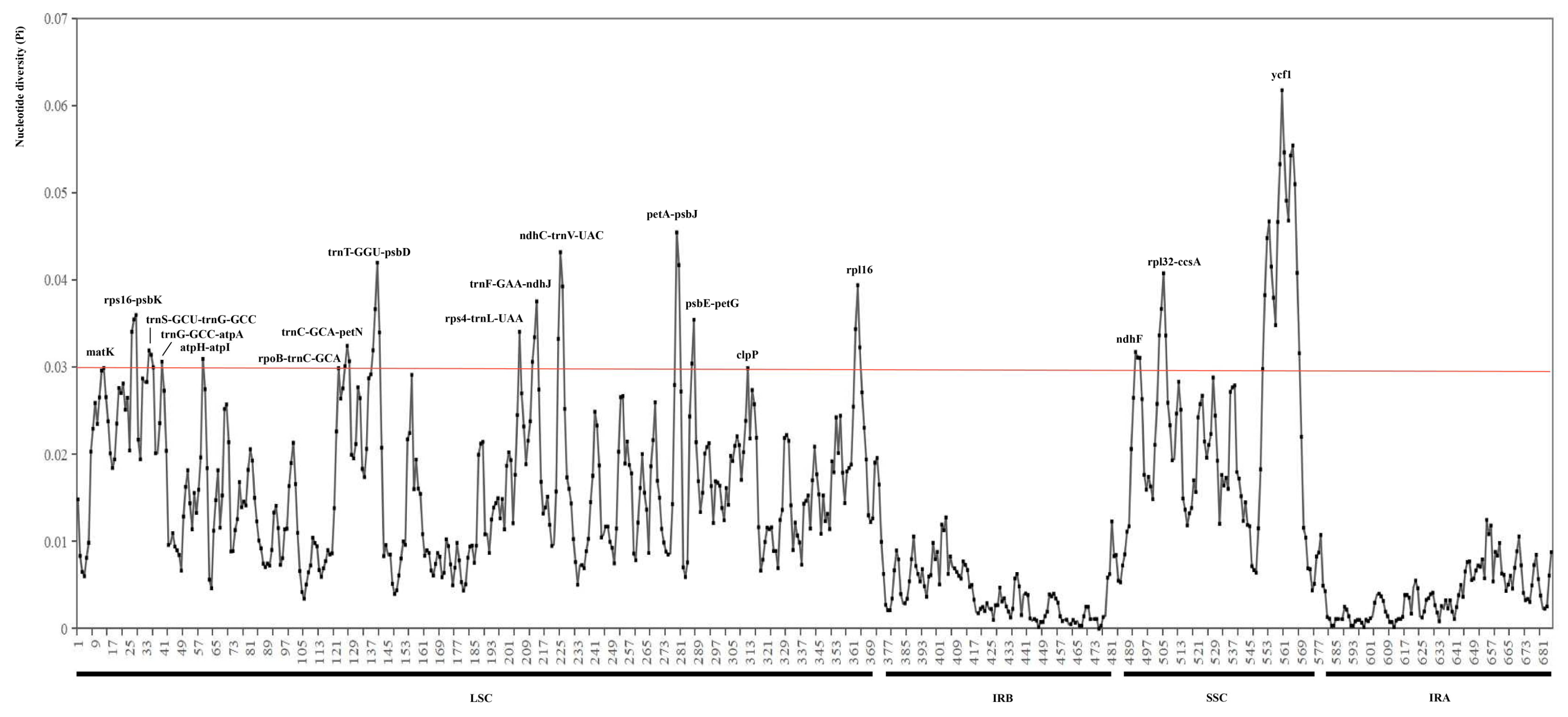
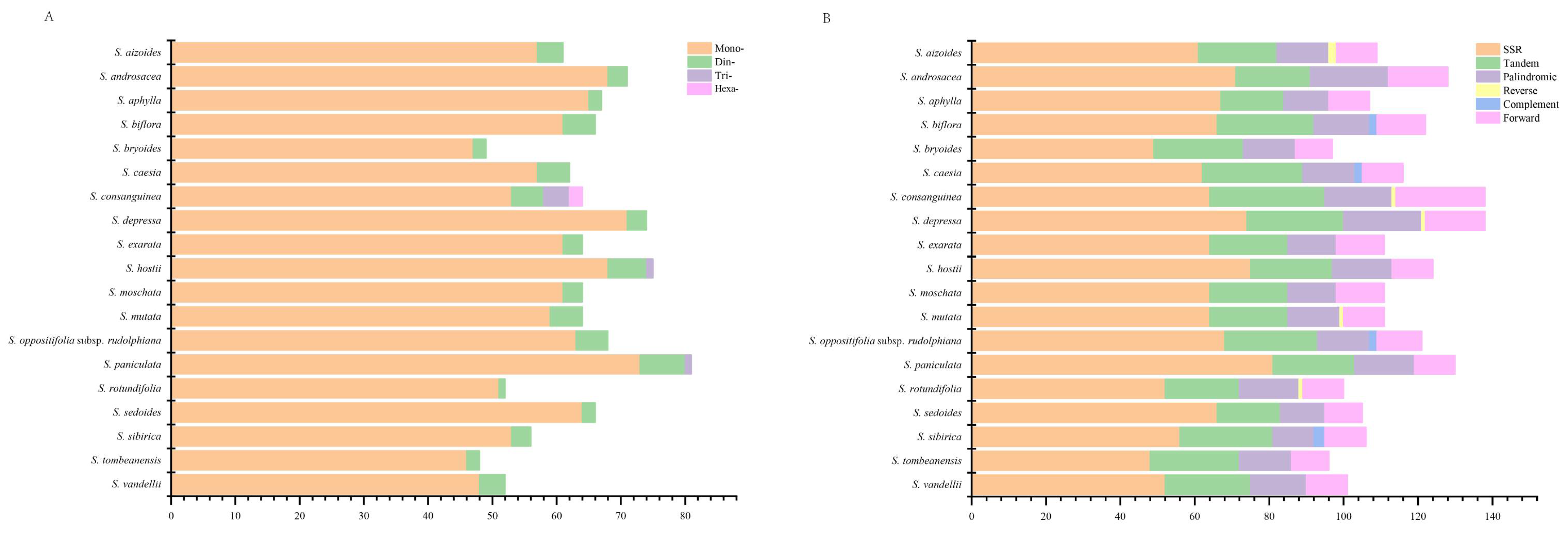
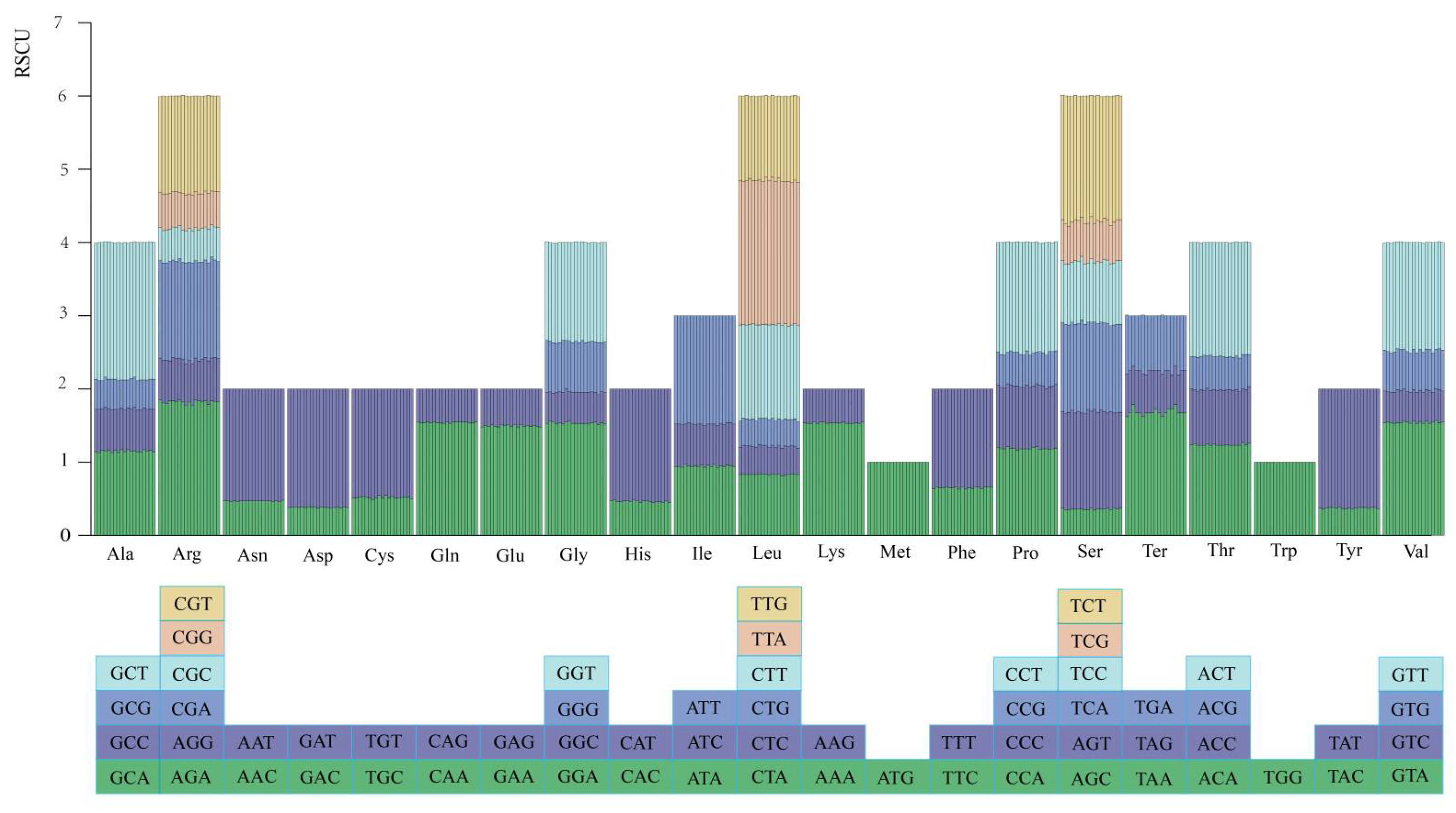
| Species | Length (bp) | GC Content (%) | Gene (Unique Gene) Number | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | LSC | SSC | IR | Total | LSC | SSC | IR | Total | CDS | tRNA | rRNA | |
| S. aizoides | 151,971 | 83,379 | 17,234 | 25,679 | 37.75 | 35.81 | 31.99 | 42.82 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. androsacea | 149,600 | 81,766 | 17,078 | 25,378 | 37.69 | 35.72 | 31.71 | 42.88 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. aphylla | 149,217 | 81,362 | 16,989 | 25,433 | 37.77 | 35.81 | 31.90 | 42.86 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. biflora | 151,552 | 82,974 | 17,194 | 25,692 | 37.76 | 35.88 | 31.84 | 42.78 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. bryoides | 152,082 | 83,432 | 17,236 | 25,707 | 37.76 | 35.77 | 32.09 | 42.90 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. caesia | 151,360 | 83,447 | 16,985 | 25,464 | 37.74 | 35.79 | 32.08 | 42.82 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. consanguinea | 150,047 | 78,948 | 16,659 | 27,220 | 37.84 | 36.16 | 32.10 | 42.04 | 134 (113) | 89 (79) | 37 (30) | 8 (4) |
| S. depressa | 149,685 | 81,790 | 17,107 | 25,394 | 37.67 | 35.70 | 31.70 | 42.86 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. exarata | 150,001 | 82,182 | 16,927 | 25,446 | 37.77 | 35.78 | 32.00 | 42.92 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. hostii | 151,913 | 83,227 | 17,274 | 25,706 | 37.78 | 35.85 | 31.94 | 42.87 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. moschata | 149,998 | 82,179 | 16,927 | 25,446 | 37.77 | 35.78 | 31.99 | 42.92 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. mutata | 152,017 | 83,427 | 17,234 | 25,678 | 37.74 | 35.80 | 31.98 | 42.82 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. oppositifolia subsp. rudolphiana | 151,875 | 83,278 | 17,195 | 25,701 | 37.71 | 35.81 | 31.82 | 42.77 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. paniculata | 152,098 | 83,439 | 17,257 | 25,701 | 37.77 | 35.83 | 31.95 | 42.86 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. rotundifolia | 152,282 | 83,618 | 17,352 | 25,656 | 37.80 | 35.90 | 32.05 | 42.85 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. sedoides | 149,363 | 81,479 | 16,996 | 25,444 | 37.76 | 35.81 | 31.85 | 42.85 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. sibirica | 151,420 | 82,833 | 16,987 | 25,800 | 37.66 | 35.67 | 31.81 | 42.77 | 131 (112) | 86 (79) | 37 (30) | 8 (4) |
| S. tombeanensis | 152,092 | 83,444 | 17,234 | 25,707 | 37.76 | 35.76 | 32.09 | 42.91 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| S. vandellii | 151,925 | 83,273 | 17,226 | 25,713 | 37.76 | 35.79 | 32.08 | 42.86 | 131 (113) | 86 (79) | 37 (30) | 8 (4) |
| Category | Gene Group | Gene Name |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, B, C, I, J, ycf3 a |
| Subunits of photosystem II | psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z | |
| Subunits of NADH dehydrogenase | ndhA b, B b,c, C, D, E, F, G, H, I, J, K | |
| Subunits of cytochrome b/f complex | petA, B b, D b, G, L, N | |
| Subunits of ATP synthase | atpA, B, E, F b, H, I | |
| Large subunit of rubisco | rbcL | |
| Self-replication | Proteins of large ribosomal subunit | rpl2 c, 14, 16 b, 20, 22 e, 23 c, 32, 33, 36 |
| Proteins of small ribosomal subunit | rps2, 3 e, 4, 7 c, 8, 11, 12 b,c,d, 14, 15, 16 b, 18, 19 e | |
| Subunits of RNA polymerase | rpoA, B, C1 b, C2 | |
| Ribosomal RNAs | rrn4.5 c, 5 c, 16 c, 23 c | |
| Transfer RNAs | trnA(UGC) b,c, C(GCA), D(GUC), E(UUC), F(GAA), fM(CAU), G(GCC) b, G(UCC), H(GUG), I(CAU) c, I(GAU) b,c, K(UUU) b, L(CAA) c, L(UAA) b, L(UAG), M(CAU), N(GUU) c, P(UGG), Q(UUG), R(ACG) c, R(UCU), S(GCU), S(GGA), S(UGA), T(GGU), T(UGU), V(GAC) c, V(UAC) b, W(CCA), Y(GUA) | |
| Other genes | Maturase | matK |
| Protease | clpP a | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | infA | |
| Proteins of unknown function | ycf1 c, ycf2 c, ycf4 |
| Taxa | Locality | Latitude (N) | Longitude (E) | Altitude (m) | GenBank Accession No. |
|---|---|---|---|---|---|
| Saxifraga sect. Ciliatae Haw. | |||||
| S. consanguinea W. W. Sm. | Dari, Qinghai, China | 33.44068° | 98.66934° | 4556 | PV426734 |
| Sect. Cotylea Tausch | |||||
| S. rotundifolia L. | Val di Loriva, Italy | 45.82570° | 10.61749° | 1900 | PV426742 |
| Sect. Ligulatae Haw. | |||||
| S. hostii Tausch | Passo di Maniva, Italy | 45.81642° | 10.41266° | 1670 | PV426737 |
| S. paniculata Mill. | Colfosco Calfosch, Italy | 46.55071° | 11.83100° | 1853 | PV426741 |
| Sect. Mesogyne Sternb. | |||||
| S. sibirica L. | Shennongjia, Hubei, China | 31.44257° | 110.27239° | 2875 | PV426744 |
| Sect. Porphyrion Tausch | |||||
| S. aizoides L. | Grossglockner, Austria | 47.07452° | 12.74526° | 2084 | PV423509 |
| S. biflora All. | Grossglockner, Austria | 47.07452° | 12.74526° | 2084 | PV426731 |
| S. caesia L. | Manira Pass, Italy | 45.80359° | 10.41020° | 1636 | PV426733 |
| S. mutata L. | Magasa, Italy | 45.78538° | 10.62095° | 940 | PV426739 |
| S. oppositifolia L. subsp. rudolphiana (Hornsch. ex W. D. J. Koch) Engl. & Irmsch. | Grossglockner, Austria | 47.07452° | 12.74526° | 2084 | PV426740 |
| S. tombeanensis Boiss. ex Engl. | Arabba, Italy | 46.47297° | 11.87086° | 2521 | PV426745 |
| S. vandellii Sternb. | Presolana, Italy | 45.96681° | 10.04827° | 2040 | PV426746 |
| Sect. Saxifraga | |||||
| S. androsacea L. | Col Rodela, Italy | 46.49837° | 11.74894° | 2300 | PV426729 |
| S. aphylla Sternb. | Innsbruck, Austria | 47.31284° | 11.38437° | 2300 | PV426730 |
| S. depressa Sternb. | Arabba, Italy | 46.47297° | 11.87086° | 2521 | PV426735 |
| S. exarata Vill. | Lago Bianco, Switzerland | 46.40780° | 10.02137° | 2276 | PV426736 |
| S. moschata Wulfen | Rudolfshectte, Austria | 47.13442° | 12.62523° | 2313 | PV426738 |
| S. sedoides L. | Sassolungo, Italy | 46.51657° | 11.73929° | 2685 | PV426743 |
| Sect. Trachyphyllum (Gaudin) W. D. J. Koch | |||||
| S. bryoides L. | Grossglockner, Austria | 47.12626° | 12.81354° | 2121 | PV426732 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leng, Z.; Pang, Z.; He, Z.; Gao, Q. Comparative Analysis of Chloroplast Genomes of 19 Saxifraga Species, Mostly from the European Alps. Int. J. Mol. Sci. 2025, 26, 6015. https://doi.org/10.3390/ijms26136015
Leng Z, Pang Z, He Z, Gao Q. Comparative Analysis of Chloroplast Genomes of 19 Saxifraga Species, Mostly from the European Alps. International Journal of Molecular Sciences. 2025; 26(13):6015. https://doi.org/10.3390/ijms26136015
Chicago/Turabian StyleLeng, Zhenning, Zhe Pang, Zaijun He, and Qingbo Gao. 2025. "Comparative Analysis of Chloroplast Genomes of 19 Saxifraga Species, Mostly from the European Alps" International Journal of Molecular Sciences 26, no. 13: 6015. https://doi.org/10.3390/ijms26136015
APA StyleLeng, Z., Pang, Z., He, Z., & Gao, Q. (2025). Comparative Analysis of Chloroplast Genomes of 19 Saxifraga Species, Mostly from the European Alps. International Journal of Molecular Sciences, 26(13), 6015. https://doi.org/10.3390/ijms26136015






