Rare Evolutionary Events Support the Phylogenetic Placement of Orthonectida Within Annelida
Abstract
1. Introduction
2. Results and Discussion
2.1. Organization of Orthonectid Mitochondrial Genomes
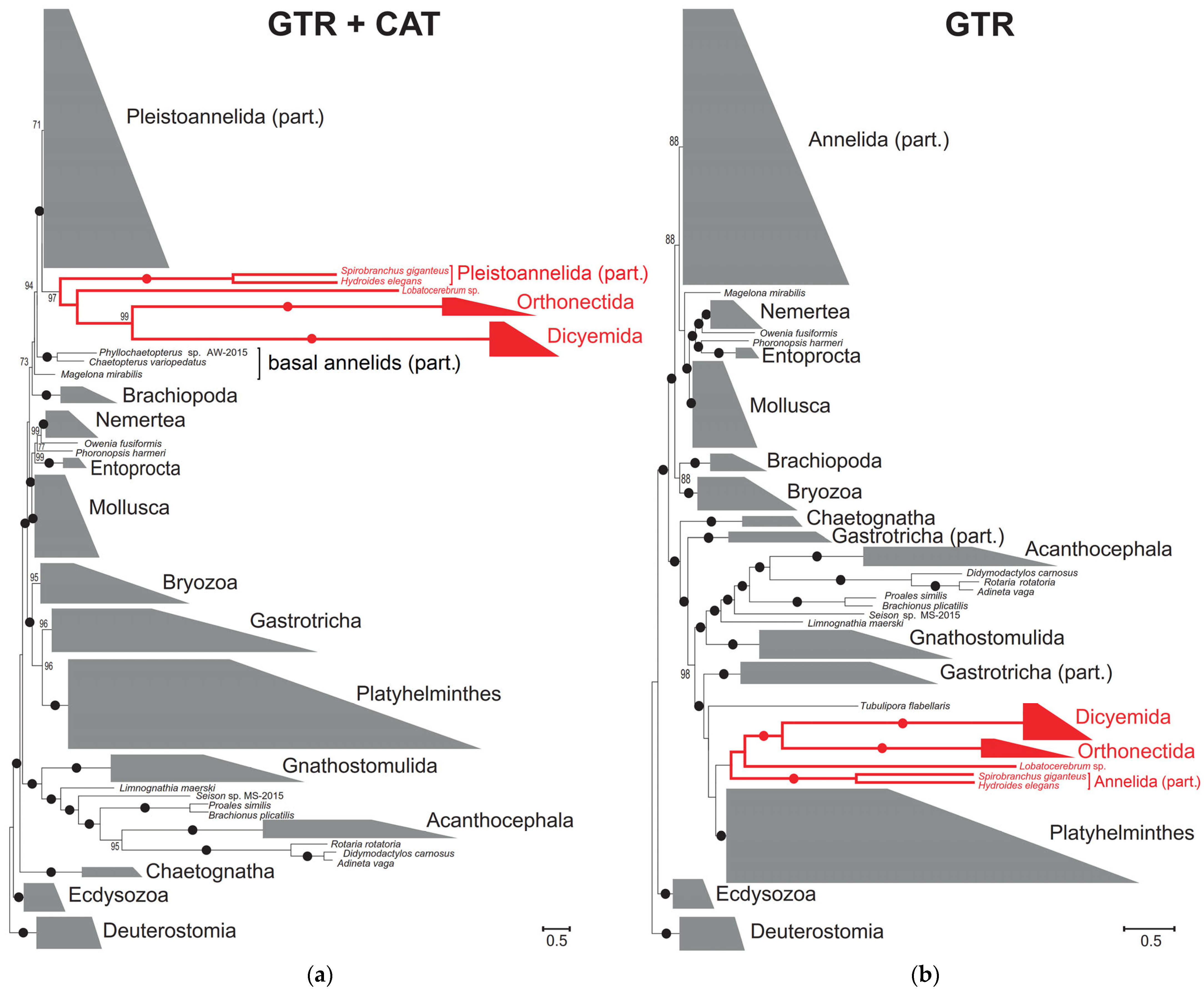
2.2. Bayesian Approach to Orthonectid Phylogeny
2.3. Synapomorphies of Annelida and Orthonectida
2.4. Placement of Orthonectida in Annelida
2.5. Synapomorphies of Orthonectida and Dicyemida
2.6. Orthonectida and Lobatocerebrida
2.7. Morphological Similarities Between Orthonectida and Annelida
2.8. In Lieu of a Conclusion: Prospects for Optimizing
3. Materials and Methods
3.1. Mitochondrial Genome Assembly and Annotation
3.2. Determination of the Primary Structure of Non-Coding Region in the Mitogenome of I. linei
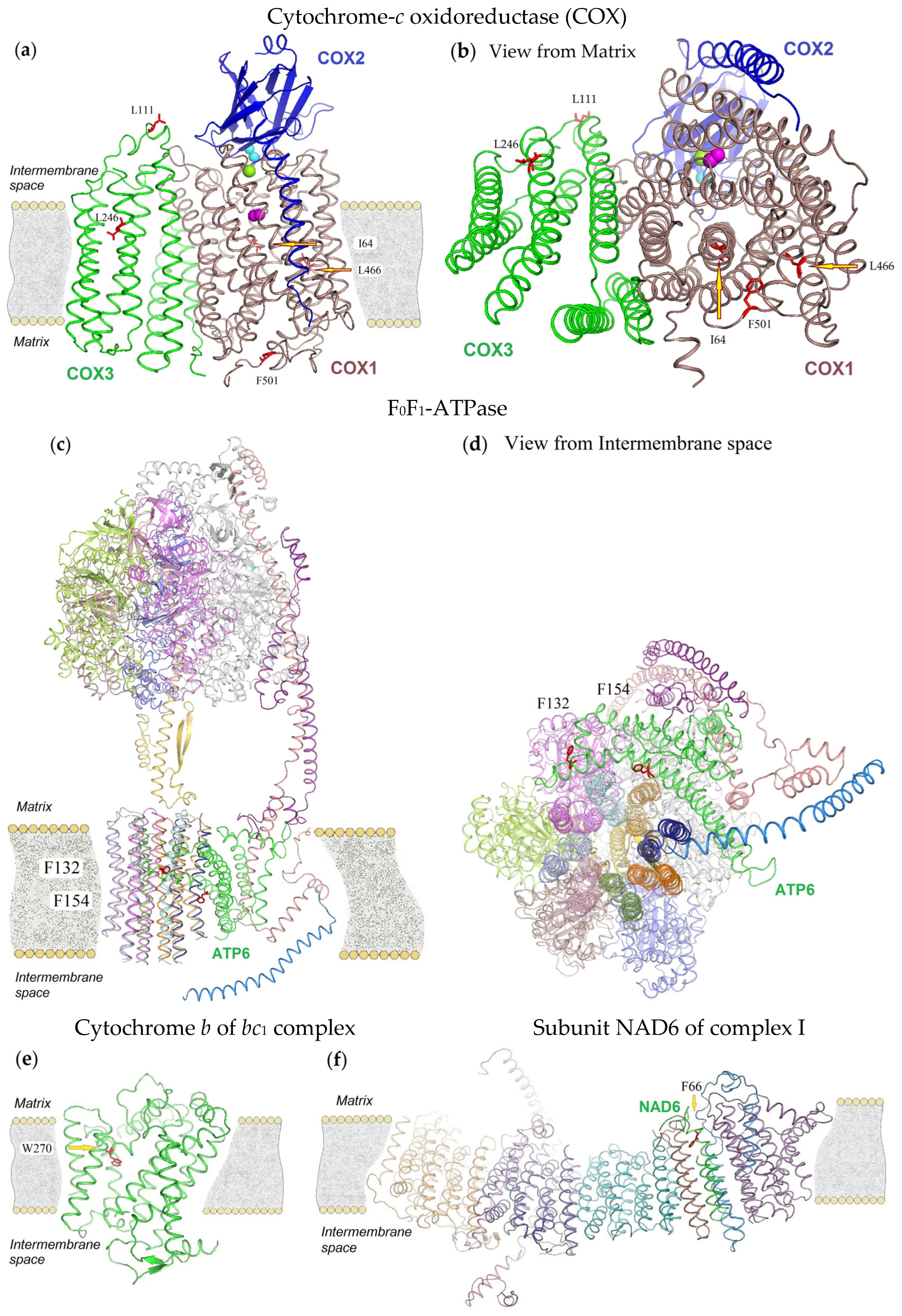
3.3. 3D-Modeling of Mitochondrial Proteins
3.4. Phylogenetic Analysis
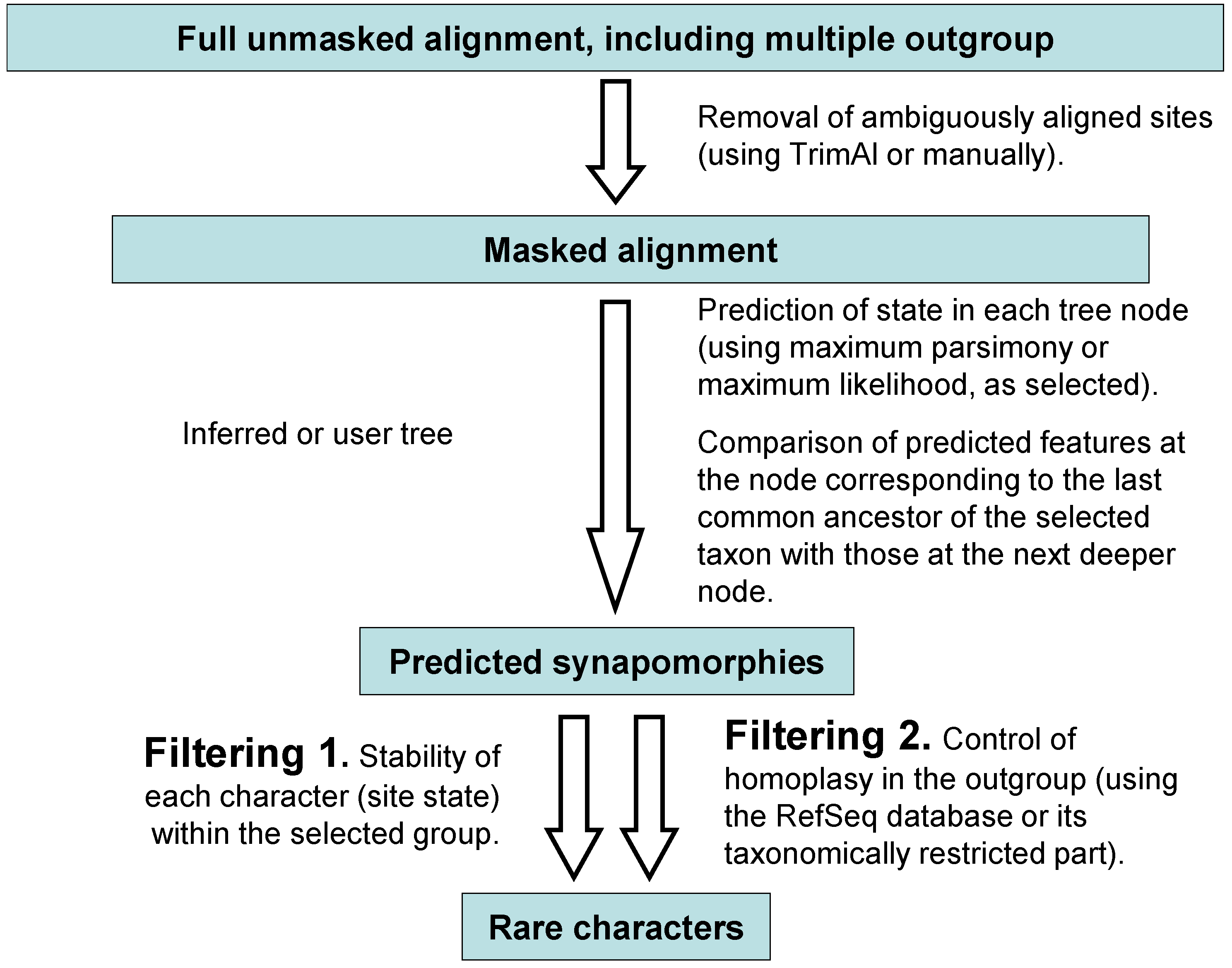
3.5. Synapomorphy Search
- h = (h1, h2, …, hn−1, hn) # Each value of the vector corresponds to the homoplasy frequency of each synapomorphy according to the RefSeq database.
- w = 1 − h # Each value of the vector corresponds to the probability of non-emergence of each synapomorphy.
- ak = combn (h,k) # Matrices contain all cases of emergence of k synapomorphies, where k is a number from 1 to n.
- bk = combn (w,k) # Matrices contain all cases of non-emergence of k synapomorphies, where k is a number from 1 to n.
- νk = apply (ak,2,prod) # The vector of probabilities of emergence of k synapomorphies for all possible subsets of k out of n synapomorphies.
- yk = apply (bk,2,prod) # The vector of probabilities of non-emergence of k synapomorphies for all possible subsets of k out of n synapomorphies.
- pk = νk × yn−k # The probabilities of independent emergence of exactly k out of n synapomorphies.
- qk = sum (pk) # The probability of independent emergence of exactly k out of n synapomorphies.
- xk = qk + qi+1 + … + qn−1 + qn # The total probability of independent emergence of k or more synapomorphies.
3.6. Probability of Loss of Synapomorphies
- s = (s1, s2, …, sn−1, sn) # Each value of vector corresponds to the frequency of occurrence of each synapomorphy in a given group.
- a = (ai), ai = si^r # The probabilities of remaining each synapomorphy in orthonectids, adjusted for the increased rate of evolution of orthonectids compared to the average representative of this group. Here r is a coefficient reflecting how much times the rate of evolution for orthonectids is higher than the rate of evolution for average representative of this group (see Table 1).
- b = (bi), bi = 1 − ai # The probabilities of loss for each synapomorphy in orthonectids.
- p = prodi∈M (bi) # The a priori probability of loss for a specific subset M of synapomorphies in orthonectids.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kozloff, E.N. The genera of the phylum Orthonectida. Cah. Biol. Mar. 1992, 33, 377–406. [Google Scholar]
- Slyusarev, G.S. Chapter: Ortonectida. In Handbook of Zoology. Miscellaneous Invertebrates; Schmidt-Rhaesa, A., Ed.; De Gruyter: Berlin, Germany; Boston, MA, USA, 2018; pp. 11–40. [Google Scholar] [CrossRef]
- Slyusarev, G.S.; Starunov, V.V. The structure of the muscular and nervous systems of the female Intoshia linei (Orthonectida). Org. Divers. Evol. 2016, 16, 65–71. [Google Scholar] [CrossRef]
- Slyusarev, G.S.; Bondarenko, N.I.; Skalon, E.K.; Rappoport, A.K.; Radchenko, D.; Starunov, V.V. The structure of the muscular and nervous systems of the orthonectid Rhopalura litoralis (Orthonectida) or what parasitism can do to an annelid. Org. Divers. Evol. 2021, 22, 35–45. [Google Scholar] [CrossRef]
- Slyusarev, G.S.; Skalon, E.K.; Starunov, V.V. Evolution of Orthonectida body plan. Evol. Dev. 2023, 26, e12462. [Google Scholar] [CrossRef]
- Mikhailov, K.V.; Slyusarev, G.S.; Nikitin, M.A.; Logacheva, M.D.; Penin, A.A.; Aleoshin, V.V.; Panchin, Y.V. The genome of Intoshia linei affirms orthonectids as highly simplified spiralians. Curr. Biol. 2016, 26, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.M.; Kanda, M.; Satoh, N.; Furuya, H. The phylogenetic position of dicyemid mesozoans offers insights into spiralian evolution. Zoological. Lett. 2017, 3, 6. [Google Scholar] [CrossRef]
- Schiffer, P.; Robertson, H.; Telford, M.J. Orthonectids are highly degenerate annelid worms. Curr. Biol. 2018, 28, 1970–1974.e3. [Google Scholar] [CrossRef]
- Zverkov, O.A.; Mikhailov, K.V.; Isaev, S.V.; Rusin, L.Y.; Popova, O.V.; Logacheva, M.D.; Penin, A.A.; Moroz, L.L.; Panchin, Y.V.; Lyubetsky, V.A.; et al. Dicyemida and Orthonectida: Two Stories of Body Plan Simplification. Front. Genet. 2019, 10, 443. [Google Scholar] [CrossRef]
- Drábková, M.; Kocot, K.M.; Halanych, K.M.; Oakley, T.H.; Moroz, L.L.; Cannon, J.T.; Kuris, A.; Garcia-Vedrenne, A.E.; Pankey, M.S.; Ellis, E.A.; et al. Different phylogenomic methods support monophyly of enigmatic ‘Mesozoa’ (Dicyemida + Orthonectida, Lophotrochozoa). Proc. Biol. Sci. 2022, 289, 20220683. [Google Scholar] [CrossRef]
- Bondarenko, N.; Bondarenko, A.; Starunov, V.; Slyusarev, G. Comparative analysis of the mitochondrial genomes of Orthonectida: Insights into the evolution of an invertebrate parasite species. Mol. Genet. Genom. 2019, 294, 715–727. [Google Scholar] [CrossRef]
- Felsenstein, J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978, 27, 401–410. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P. Is the Felsenstein zone a fly trap? Syst. Biol. 1997, 46, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Bergsten, J. A review of long-branch attraction. Cladistics 2005, 21, 163–193. [Google Scholar] [CrossRef] [PubMed]
- Susko, E.; Roger, A.J. Long Branch Attraction Biases in Phylogenetics. Syst. Biol. 2021, 70, 838–843. [Google Scholar] [CrossRef]
- Gao, J.W.; Yuan, X.P.; Wu, H.; Xiang, C.Y.; Xie, M.; Song, R.; Chen, Z.Y.; Wu, Y.A.; Ou, D.S. Mitochondrial phylogenomics of Acanthocephala: Nucleotide alignments produce long-branch attraction artefacts. Parasit. Vectors 2022, 15, 376. [Google Scholar] [CrossRef]
- Rokas, A.; Holland, P.W. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 2000, 15, 454–459. [Google Scholar] [CrossRef]
- Venkatesh, B.; Erdmann, M.V.; Brenner, S. Molecular synapomorphies resolve evolutionary relationships of extant jawed vertebrates. Proc. Natl. Acad. Sci. USA 2001, 98, 11382–11387. [Google Scholar] [CrossRef]
- Luan, P.T.; Ryder, O.A.; Davis, H.; Zhang, Y.P.; Yu, L. Incorporating indels as phylogenetic characters: Impact for interfamilial relationships within Arctoidea (Mammalia: Carnivora). Mol. Phylogenet. Evol. 2013, 66, 748–756. [Google Scholar] [CrossRef]
- Tan, M.H.; Gan, H.M.; Lee, Y.P.; Linton, S.; Grandjean, F.; Bartholomei-Santos, M.L.; Miller, A.D.; Austin, C.M. ORDER within the chaos: Insights into phylogenetic relationships within the Anomura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements. Mol. Phylogenet. Evol. 2018, 127, 320–331. [Google Scholar] [CrossRef]
- Bi, X.P.; Zhang, G.J. Ancestral developmental potentials in early bony fish contributed to vertebrate water-to-land transition. Zool. Res. 2021, 42, 135–137. [Google Scholar] [CrossRef]
- Parey, E.; Louis, A.; Montfort, J.; Bouchez, O.; Roques, C.; Iampietro, C.; Lluch, J.; Castinel, A.; Donnadieu, C.; Desvignes, T.; et al. Genome structures resolve the early diversification of teleost fishes. Science 2023, 379, 572–575. [Google Scholar] [CrossRef]
- Rusin, L.Y.; Aleshin, V.V.; Vladychenskaya, N.S.; Milyutina, I.A.; Kedrova, O.S.; Petrov, N.B. Trefusiidae are a subtaxon of marine enoplida (Nematoda): Evidence from primary structure of hairpin 35 and 48 loops of SSU rRNA gene. Mol. Biol. 2001, 35, 778–784. [Google Scholar] [CrossRef]
- Aleshin, V.V.; Konstantinova, A.V.; Mikhailov, K.V.; Nikitin, M.A.; Petrov, N.B. Do we need many genes for phylogenetic inference? Biochemistry 2007, 72, 1313–1323. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.E.; Schiffer, P.H.; Telford, M.J. The mitochondrial genomes of the mesozoans Intoshia linei, Dicyema sp. and Dicyema japonicum. Parasitol. Open. 2018, 4, e16. [Google Scholar] [CrossRef]
- Wei, S.J.; Shi, M.; He, J.H.; Sharkey, M.; Chen, X.X. The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome 2009, 52, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Le, T.H.; Blair, D.; McManus, D.P. Mitochondrial genomes of parasitic flatworms. Trends Parasitol. 2002, 18, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Stunkard, H.W. The life-history and systematic relations of the Mesozoa. Q. Rev. Biol. 1954, 29, 230–244. [Google Scholar] [CrossRef]
- Ax, P. Multicellular Animals: A New Approach to the Phylogenetic Order in Nature; Springer: Berlin, Germany, 1996; pp. 60–87. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Fisher, R.P.; Topper, J.N.; Clayton, D.A. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell 1987, 50, 247–258. [Google Scholar] [CrossRef]
- Weigert, A.; Golombek, A.; Gerth, M.; Schwarz, F.; Struck, T.H.; Bleidorn, C. Evolution of mitochondrial gene order in Annelida. Mol. Phylogenet. Evol. 2016, 94 Pt A, 196–206. [Google Scholar] [CrossRef]
- Laumer, C.E.; Bekkouche, N.; Kerbl, A.; Goetz, F.; Neves, R.C.; Sørensen, M.V.; Kristensen, R.M.; Hejnol, A.; Dunn, C.W.; Giribet, G.; et al. Spiralian phylogeny informs the evolution of microscopic lineages. Curr. Biol. 2015, 25, 2000–2006. [Google Scholar] [CrossRef]
- Telford, M.J.; Budd, G.E.; Philippe, H. Phylogenomic Insights into Animal Evolution. Curr. Biol. 2015, 25, R876–R887. [Google Scholar] [CrossRef]
- Kocot, K.M.; Struck, T.H.; Merkel, J.; Waits, D.S.; Todt, C.; Brannock, P.M.; Weese, D.A.; Cannon, J.T.; Moroz, L.L.; Lieb, B.; et al. Phylogenomics of Lophotrochozoa with consideration of systematic error. Syst. Biol. 2017, 66, 256–282. [Google Scholar] [CrossRef]
- Marlétaz, F.; Peijnenburg, K.T.C.A.; Goto, T.; Satoh, N.; Rokhsar, D.S. A New Spiralian Phylogeny Places the Enigmatic Arrow Worms among Gnathiferans. Curr. Biol. 2019, 29, P312–P318. [Google Scholar] [CrossRef] [PubMed]
- Spillane, J.L.; LaPolice, T.M.; MacManes, M.D.; Plachetzki, D.C. Signal, bias, and the role of transcriptome assembly quality in phylogenomic inference. BMC Ecol. Evol. 2021, 21, 43. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Noto, T.; Yazaki, K.; Endoh, H. Developmentally regulated extrachromosomal circular DNA formation in the mesozoan Dicyema japonicum. Chromosoma 2003, 111, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Bleidorn, C.; Eeckhaut, I.; Podsiadlowski, L.; Schult, N.; McHugh, D.; Halanych, K.M.; Milinkovitch, M.C.; Tiedemann, R. Mitochondrial genome and nuclear sequence data support myzostomida as part of the annelid radiation. Mol. Biol. Evol. 2007, 24, 1690–1701. [Google Scholar] [CrossRef]
- Struck, T.H.; Golombek, A.; Hoesel, C.; Dimitrov, D.; Elgetany, A.H. Mitochondrial genome evolution in Annelida—A systematic study on conservative and variable gene orders and the hactors influencing its evolution. Syst. Biol. 2023, 72, 925–945. [Google Scholar] [CrossRef]
- Skulachev, V.P. Chapter: Primary ΔµH Generators. In Membrane Bioenergetics; Springer: Berlin/Heidelberg, Germany, 1988; pp. 35–144. [Google Scholar] [CrossRef]
- Timón-Gómez, A.; Nývltová, E.; Abriata, L.A.; Vila, A.J.; Hosler, J.; Barrientos, A. Mitochondrial Cytochrome c Oxidase Biogenesis: Recent Developments. Semin. Cell Dev. Biol. 2018, 76, 163–178. [Google Scholar] [CrossRef]
- Klink, G.V.; Bazykin, G.A. Parallel evolution of metazoan mitochondrial proteins. Genome Biol. Evol. 2017, 9, 1341–1350. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Lang, B.F. Poriferan mtDNA and animal phylogeny based on mitochondrial gene arrangements. Syst. Biol. 2005, 54, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Bleidorn, C.; Braband, A.; Dambach, J.; Donath, A.; Fritzsch, G.; Golombek, A.; Hadrys, H.; Jühling, F.; Meusemann, K.; et al. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol. Phylogenet. Evol. 2013, 69, 352–364. [Google Scholar] [CrossRef]
- Rieger, R.M. A new group of interstitial worms, Lobatocerebridae nov. fam. (Annelida) and its significance for metazoan phylogeny. Zoomorphologie 1980, 95, 41–84. [Google Scholar] [CrossRef]
- Rieger, R.M. Fine structure of the body wall, nervous system, and digestive tract in the Lobatocerebridae Rieger and the organization of the gliointerstitial system in Annelida. J. Morphol. 1981, 167, 139–165. [Google Scholar] [CrossRef]
- Filippova, A.V.; Purschke, G.; Tzetlin, A.B.; Müller, M.C.M. Musculature in polychaetes: Comparison of Myrianida prolifera (Syllidae) and Sphaerodoropsis sp. (Sphaerodoridae). Invertebr. Biol. 2010, 129, 184–198. [Google Scholar] [CrossRef]
- Law, C.J.; Dorgan, K.M.; Rouse, G.W. Relating divergence in polychaete musculature to different burrowing behaviors: A study using opheliidae (Annelida). J. Morphol. 2014, 275, 548–571. [Google Scholar] [CrossRef] [PubMed]
- Kerbl, A.; Bekkouche, N.; Sterrer, W.; Worsaae, K. Detailed reconstruction of the nervous and muscular system of Lobatocerebridae with an evaluation of its annelid affinity. BMC Evol. Biol. 2015, 15, 277. [Google Scholar] [CrossRef]
- Kocot, K.M. On 20 years of Lophotrochozoa. Org. Divers. Evol. 2016, 16, 329–343. [Google Scholar] [CrossRef]
- Storh, V. Integument. In The Ultrastructure of Polychaeta. Microfauna Marina; Westheide, W., Hermans, C.O., Eds.; VCH Publishers, Inc.: Hoboken, NJ, USA; Gustav Fischer Verlag: New York, NY, USA; Stuttgart, Germany, 1988; Volume 4, pp. 13–36. [Google Scholar]
- Slyusarev, G.S. Fine structure and development of the cuticle of Intoshia variabili (Orthonectida). Acta Zool. 2000, 81, 1–8. [Google Scholar] [CrossRef]
- Slyusarev, G.S.; Manylov, O.G. General morphology of muscle system in the female orthonectid, Intoshia variabili (Orthonectida). Cah. Biol. Mar. 2001, 42, 239–242. [Google Scholar]
- Slyusarev, G.S. Phylum Orthonectida: Morphology, biology, and relationships to other multicellular animals. Zh. Obshch. Biol. 2008, 69, 403–427. [Google Scholar]
- Ruppert, E.E.; Fox, R.S.; Barnes, R.D. Invertebrate Zoology, A Functional Evolutionary Approach, 7th ed.; Thomson-Brooks/Cole: Belmont, CA, USA, 2004. [Google Scholar]
- Borisanova, A.O.; Yushin, V.V.; Malakhov, V.V.; Temereva, E.N. The fine structure of the cuticle of kamptozoans is similar to that of annelids. Zoomorphology 2015, 134, 165–181. [Google Scholar] [CrossRef]
- Scherholz, M.; Redl, E.; Wollesen, T.; Todt, C.; Wanninger, A. From complex to simple: Myogenesis in an aplacophoran mollusk reveals key traits in aculiferan evolution. BMC Evol. Biol. 2015, 15, 201. [Google Scholar] [CrossRef]
- Temereva, E.N.; Malakhov, V.V. The evidence of metamery in adult brachiopods and phoronids. Invertebr. Zool. 2011, 8, 91–112. [Google Scholar] [CrossRef]
- Schuchert, P.; Rieger, R.M. Ultrastructural observations on the dwarf male of Bonellia viridis (Echiura). Acta Zool. 1990, 71, 5–16. [Google Scholar] [CrossRef]
- Kerbl, A.; Fofanova, E.G.; Mayorova, T.D.; Voronezhskaya, E.E.; Worsaae, K. Comparison of neuromuscular development in two dinophilid species (Annelida) suggests progenetic origin of Dinophilus gyrociliatus. Front. Zool. 2016, 13, 49. [Google Scholar] [CrossRef]
- Worsaae, K.; Rouse, G.W. The simplicity of males: Dwarf males of four species of Osedax (Siboglinidae; Annelida) investigated by confocal laser scanning microscopy. J. Morphol. 2010, 271, 127–142. [Google Scholar] [CrossRef]
- Vortsepneva, E.; Tzetlin, A.; Purschke, G.; Mugue, N.; Haß-Cordes, E.; Zhadan, A. The parasitic polychaete known as Asetocalamyzas laonicola (Calamyzidae) is in fact the dwarf male of the spionid Scolelepis laonicola (comb. nov.). Invertebr. Biol. 2008, 127, 403–416. [Google Scholar] [CrossRef]
- Slyusarev, G.S.; Miller, D.M. Fine structure of the mature Plasmodium of Intoshia variabili (Phylum Orthonectida), a parasite of the platyhelminth Macrorhynchus crocea. Acta Zool. 1998, 79, 319–327. [Google Scholar] [CrossRef]
- Slyusarev, G.S.; Ferraguti, M. Sperm structure of Rhopalura littoralis (Orthonectida). Invertebr. Biol. 2002, 121, 91–94. [Google Scholar] [CrossRef]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Putz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef]
- Abeel, T.; van Parys, T.; Saeys, Y.; Galagan, J.; van de Peer, Y. GenomeView: A next-generation genome browser. Nucleic Acids Res. 2012, 40, e12. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS_MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Aoyama, H.; Muramoto, K.; Shinzawa-Itoh, K.; Hirata, K.; Yamashita, E.; Tsukihara, T.; Ogura, T.; Yoshikawa, S. A peroxide bridge between Fe and Cu ions in the O2 reduction site of fully oxidized cytochrome c oxidase could suppress the proton pump. Proc. Nat. Acad. Sci. USA 2009, 106, 2165–2169. [Google Scholar] [CrossRef]
- Spikes, T.E.; Montgomery, M.G.; Walker, J.E. Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes. Proc. Nat. Acad. Sci. USA 2021, 118, e2021012118. [Google Scholar] [CrossRef]
- Hong, D.W.; Leung, S.C.; Amporndanai, K.; Davies, J.; Priestley, R.S.; Nixon, G.L.; Berry, N.G.; Samar Hasnain, S.; Antonyuk, S.; Ward, S.A.; et al. Potent Antimalarial 2-Pyrazolyl Quinolone bc1 (Qi) Inhibitors with Improved Drug-like Properties. ACS Med. Chem. Lett. 2018, 12, 1205–1210. [Google Scholar] [CrossRef]
- Agip, A.N.A.; Blaza, J.N.; Bridges, H.R.; Viscomi, C.; Rawson, S.; Muench, S.P.; Hirst, J. Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol. 2018, 25, 548–556. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Tsuji, J.; Fu, S.C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom. 2015, 14, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewick, J.; Yamada, K.D. AFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]

| Feature | Gene | Amino Acid Motif 1 | Occurrence Among Annelida, % | Occurrence Outside Annelida, % |
|---|---|---|---|---|
| 1 | atp6 | DGAPD-W-LNPFL | 83 | 0.6 |
| 2 | atp6 | PLTLS-F-RLAAN | 87 | 0.7 |
| 3 | cox1 | TAHAF-L-MIFFL | 63 | 3.0 |
| 4 | cox1 | LSFVA-L-MLFIF | 83 | 3.0 |
| 5 | cox1 | DPILP-L-DFHNL | 81 | 1.3 |
| 6 | cox3 | TPEIG-C-SWPPT | 97 | 1.4 |
| 7 | cox3 | VDVVW-I-CLYLC | 92 | 2.7 |
| 8 | cytb | EWYFL-W-MYAIL | 94 | 0.1 |
| 9 | nad6 | VMFAY-F-LALTP | 99 | 1.1 |
| Taxon | Protein, Amino Acid Motif 1 and Occurence, % | |||
|---|---|---|---|---|
| COX1 | CYTB | ATP6 | NAD6 | |
| LSNMA-L-TLFMW | DWFLL-W-AYAIL | PLTLS-F-RICAI | SIYIF-F-ISSGG | |
| Annelida | 83 | 94 | 87 | 99 |
| Basal branched annelids (Owenia + Magelona + Chaetopteridae) | 0 | 0 | 0 | 33 |
| Amphinomidae + Sipuncula | 100 | 100 | 86 | 100 |
| Crown annelids (Pleistoannelida) | 94 | 97 | 96 | 100 |
| Metazoa without Annelida | 2.7 | 0.1 | 0.7 | 2.5 |
| Number of Synapomorphies (n) | Average Probability of Independent Emergence of n Synapomorphies |
|---|---|
| 1 | 0.014 |
| 2 | 2.2 × 10−4 |
| 3 | 3.13 × 10−6 |
| 4 | 4.13 × 10−8 |
| 5 | 4.95 × 10−10 |
| 6 | 5.23 × 10−12 |
| 7 | 4.87 × 10−14 |
| 8 | 3.78 × 10−16 |
| 9 | 2.04 × 10−18 |
| Taxon | Number of Species | Number of Taxon Synapomorphies (n) | Number of Taxon Synapomorphies in Orthonectids (i) |
|---|---|---|---|
| Annelida | 157 | 9 | 4 |
| Clitellata | 65 | 12 | 3 |
| Hirudinea | 29 | 45 | 3 |
| Siboglinidae | 18 | 102 | 4 |
| Taxon | Probability of Emergence | Probability of Loss |
|---|---|---|
| Annelida | 1.9 × 10−5 | 0.33 |
| Clitellata | 0.0012 | 0.015 |
| Hirudinea | 0.014 | 2.3 × 10−6 |
| Siboglinidae | 0.017 | 3.3 × 10−11 |
| Feature | Gene | Amino Acid Motif 1 | Occurrence Outside Mesozoa, % |
|---|---|---|---|
| 1 | atp6 | ILLFL-Y-DVMVC | 1.4 |
| 2 | cox1 | LITAH-G-LIMIF | 2.5 |
| 3 | cox1 | YLFSS-S-YSVDF | 2 |
| 4 | cox1 | ISSIN-S-SINFF | 2 |
| 5 | cox1 | HPEVY-V-LILPG | 2 |
| 6 | cox1 | LFSQM-S-MIYAM | 2.4 |
| 7 | cox1 | MIYAM-G-MIMFL | 0.3 |
| 8 | cox1 | SMGAV-Y-LILGS | 0.4 |
| 9 | cox1 | LLTLG-S-NMCFL | 0.04 |
| 10 | cox2 | VLPYN-K-MCSLM | 0.5 |
| 11 | cox3 | MLFFI-F-SEIMF | 2.3 |
| 12 | cytb | MLGLF-M-FLQTI | 1.1 |
| 13 | cytb | FYIQN-E-IFFGW | 2.4 |
| 14 | cytb | IGVSF-I-FILIY | 0.8 |
| 15 | cytb | MHMFR-S-IYFKL | 1.3 |
| 16 | cytb | LWFSG-M-LMFLL | 0.8 |
| 17 | cytb | LIIII-S-FLGYS | 0.7 |
| 18 | cytb | AKVIT-S-LFTII | 3 |
| 19 | cytb | LILWG-D-FTVAG | 0.3 |
| 20 | cytb | LWGDF-T-VAGPT | 1 |
| 21 | cytb | GSNNK-F-GLKNT | 0.1 |
| 22 | cytb | DSFME-S-NKLVT | 1 |
| 23 | cytb | MILLS-Y-CGGAI | 1.9 |
| 24 | nd1 | FSFMT-I-LMSFY | 2.4 |
| 25 | nd1 | FMMMI-S-FFSKS | 0.9 |
| 26 | nd1 | MFWNF-L-LPIIL | 2.2 |
| 27 | nd2 | NGFSS-L-FLFLS | 2.4 |
| 28 | nd3 | SLKYF-K-IIMLF | 0.1 |
| 29 | nd4 | LAHVE-S-PTEGS | 0.5 |
| 30 | nd5 | AFFKS-S-LFLSF | 0.4 |
| 31 | nd5 | FSKEK-M-FHMGY | 0.9 |
| Groups | Expected Number of Features | Observed Number of Features | Observed/ Expected Ratio |
|---|---|---|---|
| Orthonectida+Dicyemida | 7 | 22 | 3.1 |
| Orthonectida+Neodermata | 5 | 12 | 2.4 |
| Dicyemida+Neodermata | 6 | 15 | 2.5 |
| Feature | Gene | Amino Acid Motif 1 | Mesozoan Groups with Feature | Occurrence Outside Mesozoa, % |
|---|---|---|---|---|
| 1 | cox1 | SSYPF-S-SSYSM | Orthonectids | 1.2 |
| 2 | cox1 | FTFGG-F-TGLYL | Orthonectids | 0.3 |
| 3 | cox2 | YVLLE-S-CIIET | Both | 0.9 |
| 4 | cox2 | VKLIA-N-QWFWT | Both | 0.7 |
| 5 | cytb | YSIHS-V-GVSII | Dicyemids | 0.1 |
| 6 | nd1 | SEYIL-M-SIMAI | Orthonectids | 2.3 |
| 7 | nd3 | FLLFN-F-LFLGL | Both | 2.2 |
| 8 | nd3 | IFGLY-Y-ELGWG | Both | 0.1 |
| 9 | nd4 | LFFYF-K-DMSMI | Orthonectids | 0.6 |
| 10 | nd5 | IALST-M-NHLSI | Both | 0.6 |
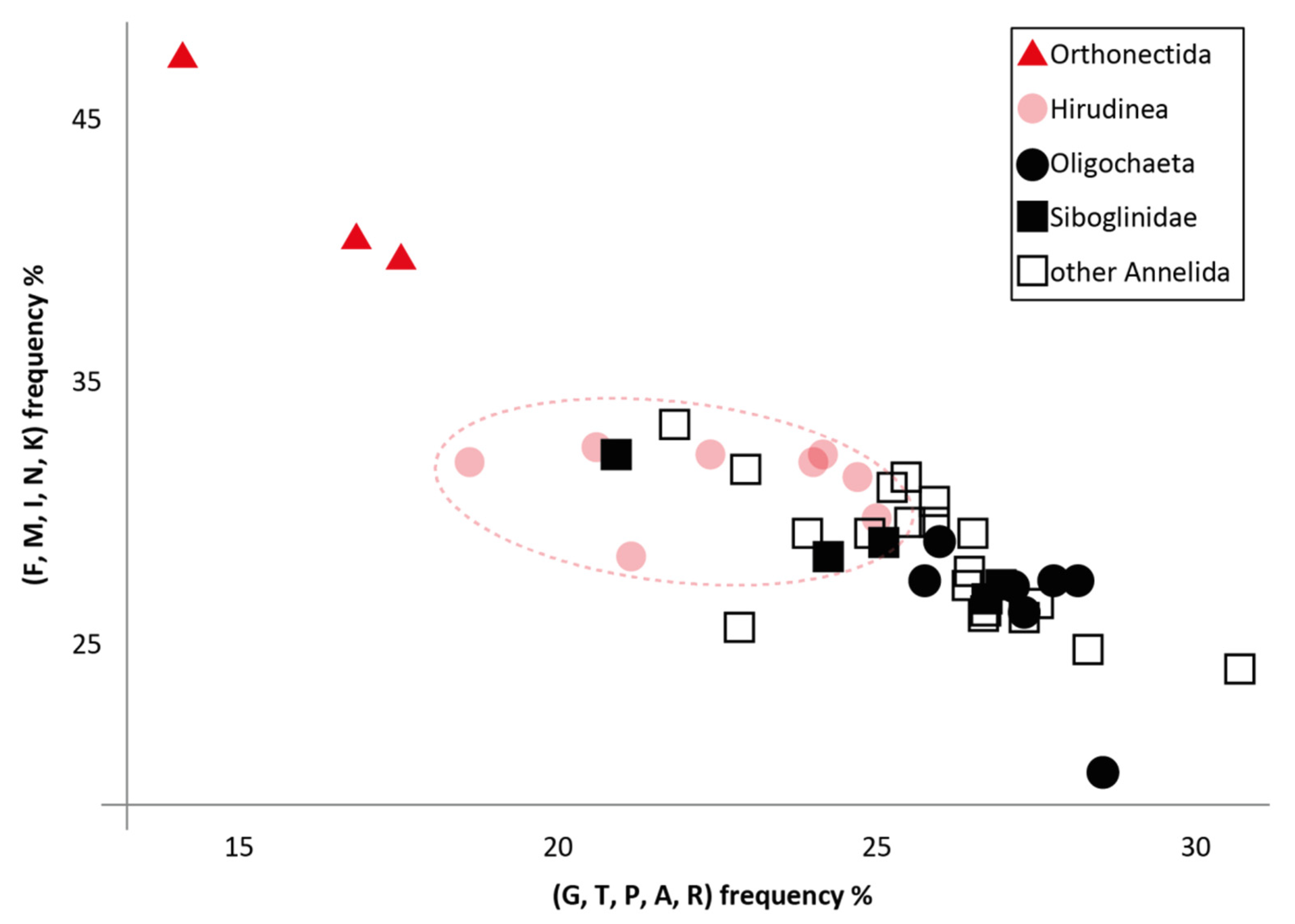
| Taxon | Average Distance from Common Ancestor | Ratio of Evolution Rates for Orthonectids and Individual Taxa |
|---|---|---|
| Annelida | 0.87 | 18.83 |
| Clitellata | 0.44 | 37.23 |
| Hirudinea | 0.65 | 25.2 |
| Siboglinidae | 0.31 | 52.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikolaeva, O.V.; Mikhailov, K.V.; Muntyan, M.S.; Zverkov, O.A.; Spirin, S.A.; Lyubetsky, V.A.; Slyusarev, G.S.; Aleoshin, V.V. Rare Evolutionary Events Support the Phylogenetic Placement of Orthonectida Within Annelida. Int. J. Mol. Sci. 2025, 26, 5983. https://doi.org/10.3390/ijms26135983
Nikolaeva OV, Mikhailov KV, Muntyan MS, Zverkov OA, Spirin SA, Lyubetsky VA, Slyusarev GS, Aleoshin VV. Rare Evolutionary Events Support the Phylogenetic Placement of Orthonectida Within Annelida. International Journal of Molecular Sciences. 2025; 26(13):5983. https://doi.org/10.3390/ijms26135983
Chicago/Turabian StyleNikolaeva, Olga V., Kirill V. Mikhailov, Maria S. Muntyan, Oleg A. Zverkov, Sergey A. Spirin, Vassily A. Lyubetsky, Georgy S. Slyusarev, and Vladimir V. Aleoshin. 2025. "Rare Evolutionary Events Support the Phylogenetic Placement of Orthonectida Within Annelida" International Journal of Molecular Sciences 26, no. 13: 5983. https://doi.org/10.3390/ijms26135983
APA StyleNikolaeva, O. V., Mikhailov, K. V., Muntyan, M. S., Zverkov, O. A., Spirin, S. A., Lyubetsky, V. A., Slyusarev, G. S., & Aleoshin, V. V. (2025). Rare Evolutionary Events Support the Phylogenetic Placement of Orthonectida Within Annelida. International Journal of Molecular Sciences, 26(13), 5983. https://doi.org/10.3390/ijms26135983







