The Involvement of LvSRSF2 in Circular RNA Biogenesis and Its Role in Immunity Against White Spot Syndrome Virus (WSSV) in Litopenaeus vannamei
Abstract
1. Introduction
2. Results
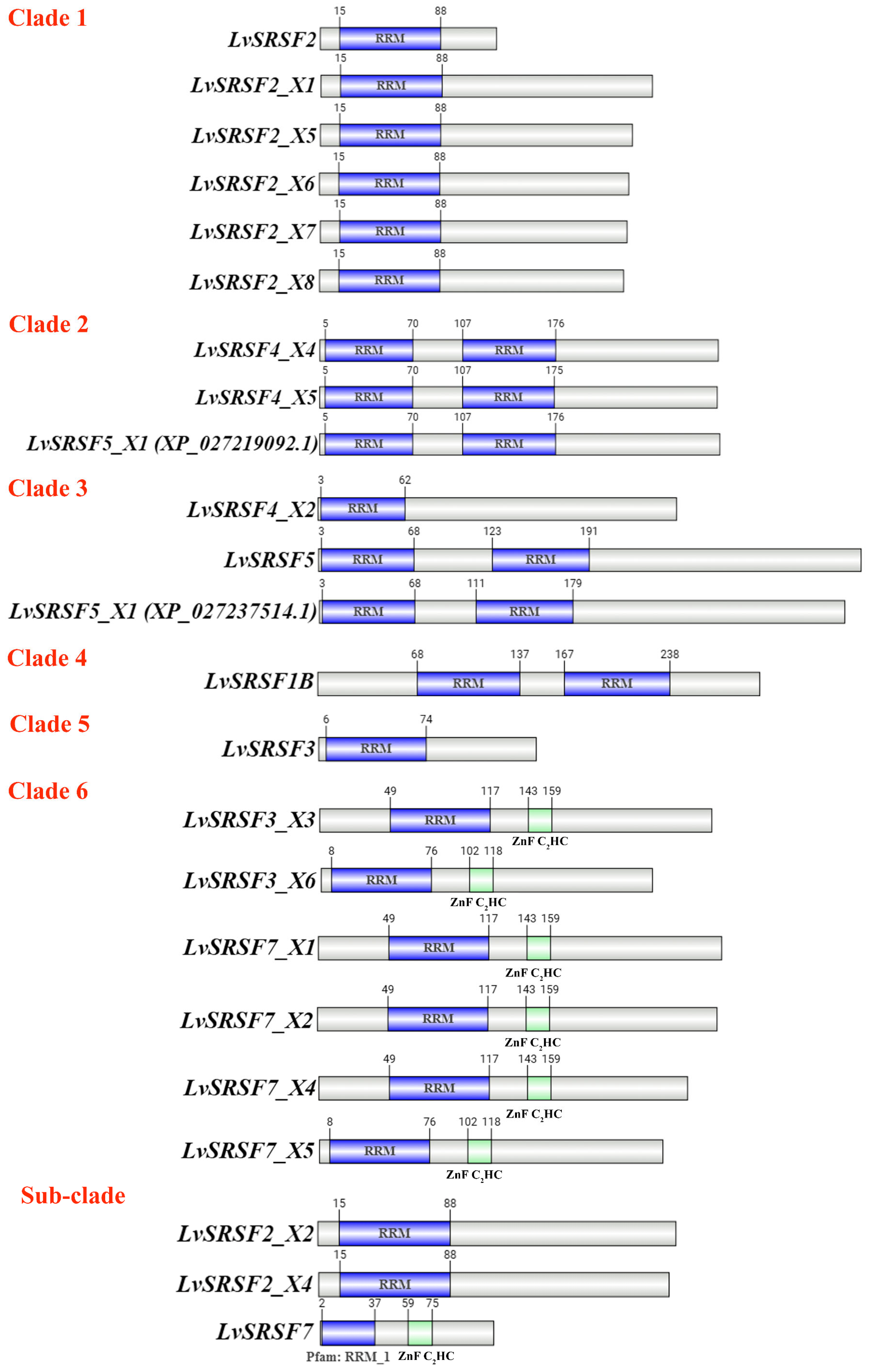
2.1. Sequence and Phylogenetic Analysis of LvSRSFs
2.2. Tissue Distribution Analysis of LvSRSF Genes
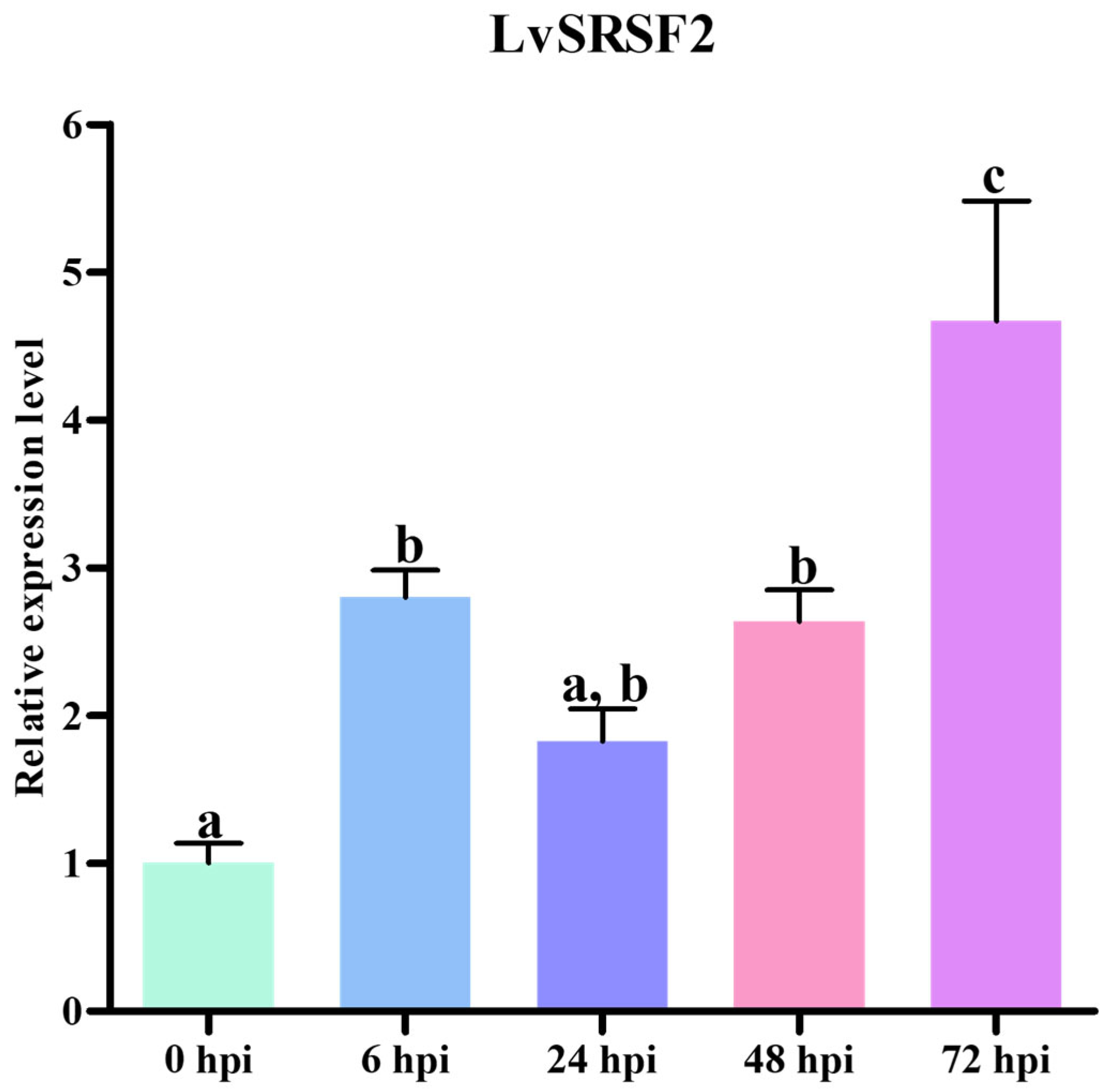
2.3. LvSRSF2 Expression Levels During WSSV Infection
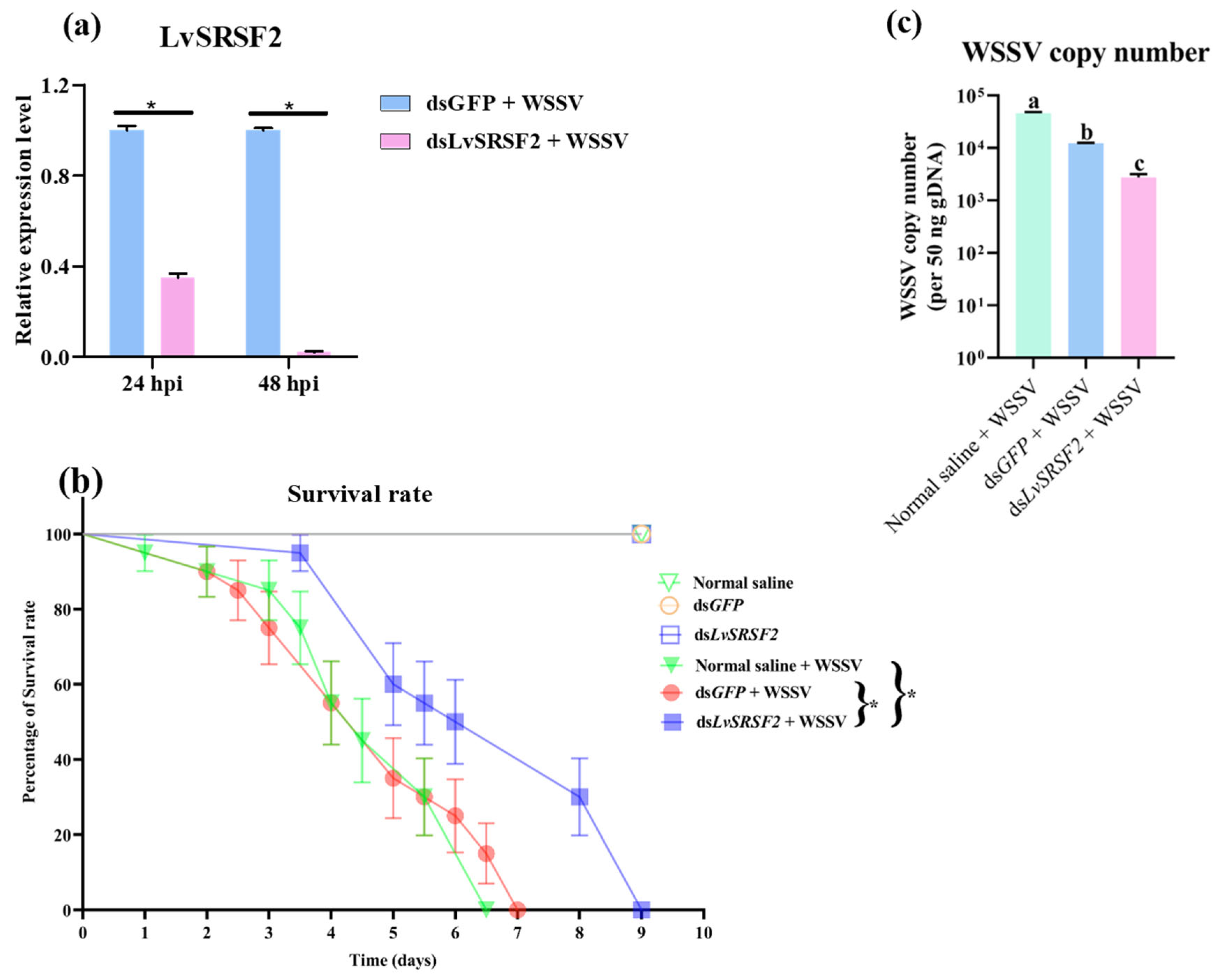
2.4. Effects of LvSRSF2 Knockdown on circRNA and liRNA Expression
2.5. Effects of LvSRSF2 Knockdown on WSSV Copy Number and Survival Rate Following WSSV Infection
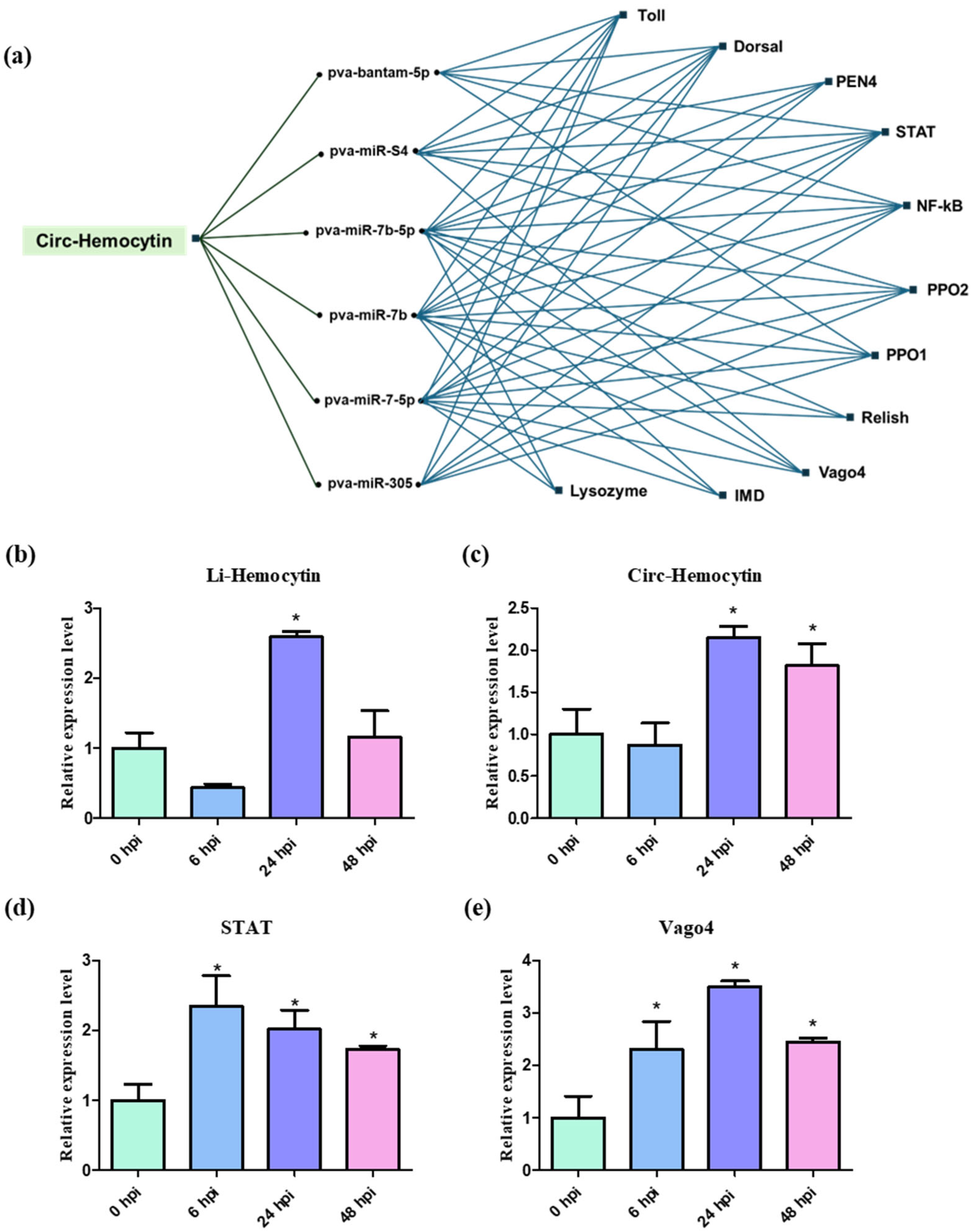
2.6. miRNA–circRNA Networks and Expression of Immune-Related circRNAs and Linear Counterparts
2.7. Effects of siRNA Knockdown on circRNA and WSSV Copy Number
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Shrimp Samples
4.3. Phylogenetic Tree and Sequence Study of LvSRSFs
4.4. Organ Distribution Analysis of LvSRSF Genes
4.5. LvSRSF Response Following WSSV Infection Using qPCR
4.6. Double-Stranded RNA (dsRNA) Preparation
4.7. Knockdown of LvSRSF2 Expression In Vivo by Double-Stranded RNA
4.8. Mortality Assays and Quantification of WSSV Copy Number Following LvSRSF2 Knockdown and WSSV Infection
4.9. The miRNAs-circRNAs Network and the Expression of circRNAs and Their Immune Response to WSSV Infection
4.10. siRNA-Induced circRNA Silencing
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| °C | Degree Celsius |
| µg | Microgram |
| µL | Microliter |
| AMPs | Antimicrobial peptides |
| bp | Base pair |
| cDNA | Complementary DNA |
| circ-Alpha2 | Circular RNA of Alpha2 |
| circ-Anillin | Circular RNA of Anillin |
| circ-Calpain | Circular RNA of Calpain |
| circ-Hemocytin | Circular RNA of Hemocytin |
| circ-Nephrin | Circular RNA of Nephrin |
| circPLCE1 | Circular phospholipase C epsilon 1 |
| circRNAs | Circular RNAs |
| circ-Toll | Circular RNA of Toll |
| cm | Centimeter |
| d | Days |
| DECs | Differentially expressed circRNAs |
| DEPC | Diethyl pyrocarbonate |
| DI water | Deionized water |
| DNA | Deoxyribonucleic acid |
| dsGFP | dsRNA that specifically targeted the green fluorescent protein |
| dsLvSRSF2 | dsRNA that specifically targeted the Serine/arginine splicing factor isoform 2 of L. vannamei |
| dsRNAs | Double-stranded RNA |
| EDTA | Ethylenediaminetetraacetic acid |
| EF-1α | Elongation factor 1-alpha |
| g | Gram |
| h | Hours |
| HBV | Hepatitis B virus |
| HIV-1 | Human immunodeficiency virus 1 |
| hpi | Hours post injection |
| HPV-16 | Human papillomavirus |
| HSV-1 | Herpes simplex virus type 1 |
| ICP0 | Infected cell polypeptide 0 |
| ICP27 | Infected cell polypeptide 27 |
| IPTG | Isopropyl β-D-thiogalactoside |
| IRF | Interferon regulatory factor |
| JTT | The Jones–Taylor–Thornton |
| li-Alpha2 | Linear RNA of Alpha2 |
| li-Anillin | Linear RNA of Anillin |
| li-Calpain | Linear RNA of Calpain |
| li-Hemocytin | Linear RNA of Hemocytin |
| li-Nephrin | Linear RNA of Nephrin |
| liRNAs | Linear RNAs |
| li-Toll | Linear RNA of Toll |
| LvSRSF1B | Serine/arginine splicing factor isoform 1B of L. vannamei |
| LvSRSF2 | Serine/arginine splicing factor isoform 2 of L. vannamei |
| LvSRSF2 X1 | Serine/arginine splicing factor isoform 2 X1 of L. vannamei |
| LvSRSF2 X2 | Serine/arginine splicing factor isoform 2 X2. of L. vannamei |
| LvSRSF2 X4 | Serine/arginine splicing factor isoform 2 X4 of L. vannamei |
| LvSRSF2 X5 | Serine/arginine splicing factor isoform 2 X5 of L. vannamei |
| LvSRSF2 X6 | Serine/arginine splicing factor isoform 2 X6 of L. vannamei |
| LvSRSF2 X7 | Serine/arginine splicing factor isoform 2 X7 of L. vannamei |
| LvSRSF2 X8 | Serine/arginine splicing factor isoform 2 X8 of L. vannamei |
| LvSRSF3 X1 | Serine/arginine splicing factor isoform 3 X1 of L. vannamei |
| LvSRSF3 X3 | Serine/arginine splicing factor isoform 3 X3 of L. vannamei |
| LvSRSF3 X6 | Serine/arginine splicing factor isoform 3 X6 of L. vannamei |
| LvSRSF4 X1 | Serine/arginine splicing factor isoform 4 X1 of L. vannamei |
| LvSRSF4 X2 | Serine/arginine splicing factor isoform 4 X2 of L. vannamei |
| LvSRSF4 X4 | Serine/arginine splicing factor isoform 4 X4 of L. vannamei |
| LvSRSF4 X5 | Serine/arginine splicing factor isoform 4 X5 of L. vannamei |
| LvSRSF5 | Serine/arginine splicing factor isoform 5 of L. vannamei |
| LvSRSF5 X1 | Serine/arginine splicing factor isoform 5 X1 of L. vannamei |
| LvSRSF5 X2 | Serine/arginine splicing factor isoform 5 X2 of L. vannamei |
| LvSRSF7 | Serine/arginine splicing factor isoform 7 of L. vannamei |
| LvSRSF7 X1 | Serine/arginine splicing factor isoform 7 X1 of L. vannamei |
| LvSRSF7 X2 | Serine/arginine splicing factor isoform 7 X2 of L. vannamei |
| LvSRSF7 X3 | Serine/arginine splicing factor isoform 7 X3 of L. vannamei |
| LvSRSF7 X4 | Serine/arginine splicing factor isoform 7 X4 of L. vannamei |
| LvSRSF7 X5 | Serine/arginine splicing factor isoform 7 X5 of L. vannamei |
| LvSRSF7 X6 | Serine/arginine splicing factor isoform 7 X6 of L. vannamei |
| LvSRSFs | Serine/arginine splicing factors of L. vannamei |
| M | Molar |
| min | Minutes |
| miRNAs | MicroRNAs |
| mL | Milliliter |
| mM | Micromolar |
| NaCl | Sodium chloride |
| ncRNAs | Non-coding RNAs |
| NJ | The neighbor-joining |
| nm | Nanometer |
| O.D. | Optical density |
| PAMPs | Pathogen-associated molecular patterns |
| PBS | Phosphate-buffered saline |
| PCR | Polymerase chain reaction |
| ppt | Parts per thousand |
| PRRs | Pattern recognition receptors |
| qPCR | Quantitative real-time PCR |
| qRT-PCR | Quantitative reverse transcription PCR |
| RBPs | RNA-binding proteins |
| RNA | Ribonucleic acid |
| RNF125 | Ring Finger Protein 125 |
| rpm | Revolutions per minute |
| RRM | RNA recognition motif |
| s | Seconds |
| SD | Standard deviations |
| SE | Standard error |
| siRNAs | Small interfering RNAs |
| SRSF1 | Serine/arginine splicing factor isoform 1 |
| SRSF10 | Serine/arginine splicing factor isoform 10 |
| SRSF2 | Serine/arginine splicing factor isoform 2 |
| SRSF3 | Serine/arginine splicing factor isoform 3 |
| SRSF5 | Serine/arginine splicing factor isoform 5 |
| SRSF9 | Serine/arginine splicing factor isoform 9 |
| SRSFs | Serine/arginine splicing factors |
| TLRs | Toll-like receptors |
| U | Unit |
| UV | Ultraviolet |
| Vif | HIV-1 viral infectivity factor |
| w/v | Weight per volume |
| WSD | White spot disease |
| WSSV | White spot syndrome virus |
| YHV | Yellow head virus |
References
- Millard, R.; Dong, X. White spot syndrome virus and disease. Aquac. Pathophysiol. 2022, 2, 103–115. [Google Scholar]
- van Hulten, M.C.; Vlak, J.M. Identification and phylogeny of a protein kinase gene of white spot syndrome virus. Virus Genes 2001, 22, 201–207. [Google Scholar] [CrossRef]
- Kulkarni, A.; Krishnan, S.; Anand, D.; Kokkattunivarthil Uthaman, S.; Otta, S.K.; Karunasagar, I.; Kooloth Valappil, R. Immune responses and immunoprotection in crustaceans with special reference to shrimp. Rev. Aquac. 2021, 13, 431–459. [Google Scholar] [CrossRef]
- Limkul, S.; Phiwthong, T.; Massu, A.; Boonanuntanasarn, S.; Teaumroong, N.; Somboonwiwat, K.; Boonchuen, P. Transcriptome-based insights into the regulatory role of immune-responsive circular RNAs in Litopanaeus vannamei upon WSSV infection. Fish Shellfish Immunol. 2023, 132, 108499. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-coding RNAs and their integrated networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, C.; Wang, X.; Shan, G. Circular RNAs in eukaryotic cells. Curr. Genom. 2015, 16, 312–318. [Google Scholar] [CrossRef]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef]
- Chen, J.; Chan, A.W.; To, K.F.; Chen, W.; Zhang, Z.; Ren, J.; Ko, B.C. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3β/β-catenin signaling. Hepatology 2013, 57, 2287–2298. [Google Scholar] [CrossRef]
- Guo, H.; Zhou, T.; Jiang, D.; Cuconati, A.; Xiao, G.H.; Block, T.M.; Guo, J.T. Regulation of hepatitis B virus replication by the phosphatidylinositol 3-kinase-akt signal transduction pathway. J. Virol. 2007, 81, 10072–10080. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, L.; Zhang, Y.; Li, H. Circ-ATP5H induces hepatitis B virus replication and expression by regulating miR-138-5p/TNFAIP3 axis. Cancer Manag. Res. 2020, 12, 11031–11040. [Google Scholar] [CrossRef]
- Wang, M.; Gu, B.; Yao, G.; Li, P.; Wang, K. Circular RNA expression profiles and the pro-tumorigenic function of circRNA_10156 in hepatitis B virus-related liver cancer. Int. J. Med. Sci. 2020, 17, 1351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Z. Circular RNA hsa_circ_0004812 impairs IFN-induced immune response by sponging miR-1287-5p to regulate FSTL1 in chronic hepatitis B. Virol. J. 2020, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Massu, A.; Mahanil, K.; Limkul, S.; Phiwthong, T.; Boonanuntanasarn, S.; Teaumroong, N.; Somboonwiwat, K.; Boonchuen, P. Identification of immune-responsive circular RNAs in shrimp (Litopenaeus vannamei) upon yellow head virus infection. Fish Shellfish Immunol. 2024, 144, 109246. [Google Scholar] [CrossRef] [PubMed]
- Gianazza, E.; Eberini, I.; Palazzolo, L.; Miller, I. Hemolymph proteins: An overview across marine arthropods and molluscs. J. Proteom. 2021, 245, 104294. [Google Scholar] [CrossRef]
- Ma, H.; Wang, B.; Zhang, J.; Li, F.; Xiang, J. Multiple forms of alpha-2 macroglobulin in shrimp Fenneropenaeus chinesis and their transcriptional response to WSSV or Vibrio pathogen infection. Dev. Comp. Immunol. 2010, 34, 677–684. [Google Scholar] [CrossRef]
- Field, C.M.; Alberts, B.M. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 1995, 131, 165–178. [Google Scholar] [CrossRef]
- Wang, P.; Qian, W.; Wang, W.; Guo, M.; Xia, Q.; Cheng, D. Identification and Characterization of the Anillin Gene in Silkworm. DNA Cell Biol. 2019, 38, 532–540. [Google Scholar] [CrossRef]
- Arai, I.; Ohta, M.; Suzuki, A.; Tanaka, S.; Yoshizawa, Y.; Sato, R. Immunohistochemical analysis of the role of hemocytin in nodule formation in the larvae of the silkworm, Bombyx mori. J. Insect Sci. 2013, 13, 125. [Google Scholar] [CrossRef]
- Kotani, E.; Yamakawa, M.; Iwamoto, S.-I.; Tashiro, M.; Mori, H.; Sumida, M.; Matsubara, F.; Taniai, K.; Kadono-Okuda, K.; Kato, Y.; et al. Cloning and expression of the gene of hemocytin, an insect humoral lectin which is homologous with the mammalian von Willebrand factor. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1995, 1260, 245–258. [Google Scholar] [CrossRef]
- Ni, W.; Bao, J.; Mo, B.; Liu, L.; Li, T.; Pan, G.; Chen, J.; Zhou, Z. Hemocytin facilitates host immune responses against Nosema bombycis. Dev. Comp. Immunol. 2020, 103, 103495. [Google Scholar] [CrossRef]
- Fukasawa, H.; Bornheimer, S.; Kudlicka, K.; Farquhar, M.G. Slit diaphragms contain tight junction proteins. J. Am. Soc. Nephrol. 2009, 20, 1491–1503. [Google Scholar] [CrossRef]
- Pavenstadt, H.; Kriz, W.; Kretzler, M. Cell biology of the glomerular podocyte. Physiol. Rev. 2003, 83, 253–307. [Google Scholar] [CrossRef] [PubMed]
- Khodabandeh, S.; Charmantier, G.; Charmantier-Daures, M. Ultrastructural studies and Na+, K+-ATPase immunolocalization in the antennal urinary glands of the lobster Homarus gammarus (Crustacea, Decapoda). J. Histochem. Cytochem. 2005, 53, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Armelloni, S.; Edefonti, A.; Messa, P.; Rastaldi, M.P. Fifteen years of research on nephrin: What we still need to know. Nephrol. Dial. Transplant. 2013, 28, 767–770. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Chtarbanova, S.; Imler, J.L. Microbial sensing by Toll receptors: A historical perspective. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 1734–1738. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.H.; Liang, J.P.; Gu, Z.H.; Wan, D.H.; Weng, S.P.; Yu, X.Q.; He, J.G. Molecular cloning, characterization and expression analysis of two novel Tolls (LvToll2 and LvToll3) and three putative Spätzle-like Toll ligands (LvSpz1–3) from Litopenaeus vannamei. Dev. Comp. Immunol. 2012, 36, 359–371. [Google Scholar] [CrossRef]
- Feng, J.; Zhao, L.; Jin, M.; Li, T.; Wu, L.; Chen, Y.; Ren, Q. Toll receptor response to white spot syndrome virus challenge in giant freshwater prawns (Macrobrachium rosenbergii). Fish Shellfish. Immunol. 2016, 57, 148–159. [Google Scholar] [CrossRef]
- Huang, Y.; Li, T.; Jin, M.; Yin, S.; Hui, K.M.; Ren, Q. Newly identified PcToll4 regulates antimicrobial peptide expression in intestine of red swamp crayfish Procambarus clarkii. Gene 2017, 610, 140–147. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chen, X.X.; Lin, F.Y.; Chen, Q.F.; Ma, X.Y.; Liu, H.P. CqToll participates in antiviral response against white spot syndrome virus via induction of anti-lipopolysaccharide factor in red claw crayfish Cherax quadricarinatus. Dev. Comp. Immunol. 2017, 74, 217–226. [Google Scholar] [CrossRef]
- Deepika, A.; Sreedharan, K.; Paria, A.; Makesh, M.; Rajendran, K.V. Toll-pathway in tiger shrimp (Penaeus monodon) responds to white spot syndrome virus infection: Evidence through molecular characterisation and expression profiles of MyD88, TRAF6 and TLR genes. Fish Shellfish Immunol. 2014, 41, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Guanzon, D.A.V.; Maningas, M.B.B. Functional elucidation of LvToll 3 receptor from P. vannamei through RNA interference and its potential role in the shrimp antiviral response. Dev. Comp. Immunol. 2018, 84, 172–180. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, H.; Yang, L.; Li, X.; Wang, Z. CircPLCE1 facilitates the malignant progression of colorectal cancer by repressing the SRSF2-dependent PLCE1 pre-RNA splicing. J. Cell. Mol. Med. 2021, 25, 7244–7256. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Johansson, C.; Cardoso Palacios, C.; Mossberg, A.; Dhanjal, S.; Bergvall, M.; Schwartz, S. Eight nucleotide substitutions inhibit splicing to HPV-16 3′-splice site SA3358 and reduce the efficiency by which HPV-16 increases the life span of primary human keratinocytes. PLoS ONE 2013, 8, e72776. [Google Scholar] [CrossRef]
- Mole, S.; Faizo, A.A.A.; Hernandez-Lopez, H.; Griffiths, M.; Stevenson, A.; Roberts, S.; Graham, S.V. Human papillomavirus type 16 infection activates the host serine arginine protein kinase 1 (SRPK1)–splicing factor axis. J. Gen. Virol. 2020, 101, 523–532. [Google Scholar] [CrossRef]
- Loerch, S.; Kielkopf, C.L. Dividing and Conquering the RNA Recognition Motif Family: A Representative Case Based on hnRNP L. J. Mol. Biol. 2015, 427, 2997. [Google Scholar] [CrossRef] [PubMed]
- Auweter, S.D.; Oberstrass, F.C.; Allain, F.H.T. Solving the structure of PTB in complex with pyrimidine tracts: An NMR study of protein–RNA complexes of weak affinities. J. Mol. Biol. 2007, 367, 174–186. [Google Scholar] [CrossRef]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503. [Google Scholar] [CrossRef]
- Sawicka, K.; Bushell, M.; Spriggs, K.A.; Willis, A.E. Polypyrimidine-tract-binding protein: A multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008, 36, 641–647. [Google Scholar] [CrossRef]
- Cáceres, J.F.; Misteli, T.; Screaton, G.R.; Spector, D.L.; Krainer, A.R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997, 138, 225–238. [Google Scholar] [CrossRef]
- Cléry, A.; Krepl, M.; Nguyen, C.K.X.; Moursy, A.; Jorjani, H.; Katsantoni, M.; Okoniewski, M.; Mittal, N.; Zavolan, M.; Sponer, J.; et al. Structure of SRSF1 RRM1 bound to RNA reveals an unexpected bimodal mode of interaction and explains its involvement in SMN1 exon7 splicing. Nat. Commun. 2021, 12, 428. [Google Scholar] [CrossRef] [PubMed]
- De Silva, N.I.U.; Fargason, T.; Zhang, Z.; Wang, T.; Zhang, J. Inter-domain flexibility of human Ser/Arg-rich splicing factor 1 allows variable spacer length in cognate RNA’s bipartite motifs. Biochemistry 2022, 61, 2922–2932. [Google Scholar] [CrossRef] [PubMed]
- De Silva, N.I.U.; Lehman, N.; Fargason, T.; Paul, T.; Zhang, Z.; Zhang, J. Unearthing a novel function of SRSF1 in binding and unfolding of RNA G-quadruplexes. Nucleic Acids Res. 2024, 52, 4676–4690. [Google Scholar] [CrossRef] [PubMed]
- Juarez, I.; Su, S.; Herbert, Z.T.; Teijaro, J.R.; Moulton, V.R. Splicing factor SRSF1 is essential for CD8 T cell function and host antigen-specific viral immunity. Front. Immunol. 2022, 13, 906355. [Google Scholar] [CrossRef]
- Paz, S.; Lu, M.L.; Takata, H.; Trautmann, L.; Caputi, M. SRSF1 RNA recognition motifs are strong inhibitors of HIV-1 replication. J. Virol. 2015, 89, 6275–6286. [Google Scholar] [CrossRef]
- Sertznig, H.; Roesmann, F.; Wilhelm, A.; Heininger, D.; Bleekmann, B.; Elsner, C.; Santiago, M.; Schuhenn, J.; Karakoese, Z.; Benatzy, Y.; et al. SRSF1 acts as an IFN-I-regulated cellular dependency factor decisively affecting HIV-1 post-integration steps. Front. Immunol. 2022, 13, 935800. [Google Scholar] [CrossRef]
- Li, K.; Wang, Z. Splicing factor SRSF2-centric gene regulation. Int. J. Biol. Sci. 2021, 17, 1708. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Q.; Lu, J.; Fan, P.; Xie, W.; Qiu, W.; Wang, F.; Hu, G.; Zhang, Y. Serine/arginine-rich splicing factor 2 modulates herpes simplex virus type 1 replication via regulating viral gene transcriptional activity and pre-mRNA splicing. J. Biol. Chem. 2016, 291, 26377–26387. [Google Scholar] [CrossRef]
- Klymenko, T.; Hernandez-Lopez, H.; MacDonald, A.I.; Bodily, J.M.; Graham, S.V. Human papillomavirus E2 regulates SRSF3 (SRp20) to promote capsid protein expression in infected differentiated keratinocytes. J. Virol. 2016, 90, 5047–5058. [Google Scholar] [CrossRef]
- Li, Q.; Jiang, Z.; Ren, S.; Guo, H.; Song, Z.; Chen, S.; Gao, X.; Meng, F.; Zhu, J.; Liu, L.; et al. SRSF5-mediated alternative splicing of M gene is essential for influenza a virus replication: A host-directed target against influenza virus. Adv. Sci. 2022, 9, 2203088. [Google Scholar] [CrossRef]
- Kim, G.N.; Yu, K.L.; Kim, H.I.; You, J.C. Investigation of the effect of SRSF9 overexpression on HIV-1 production. BMB Rep. 2022, 55, 639. [Google Scholar] [CrossRef]
- Shkreta, L.; Delannoy, A.; Salvetti, A.; Chabot, B. SRSF10: An atypical splicing regulator with critical roles in stress response, organ development, and viral replication. RNA 2021, 27, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Chabrolles, H.; Auclair, H.; Vegna, S.; Lahlali, T.; Pons, C.; Michelet, M.; Couté, Y.; Belmudes, L.; Chadeuf, G.; Kim, Y.; et al. Hepatitis B virus Core protein nuclear interactome identifies SRSF10 as a host RNA-binding protein restricting HBV RNA production. PLoS Pathog. 2020, 16, e1008593. [Google Scholar] [CrossRef] [PubMed]
- Fang, A.; Bi, Z.; Ye, H.; Yan, L. SRSF10 inhibits the polymerase activity and replication of avian influenza virus by regulating the alternative splicing of chicken ANP32A. Virus Res. 2020, 286, 198063. [Google Scholar] [CrossRef]
- Li, D.; Yu, W.; Lai, M. Towards understandings of serine/arginine-rich splicing factors. Acta Pharm. Sin. B 2023, 13, 3181–3207. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Caponnetto, A.; Brex, D.; Mirabella, F.; Barbagallo, C.; Lauretta, G.; Morrone, A.; Certo, F.; Broggi, G.; Caltabiano, R.; et al. CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers 2019, 11, 194. [Google Scholar] [CrossRef]
- Zhou, X.; Li, X.; Yu, L.; Wang, R.; Hua, D.; Shi, C.; Sun, C.; Luo, W.; Rao, C.; Jiang, Z.; et al. The RNA-binding protein SRSF1 is a key cell cycle regulator via stabilizing NEAT1 in glioma. Int. J. Biochem. Cell Biol. 2019, 113, 75–86. [Google Scholar] [CrossRef]
- Mo, Y.; Liu, Y.; Lu, A.; Zhang, H.; Tang, L. Role of circRNAs in viral infection and their significance for diagnosis and treatment. Int. J. Mol. Med. 2021, 47, 88. [Google Scholar] [CrossRef]
- Chen, W.Y.; Ho, K.C.; Leu, J.H.; Liu, K.F.; Wang, H.C.; Kou, G.H.; Lo, C.F. WSSV infection activates STAT in shrimp. Dev. Comp. Immunol. 2008, 32, 1142–1150. [Google Scholar] [CrossRef]
- Limkul, S.; Phiwthong, T.; Massu, A.; Jaree, P.; Thawonsuwan, J.; Teaumroong, N.; Boonanuntanasarn, S.; Somboonwiwat, K.; Boonchuen, P. The interferon-like proteins, Vagos, in Fenneropenaeus merguiensis elicit antimicrobial responses against WSSV and VPAHPND infection. Fish Shellfish Immunol. 2022, 131, 718–728. [Google Scholar] [CrossRef]
- Phiwthong, T.; Limkul, S.; Aunkam, P.; Seabkongseng, T.; Teaumroong, N.; Tittabutr, P.; Boonchuen, P. Quaking RNA-Binding protein (QKI) mediates circular RNA biogenesis in Litopenaeus vannamei during WSSV infection. Fish Shellfish Immunol. 2025, 159, 110178. [Google Scholar] [CrossRef] [PubMed]
- Limkul, S.; Phiwthong, T.; Wanvimonsuk, S.; Seabkongseng, T.; Aunkam, P.; Jaree, P.; Luangtrakul, W.; Mahanil, K.; Teamtisong, K.; Tittabutr, P.; et al. Viral circular RNA-encoded protein, ceVP28, divulges an antiviral response in invertebrates. Proc. Natl. Acad. Sci. USA 2025, 122, e2321707122. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Li, H.; Xu, L.; Yang, F. A simple and efficient method for purification of intact white spot syndrome virus (WSSV) viral particles. Virus Res. 2005, 108, 63–67. [Google Scholar] [CrossRef]
- Seabkongseng, T.; Limkul, S.; Sriphuttha, C.; Phiwthong, T.; Aunkam, P.; Suwannathit, R.; Jaree, P.; Somboonwiwat, K.; Tittabutr, P.; Teaumroong, N.; et al. Supplementation of Bacillus velezensis S141 in feed as a probiotic enhances growth performance, pathogenic tolerances, and immune system in shrimp. Aquaculture 2025, 604, 742448. [Google Scholar] [CrossRef]
- Perdomo-Morales, R.; Montero-Alejo, V.; Rodríguez-Viera, L.; Perera, E. Evaluation of anticoagulants and hemocyte-maintaining solutions for the study of hemolymph components in the spiny lobster Panulirus argus (Latreille, 1804) (Decapoda: Achelata: Palinuridae). J. Crustac. Biol. 2020, 40, 213–217. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mendoza-Cano, F.; Sánchez-Paz, A. Development and validation of a quantitative real-time polymerase chain assay for universal detection of the White Spot Syndrome Virus in marine crustaceans. Virol. J. 2013, 10, 1–11. [Google Scholar] [CrossRef]




| Primer Name | Primer Sequences (5′ → 3′) | Annealing Temperature |
|---|---|---|
| F-LvAlpha2 Circ R-LvAlpha2 Circ | CACCATTACGCCCACTAGC TGTAAGGGTCCTCCACTAACTG | 57 °C |
| F-LvAlpha2 Li R-LvAlpha2 Li | TCATAGCCCGCGGTAAG TGTAAGGGTCCTCCACTAAC | 60 °C |
| F-LvAnillin Circ R-LvAnillin Circ | GGGCAGCAGCCTTTGATTC GCAGCGGGTAGCTTTCTTG | 60 °C |
| F-LvAnillin Li R-LvAnillin Li | ACCTGAGCGCCAACAAAG TCCTGCAACAGTGTCCATC | 60 °C |
| F-LvCalpain Circ R-LvCalpain Circ | TGAGAGTGTTCTCGGAGAAG TTCCGGAGTCCATACACTG | 60 °C |
| F-LvCalpain Li R-LvCalpain Li | TGGCTGCCATTGCGAAC ACAAGGCCGTTGTTGCAG | 57 °C |
| F- EF-1α R- EF-1α | CGCAAGAGCGACAACTATGA TGGCTTCAGGATACCAGTCT | 60 °C |
| F-LvHemocytin Circ R-LvHemocytin Circ | TCGGACGCAGAGATGATTC TAGACTTCGCCGTCGATCTC | 57 °C |
| F-LvHemocytin Li R-LvHemocytin Li | AGGCCGACTGCGAGAAGAAG TTCCTCGTGCACACGCAAG | 60 °C |
| F-LvNephrin Circ R-LvNephrin Circ | CACGGGAATCAACCCAGCTC ACAGATCTCGACGCCGTCAG | 60 °C |
| F-LvNephrin Li R-LvNephrin Li | AACCTACGCCGTGGAAG AGGATCCGGACTTCCATTG | 60 °C |
| F-LvSSRF1B R-LvSSRF1B | TCATCGACCTGAAGAACCGC ACATCTCCTGCCTCCCTCAT | 60 °C |
| F-LvSRSF2 R-LvSRSF2 | TTGGAGACGTGTACATCCCG CCATTCCACGCTCATTTTGCT | 60 °C |
| F-LvSRSF2 X1 R-LvSRSF2 X1 | TACTCGGACAATTCCAGGTCG GAGTGGCTGCCAGACCTT | 60 °C |
| F-LvSRSF2 X2 R-LvSRSF2 X2 | CGGGACAATTCCAGGTCG GAGTGGCTGCCAGACCTT | 60 °C |
| F-LvSRSF2 X4 R-LvSRSF2 X4 | CCGACGGTCGAGGTCGAG GAGTGGCTGCCAGACCTT | 60 °C |
| F-LvSRSF2 X5 R-LvSRSF2 X5 | TACTCGGACAATTCCAGGTCG TGTGACCTTGATCTAGAGTGC | 60 °C |
| F-LvSRSF2 X6 R-LvSRSF2 X6 | CGGGACAATTCCAGGTCG TGTGACCTTGATCTAGAGTGC | 60 °C |
| F-LvSRSF2 X7 R-LvSRSF2 X7 | CCGACGGTACTCGTCGAGG TGTGACCTTGATCTAGAGTGC | 60 °C |
| F-LvSRSF2 X8 R-LvSRSF2 X8 | CCGACGGTCGAGGTCGAG TGTGACCTTGATCTAGAGTGC | 60 °C |
| F-LvSRSF3 (XM_027370964.1) R-LvSRSF3 (XM_027370964.1) | GTTCTCGACGGGACCGATAC TCTGGGGAATCACTTCTGCG | 60 °C |
| F-LvSRSF4 conserved R-LvSRSF4 conserved | CCTAAGCTACCGTGTGGGTG GAACCCTCTTGGTGGAGGTG | 60 °C |
| F-LvSRSF5 R-LvSRSF5 | GCCGTGAAAGAGACCTGGAA CGGTGAAGGTCACTTGGCTT | 60 °C |
| F-LvSRSF5 X1 R-LvSRSF5 X1 | GCTTGGCTTGACAGGTACGG ATCCTTGAGATCCTGCCAGC | 60 °C |
| F-LvSRSF5 X2 R-LvSRSF5 X2 | ATGAGTTACGCCCACGTTAG AACGGGAGTGTTGTCGATCC | 60 °C |
| F-LvSRSF7 conserved R-LvSRSF7 conserved | TTTGAGGACATGCGAGATGC TGACTTCCCCGTTGATAGCTC | 60 °C |
| F-LvSRSF7 (XM_027216940.1) R-LvSRSF7 (XM_027216940.1) | TTTGGCTATAAGCGACCCCC GGATCTTGATCGGCTGTGGT | 60 °C |
| F-dsLvSRSF2 R-dsLvSRSF2 | CCTCTAGACAGAAGATTTGCGGCG CCCTCGAGTTGATTCACGTTGGGG | 60 °C |
| F-LvToll Li R-LvToll Li | CGCTTCTCTGTCCTCATTTC GGTTGCCTCGAAGTTTCAG | 60 °C |
| F-LvToll Circ R-LvToll Circ | AGGTCATCATCGCCAGCACAG ACCACCACGAGGCAAGGAAG | 60 °C |
| F-VP28 R-VP28 | AAACCTCCGCATTCCTGTGA TCCGCATCTTCTTCCTTCAT | 60 °C |
| F-STAT R-STAT | TATATCCGAATGTGCCTA ATAGTTTGTGGTGTGTTG | 60 °C |
| F-Vago4 R-Vago4 | AGCTGCTGCCCCATCATCT ATCCAATTCGTGAACTCGTCGTA | 60 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potiyanadech, W.; Sriphuttha, C.; Seabkongseng, T.; Teaumroong, N.; Tittabutr, P.; Boonchuen, P. The Involvement of LvSRSF2 in Circular RNA Biogenesis and Its Role in Immunity Against White Spot Syndrome Virus (WSSV) in Litopenaeus vannamei. Int. J. Mol. Sci. 2025, 26, 5981. https://doi.org/10.3390/ijms26135981
Potiyanadech W, Sriphuttha C, Seabkongseng T, Teaumroong N, Tittabutr P, Boonchuen P. The Involvement of LvSRSF2 in Circular RNA Biogenesis and Its Role in Immunity Against White Spot Syndrome Virus (WSSV) in Litopenaeus vannamei. International Journal of Molecular Sciences. 2025; 26(13):5981. https://doi.org/10.3390/ijms26135981
Chicago/Turabian StylePotiyanadech, Wutthipat, Cheeranan Sriphuttha, Tuangrak Seabkongseng, Neung Teaumroong, Panlada Tittabutr, and Pakpoom Boonchuen. 2025. "The Involvement of LvSRSF2 in Circular RNA Biogenesis and Its Role in Immunity Against White Spot Syndrome Virus (WSSV) in Litopenaeus vannamei" International Journal of Molecular Sciences 26, no. 13: 5981. https://doi.org/10.3390/ijms26135981
APA StylePotiyanadech, W., Sriphuttha, C., Seabkongseng, T., Teaumroong, N., Tittabutr, P., & Boonchuen, P. (2025). The Involvement of LvSRSF2 in Circular RNA Biogenesis and Its Role in Immunity Against White Spot Syndrome Virus (WSSV) in Litopenaeus vannamei. International Journal of Molecular Sciences, 26(13), 5981. https://doi.org/10.3390/ijms26135981





