Hippo Pathway Dysregulation in Thymic Epithelial Tumors (TETs): Associations with Clinicopathological Features and Patients’ Prognosis
Abstract
1. Introduction
2. Results
2.1. Immunohistochemical Expression of MST1 in TETs
2.2. Immunohistochemical Expression of SAV1 in TETs
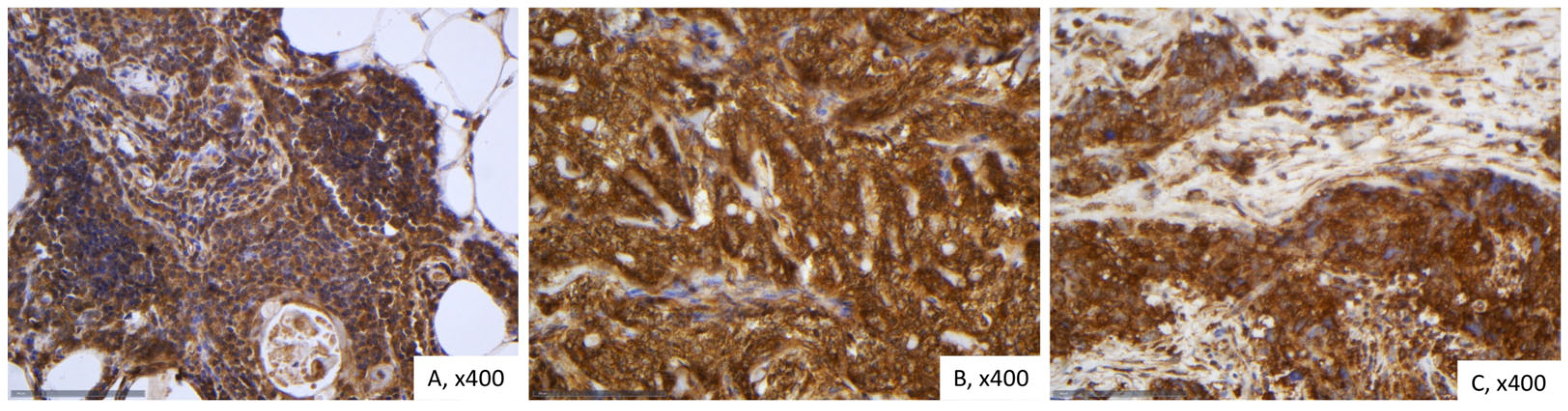
2.3. Immunohistochemical Expression of LATS1 in TETs
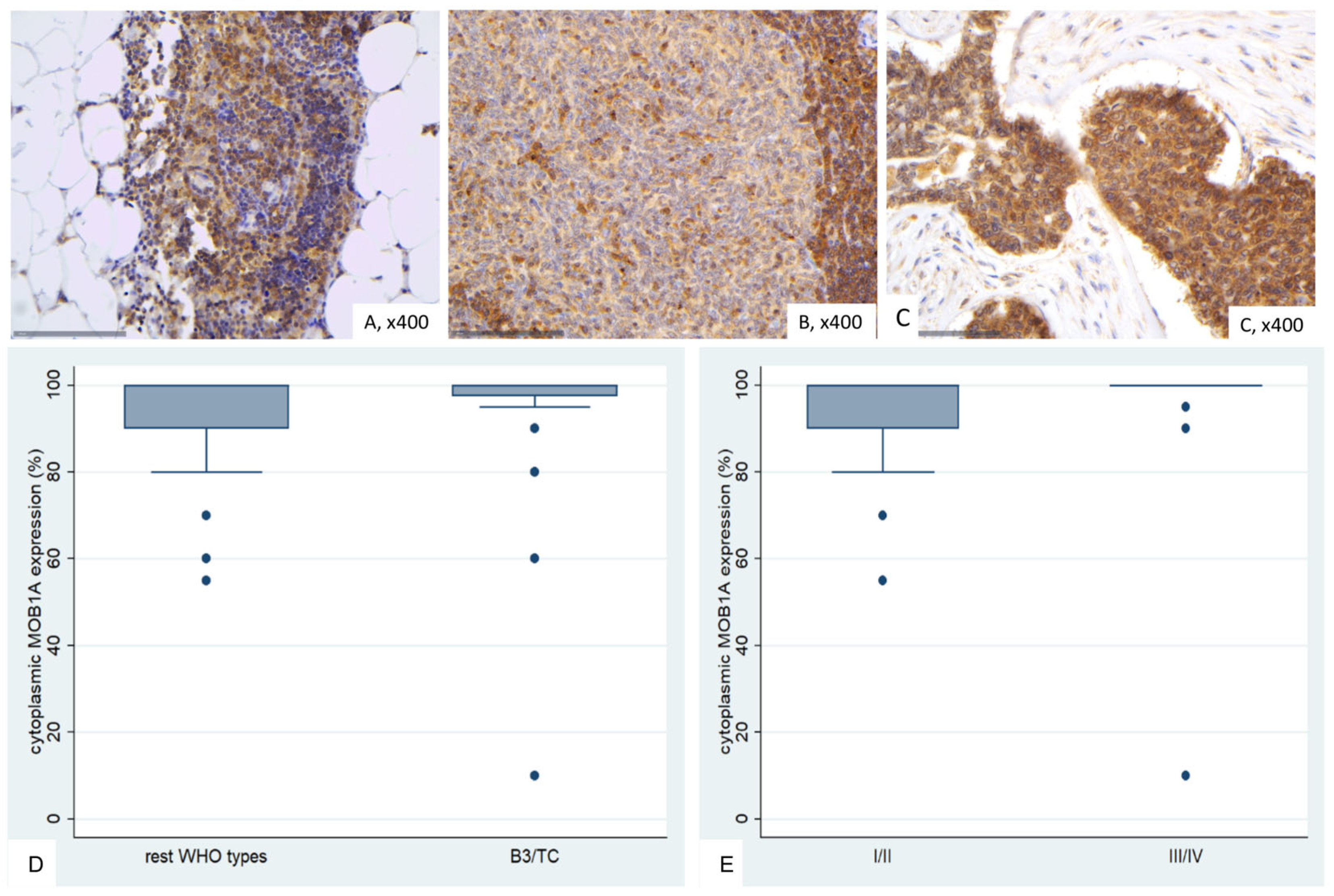
2.4. Immunohistochemical Expression of MOB1A in TETs
2.5. Immunohistochemical Expression of YAP1 in TETs
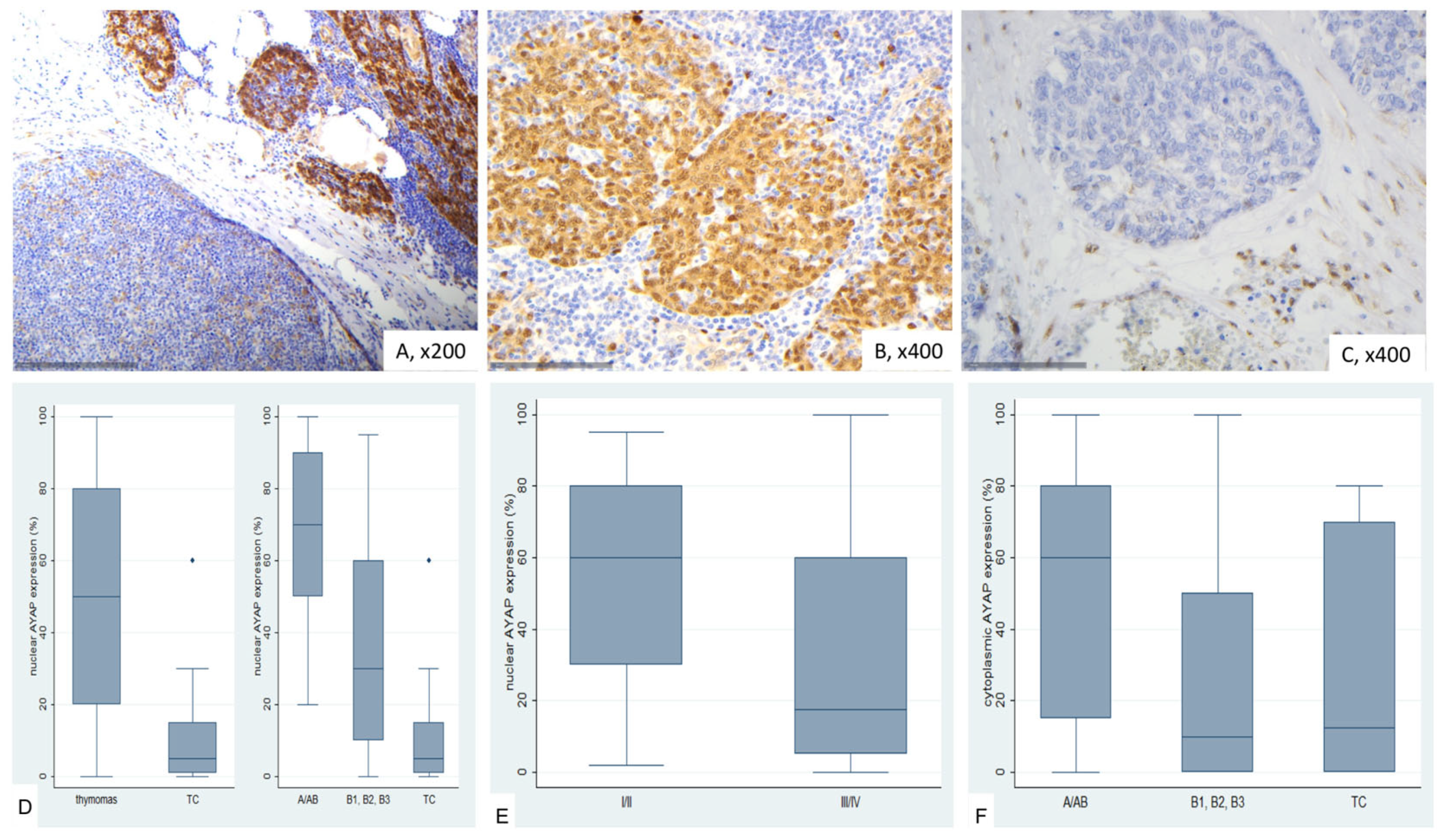
2.6. Immunohistochemical Expression of Active YAP (AYAP) in TETs
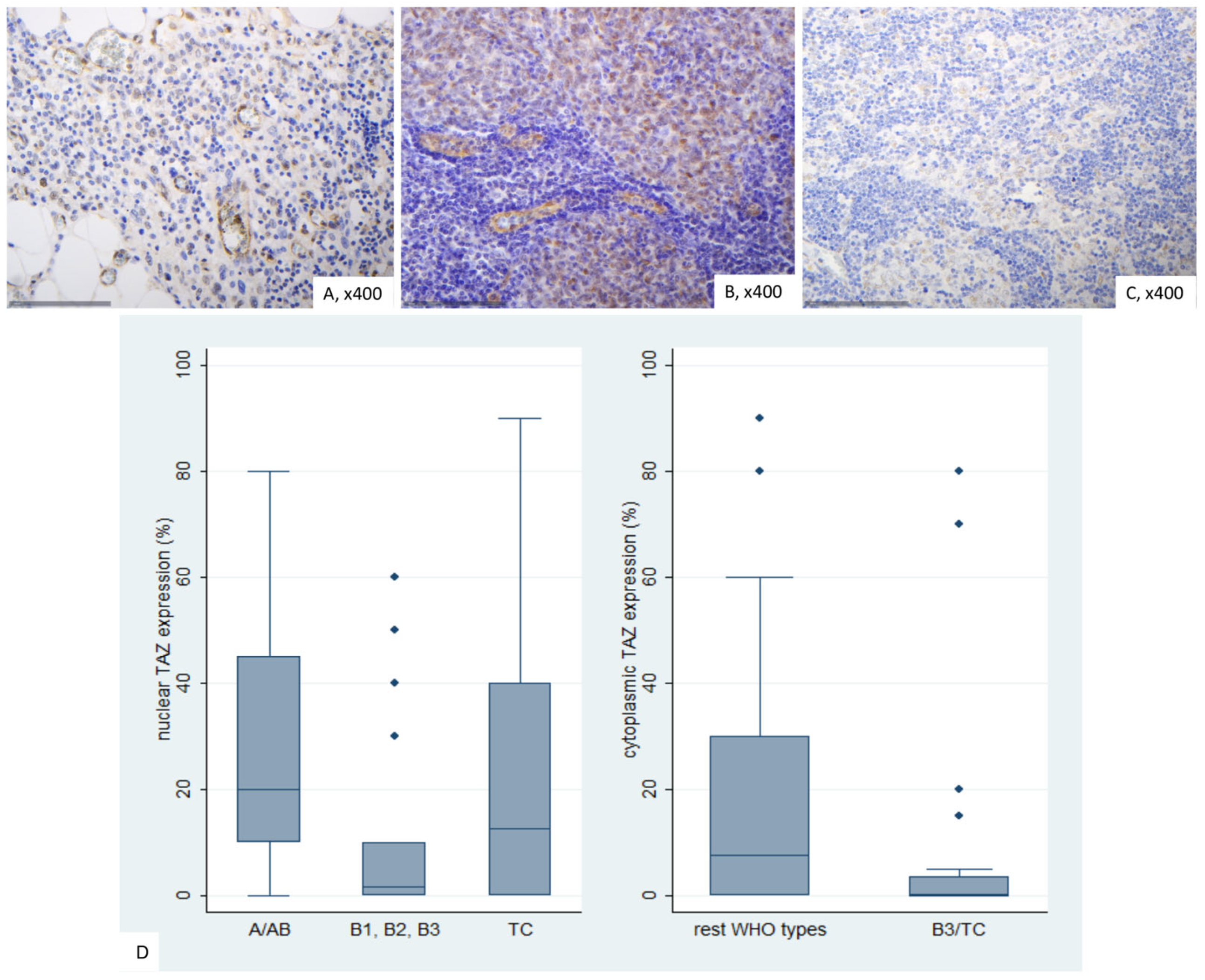
2.7. Immunohistochemical Expression of TAZ in TETs
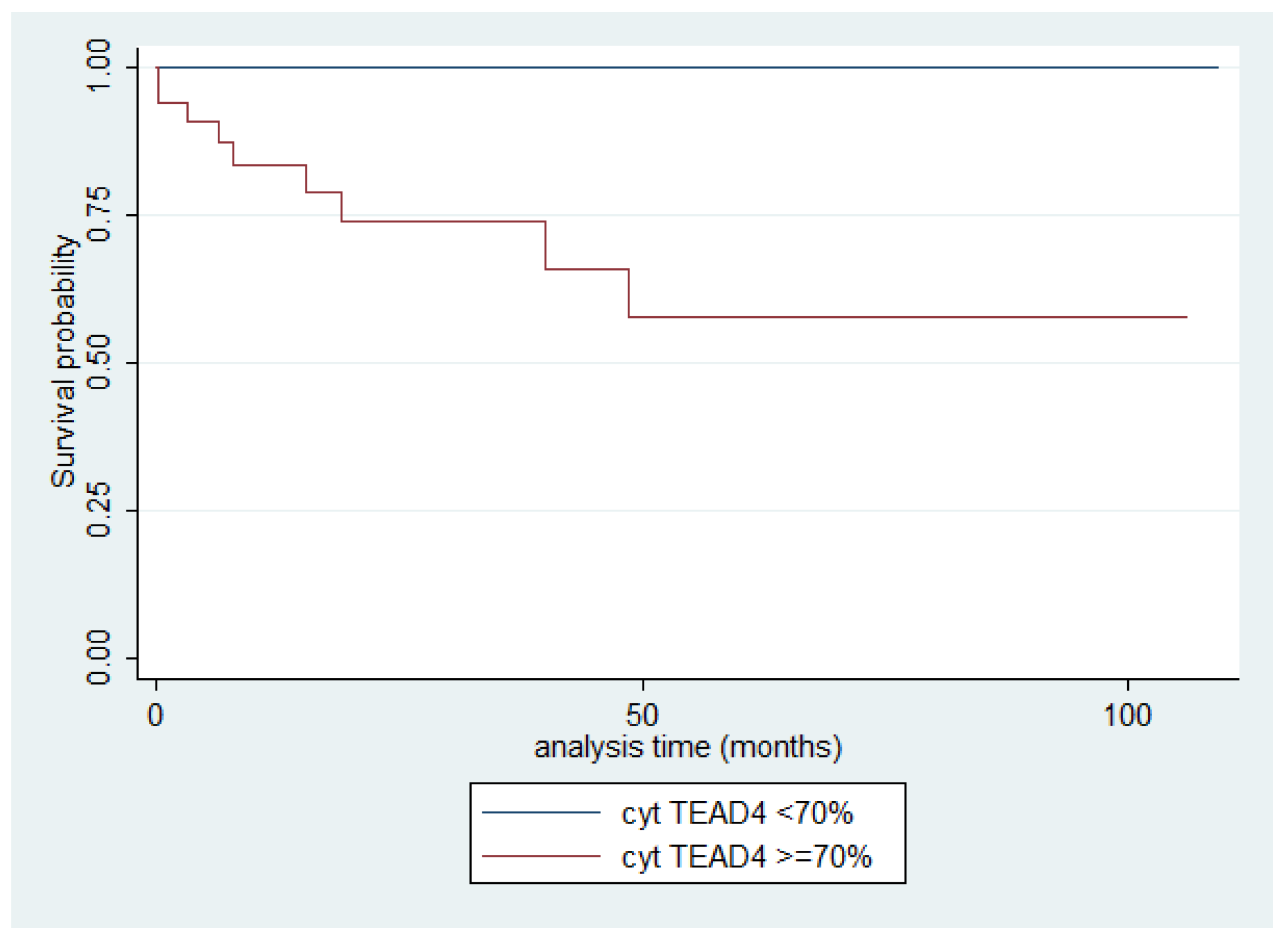
2.8. Immunohistochemical Expression of TEAD4 in TETs
2.9. Associations Between the Investigated Molecules of the Hippo Cascade
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Immunohistochemistry
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TET(s) | Thymic Epithelial Tumor(s) |
| WHO | World Health Organization |
| TC | Thymic Carcinoma |
| MST1/2 | Mammalian STE20-like kinases |
| LATS1/2 | Large tumor suppressor kinases). |
| TAZ | Transcriptional co-activator with PDZ-binding motif |
| YAP | Yes-associated protein |
| MOB1(A) | Mps one binder 1(A) |
| SAV1 | Salvador homolog 1 |
| TEAD4 | TEA domain transcription factor 4 |
| RTK | Receptor tyrosine kinase |
| AYAP | Active YAP1 |
| NSCLC | Non-small-cell lung cancer |
| OXPHOS | Mitochondrial oxidative phosphorylation |
| FFPE | Formalin-Fixed, Paraffin-Embedded |
| RT-qPCR | Reverse Transcription quantitative Polymerase Chain Reaction |
| NGS | Next-Generation Sequencing |
| WWTR1 | WW domain-containing transcription regulator 1 |
| MNT | Micronodular thymoma with lymphoid stroma |
| PMU | Paracelsus Medical University |
Appendix A
Appendix A.1. MST1
| Parameter | MST1 Cytoplasmic Expression | |
|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value |
| Age | R = −0.08 | p = 0.465 |
| Tumor size | R = 0.18 | p = 0.152 |
| Categorical/nominal variables | ||
| Median (min–max) | p-value * | |
| Gender | ||
| Male | 15 (0–100) | p = 0.370 |
| Female | 10 (1–95) | |
| WHO subtypes | ||
| Thymomas | 10 (0–95) | p = 0.014 |
| Thymic carcinomas (TC) | 70 (1–100) | |
| Masaoka–Koga stage | ||
| I–II | 10 (0–95) | p = 0.002 |
| III–IV | 45 (0–100) |
Appendix A.2. SAV1
| Parameter | SAV1 Expression | ||
|---|---|---|---|
| Numerical Variables | <100% Median (Min–Max) | 100% Median (Min–Max) | p-Value * |
| Age (years) | 74 (45–88) | 67 (21–85) | p = 0.264 |
| Tumor size (cm) | 4.5 (2.4–9) | 6, (0.9–14) | p = 0.237 |
| Categorical/nominal variables | |||
| <100% (n) | 100% (n) | p-value ** | |
| Gender | |||
| Male | 4 | 35 | p = 0.138 |
| Female | 9 | 29 | |
| WHO subtypes | |||
| Rest WHO types | 12 | 37 | p = 0.025 |
| B3/TC | 1 | 27 | |
| Masaoka–Koga stage | |||
| I–II | 11 | 36 | p = 0.027 |
| III–IV | 0 | 18 | |
Appendix A.3. LATS1
| Parameter | LATS1 Cytoplasmic Expression | |
|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value |
| Age | R = −0.08 | p = 0.788 |
| Tumor size | R = −0.07 | p = 0.566 |
| Categorical/nominal variables | ||
| Median (min–max) | p-value * | |
| Gender | ||
| Male | 90 (10–100) | p = 0.528 |
| Female | 90 (20–100) | |
| WHO subtypes | ||
| Rest WHO types | 80 (10–100) | p < 0.001 |
| B3/TC | 100 (15–100) | |
| Masaoka–Koga stage | ||
| I–II | 80 (20–100) | p = 0.007 |
| III–IV | 100 (40–100) |
Appendix A.4. MOB1A
| Parameter | MOB1A Cytoplasmic Expression | |
|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value |
| Age | R = −0.01 | p = 0.879 |
| Tumor size | R = −0.10 | p = 0.389 |
| Categorical/nominal variables | ||
| Median (min–max) | p-value * | |
| Gender | ||
| Male | 100 (10–100) | p = 0.438 |
| Female | 100 (55–100) | |
| WHO subtypes | ||
| Rest WHO types | 100 (55–100) | p = 0.063 |
| B3/TC | 100 (10–100) | |
| Masaoka–Koga stage | ||
| I–II | 100 (55–100) | p = 0.033 |
| III–IV | 100 (10–100) |
Appendix A.5. YAP1
| Parameter | YAP1 Nuclear Expression | |
|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value |
| Age | R = 0.04 | p = 0.669 |
| Tumor size | R = −0.02 | p = 0.870 |
| Categorical/nominal variables | ||
| Median (min–max) | p-value * | |
| Gender | ||
| Male | 30 (0–95) | p = 0.506 |
| Female | 40 (0–90) | |
| WHO subtypes | ||
| Thymomas | 50 (0–95) | p = 0.001 |
| TC | 5 (0–50) | |
| Masaoka–Koga stage | ||
| I–II | 60 (0–95) | p = 0.023 |
| III–IV | 12.5 (0–95) | |
| Parameter | YAP1 Cytoplasmic Expression | |
|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value |
| Age | R = 0.12 | p = 0.273 |
| Tumor size | R = −0.18 | p = 0.144 |
| Categorical/nominal variables | ||
| Median (min–max) | p-value * | |
| Gender | ||
| Male | 20 (0–100) | p = 0.470 |
| Female | 10 (0–100) | |
| WHO subtypes | ||
| Rest WHO types | 50 (0–95) | p = 0.740 |
| B3/TC | 5 (0–50) | |
| Masaoka–Koga stage | ||
| I | 5 (0–80) | p = 0.032 |
| II-IV | 30 (0–100) | |
Appendix A.6. AYAP
| Parameter | AYAP Nuclear Expression | AYAP Cytoplasmic Expression | ||
|---|---|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value | Spearman’s Correlation Coefficient | p-Value |
| Age | R = 0.01 | p = 0.905 | R = 0.12 | p = 0.281 |
| Tumor size | R = 0.04 | p = 0.740 | R = −0.18 | p = 0.131 |
| Categorical/Nominal Variables | ||||
| Median (min–max) | p-value | Median (min–max) | p-value | |
| Gender | ||||
| Male | 50 (0–100) | p = 0.299 * | 40 (0–100) | p = 0.384 * |
| Female | 35 (0–90) | 17.5 (0–100) | ||
| WHO Subtypes | ||||
| A/AB | 70 (20–100) | p = 0.001 ** | 60 (0–100) | p = 0.011 ** |
| B1, B2, B3 | 30 (0–95) | 10 (0–100) | ||
| TC | 5 (0–60) | 12.5 (0–80) | ||
| Masaoka–Koga Stage | ||||
| I–II | 60 (2–95) | p = 0.007 * | 45 (0–100) | p = 0.947 ** |
| III–IV | 17.5 (0–100) | 45 (0–95) | ||
Appendix A.7. TAZ
| Parameter | TAZ Nuclear Expression | |
|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value |
| Age | R = 0.197 | p = 0.086 |
| Tumor size | R = −0.001 | p > 0.999 |
| Categorical/nominal variables | ||
| Median (min–max) | p-value * | |
| Gender | ||
| Male | 10 (0–90) | p = 0.872 |
| Female | 10 (0–70) | |
| WHO subtypes | ||
| Thymomas A/AB | 20 (0–80) | p = 0.004 |
| B1, B2, B3 | 1.5 (0–60) | |
| TC | 12.5 (0–90) | |
| Masaoka–Koga stage | ||
| I–II | 15 (0–70) | p = 0.182 |
| III–IV | 0 (0–80) | |
| Parameter | TAZ Cytoplasmic Expression | |
|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value |
| Age | R = 0.101 | p = 0.384 |
| Tumor size | R = 0.198 | p = 0.123 |
| Categorical/nominal variables | ||
| Median (min–max) | p-value * | |
| Gender | ||
| Male | 5 (0–90) | p = 0.298 |
| Female | 0 (0–80) | |
| WHO subtypes | ||
| Rest WHO types | 7.5 (0–90) | p = 0.004 |
| B3/TC | 3.5 (0–80) | |
| Masaoka–Koga stage | ||
| I–II | 5 (0–80) | p = 0.281 |
| III–IV | 0 (0–90) | |
Appendix A.8. TEAD4
| Parameter | TEAD4 Nuclear Expression | |
|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value |
| Age | R = 0.12 | p = 0.292 |
| Tumor size | R = −0.15 | p = 0.222 |
| Categorical/nominal variables | ||
| Median (min–max) | p-value | |
| Gender | ||
| Male | 20 (0–90) | p = 0.545 * |
| Female | 12.5 (0–100) | |
| WHO subtypes | ||
| A/AB | 25 (0–90) | p = 0.005 ** |
| B1, B2, B3 | 0 (0–90) | |
| TC | 27.5 (0–100) | |
| Masaoka–Koga stage | ||
| I–II | 10 (0–90) | p = 0.957 * |
| III–IV | 7.5 (0–90) | |
| Parameter | TEAD4 Cytoplasmic Expression | |
|---|---|---|
| Numerical Variables | Spearman’s Correlation Coefficient | p-Value |
| Age | R = 0.25 | p = 0.027 |
| Tumor size | R = −0.11 | p = 0.348 |
| Categorical/nominal variables | ||
| Median (min–max) | p-value * | |
| Gender | ||
| Male | 80 (0–100) | p = 0.198 |
| Female | 60 (0–100) | |
| WHO subtypes | ||
| Thymomas | 60 (0–100) | p = 0.002 |
| TC | 100 (0–100) | |
| Masaoka–Koga stage | ||
| I–II | 60 (0–100) | p < 0.001 |
| III–IV | 95 (20–100) | |
References
- Palamaris, K.; Levidou, G.; Kordali, K.; Masaoutis, C.; Rontogianni, D.; Theocharis, S. Searching for Novel Biomarkers in Thymic Epithelial Tumors: Immunohistochemical Evaluation of Hippo Pathway Components in a Cohort of Thymic Epithelial Tumors. Biomedicines 2023, 11, 1876. [Google Scholar] [CrossRef]
- Gameiro, J.; Nagib, P.; Verinaud, L. The thymus microenvironment in regulating thymocyte differentiation. Cell Adh. Migr. 2010, 4, 382–390. [Google Scholar] [CrossRef]
- Lin, X.; Gao, Y.; Ding, Y.; Zhao, B. Editorial: Revisiting the thymus: The origin of T cells. Front. Immunol. 2023, 14, 1197066. [Google Scholar] [CrossRef]
- Dai, H.; Lan, B.; Li, S.; Huang, Y.; Jiang, G.; Tian, J. Prognostic CT features in patients with untreated thymic epithelial tumors. Sci. Rep. 2023, 13, 2910. [Google Scholar] [CrossRef] [PubMed]
- Sumi, A.; Fujimoto, K. Is there any consensus of long-term follow-up for incidental anterior mediastinal nodular lesions? Shanghai Chest 2018, 2. [Google Scholar] [CrossRef]
- Veraar, C.; Janik, S.; Thanner, J.; Veraar, C.; Mouhieddine, M.; Schiefer, A.-I.; Müllauer, L.; Dworschak, M.; Klepetko, W.; Ankersmit, H.J.; et al. Clinical prognostic scores for patients with thymic epithelial tumors. Sci. Rep. 2019, 9, 18581. [Google Scholar] [CrossRef] [PubMed]
- Elm, L.; Levidou, G. The Molecular Landscape of Thymic Epithelial Tumors: A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 1554. [Google Scholar] [CrossRef] [PubMed]
- Marx, A.; Ströbel, P.; Badve, S.S.; Chalabreysse, L.; Chan, J.K.; Chen, G.; de Leval, L.; Detterbeck, F.; Girard, N.; Huang, J.; et al. ITMIG consensus statement on the use of the WHO histological classification of thymoma and thymic carcinoma: Refined definitions, histological criteria, and reporting. J. Thorac. Oncol. 2014, 9, 596–611. [Google Scholar] [CrossRef]
- Tomiyama, N.; Johkoh, T.; Mihara, N.; Honda, O.; Kozuka, T.; Koyama, M.; Hamada, S.; Okumura, M.; Ohta, M.; Eimoto, T.; et al. Using the World Health Organization Classification of Thymic Epithelial Neoplasms to Describe CT Findings. Am. J. Roentgenol. 2002, 179, 881–886. [Google Scholar] [CrossRef]
- Tassi, V.; Vannucci, J.; Ceccarelli, S.; Gili, A.; Matricardi, A.; Avenia, N.; Puma, F. Stage-related outcome for thymic epithelial tumours. BMC Surg. 2019, 18 (Suppl. S1), 114. [Google Scholar] [CrossRef]
- Lee, G.D.; Kim, H.R.; Choi, S.H.; Kim, Y.H.; Kim, D.K.; Park, S.I. Prognostic stratification of thymic epithelial tumors based on both Masaoka-Koga stage and WHO classification systems. J. Thorac. Dis. 2016, 8, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Ke, X.; Man, J.; Zhang, B.; Wang, F.; Zhou, J. Predicting Masaoka-Koga Clinical Stage of Thymic Epithelial Tumors Using Preoperative Spectral Computed Tomography Imaging. Front. Oncol. 2021, 11, 631649. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, X.; Song, G. The Hippo Pathway: A Master Regulatory Network Important in Cancer. Cells 2021, 10, 1416. [Google Scholar] [CrossRef]
- Liu, C.; Song, Y.; Li, D.; Wang, B. Regulation of the tumor immune microenvironment by the Hippo Pathway: Implications for cancer immunotherapy. Int. Immunopharmacol. 2023, 122, 110586. [Google Scholar] [CrossRef]
- Mo, J.S.; Park, H.W.; Guan, K.L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014, 15, 642–656. [Google Scholar] [CrossRef]
- Moshe Oren, Y.A. The Hippo Signaling Pathway and Cancer; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Fu, M.; Hu, Y.; Lan, T.; Guan, K.-L.; Luo, T.; Luo, M. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct. Target. Ther. 2022, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Xu, Y.; Zhao, J.; Chen, M.; Wang, M. Hippo pathway in non-small cell lung cancer: Mechanisms, potential targets, and biomarkers. Cancer Gene Ther. 2024, 31, 652–666. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, G.; Yang, Y.; Mei, Z.; Liang, Z.; Cui, A.; Wu, T.; Liu, C.Y.; Cui, L. Increased TEAD4 expression and nuclear localization in colorectal cancer promote epithelial-mesenchymal transition and metastasis in a YAP-independent manner. Oncogene 2016, 35, 2789–2800. [Google Scholar] [CrossRef]
- Takeda, M.; Yoshida, S.; Inoue, T.; Sekido, Y.; Hata, T.; Hamabe, A.; Ogino, T.; Miyoshi, N.; Uemura, M.; Yamamoto, H.; et al. The Role of KRAS Mutations in Colorectal Cancer: Biological Insights, Clinical Implications, and Future Therapeutic Perspectives. Cancers 2025, 17, 428. [Google Scholar] [CrossRef]
- Lee, J.E.; Park, H.S.; Lee, D.; Yoo, G.; Kim, T.; Jeon, H.; Yeo, M.-K.; Lee, C.-S.; Moon, J.Y.; Jung, S.S.; et al. Hippo pathway effector YAP inhibition restores the sensitivity of EGFR-TKI in lung adenocarcinoma having primary or acquired EGFR-TKI resistance. Biochem. Biophys. Res. Commun. 2016, 474, 154–160. [Google Scholar] [CrossRef]
- Franklin, J.M.; Wu, Z.; Guan, K.L. Insights into recent findings and clinical application of YAP and TAZ in cancer. Nat. Rev. Cancer 2023, 23, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Conrad, C.; Xia, F.; Park, J.-S.; Payer, B.; Yin, Y.; Lauwers, G.Y.; Thasler, W.; Lee, J.T.; Avruch, J.; et al. Mst1 and Mst2 Maintain Hepatocyte Quiescence and Suppress Hepatocellular Carcinoma Development through Inactivation of the Yap1 Oncogene. Cancer Cell 2009, 16, 425–438. [Google Scholar] [CrossRef]
- Cao, L.; Sun, P.-L.; Yao, M.; Jia, M.; Gao, H. Expression of YES-associated protein (YAP) and its clinical significance in breast cancer tissues. Hum. Pathol. 2017, 68, 166–174. [Google Scholar] [CrossRef]
- Feng, J.; Ren, P.; Gou, J.; Li, Z. Prognostic significance of TAZ expression in various cancers: A meta-analysis. OncoTargets Ther. 2016, 9, 5235–5244. [Google Scholar] [CrossRef]
- Home, P.; Saha, B.; Ray, S.; Dutta, D.; Gunewardena, S.; Yoo, B.; Pal, A.; Vivian, J.L.; Larson, M.; Petroff, M.; et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc. Natl. Acad. Sci. USA 2012, 109, 7362–7367. [Google Scholar] [CrossRef]
- Kaneko, K.J.; DePamphilis, M.L. TEAD4 establishes the energy homeostasis essential for blastocoel formation. Development 2013, 140, 3680–3690. [Google Scholar] [CrossRef] [PubMed]
- Cinar, B.; Fang, P.K.; Lutchman, M.; Di Vizio, D.; Adam, R.M.; Pavlova, N.; Rubin, M.A.; Yelick, P.C.; Freeman, M.R. The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. EMBO J. 2007, 26, 4523–4534. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Huang, B.; Zhu, L.; Chen, K.; Liu, M.; Zhong, C. Structural and Functional Overview of TEAD4 in Cancer Biology. OncoTargets Ther. 2020, 13, 9865–9874. [Google Scholar] [CrossRef]
- Hsu, S.-C.; Lin, C.-Y.; Lin, Y.-Y.; Collins, C.C.; Chen, C.-L.; Kung, H.-J. TEAD4 as an Oncogene and a Mitochondrial Modulator. Front. Cell Dev. Biol. 2022, 10, 890419. [Google Scholar] [CrossRef]
- Qi, Y.; Yu, J.; Han, W.; Fan, X.; Qian, H.; Wei, H.; Tsai, Y.-H.S.; Zhao, J.; Zhang, W.; Liu, Q.; et al. A splicing isoform of TEAD4 attenuates the Hippo–YAP signalling to inhibit tumour proliferation. Nat. Commun. 2016, 7, ncomms11840. [Google Scholar] [CrossRef]
- Cui, J.; Zhou, Z.; Yang, H.; Jiao, F.; Li, N.; Gao, Y.; Wang, L.; Chen, J.; Quan, M. MST1 Suppresses Pancreatic Cancer Progression via ROS-Induced Pyroptosis. Mol. Cancer Res. 2019, 17, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Zhu, L.; Xiao, S.; Cui, Z.; Tang, J.; Yu, J.; Xie, M. MST1 inhibits the progression of breast cancer by regulating the Hippo signaling pathway and may serve as a prognostic biomarker. Mol. Med. Rep. 2021, 23, 383. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Mak, K.K.; Topol, L.; Yun, K.; Hu, J.; Garrett, L.; Chen, Y.; Park, O.; Chang, J.; Simpson, R.M.; et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. USA 2010, 107, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Turunen, S.P.; von Nandelstadh, P.; Öhman, T.; Gucciardo, E.; Seashore-Ludlow, B.; Martins, B.; Rantanen, V.; Li, H.; Höpfner, K.; Östling, P.; et al. FGFR4 phosphorylates MST1 to confer breast cancer cells resistance to MST1/2-dependent apoptosis. Cell Death Differ. 2019, 26, 2577–2593. [Google Scholar] [CrossRef]
- Ura, S.; Masuyama, N.; Graves, J.D.; Gotoh, Y. Caspase cleavage of MST1 promotes nuclear translocation and chromatin condensation. Proc. Natl. Acad. Sci. USA 2001, 98, 10148–10153. [Google Scholar] [CrossRef]
- Chen, K.H.; He, J.; Wang, D.L.; Cao, J.J.; Li, M.C.; Zhao, X.M.; Sheng, X.; Li, W.B.; Liu, W.J. Methylation-associated inactivation of LATS1 and its effect on demethylation or overexpression on YAP and cell biological function in human renal cell carcinoma. Int. J. Oncol. 2014, 45, 2511–2521. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Li, P.P.; Wang, R.; Zhu, Y.; Zheng, F.; Li, L.; Cui, J.J.; Wang, L.W. Expression profile and prognostic value of SAV1 in patients with pancreatic ductal adenocarcinoma. Tumour Biol. 2016, 37, 16207–16213. [Google Scholar] [CrossRef]
- Ando, N.; Tanaka, K.; Otsubo, K.; Toyokawa, G.; Ikematsu, Y.; Ide, M.; Yoneshima, Y.; Iwama, E.; Inoue, H.; Ijichi, K.; et al. Association of Mps one binder kinase activator 1 (MOB1) expression with poor disease-free survival in individuals with non-small cell lung cancer. Thorac. Cancer 2020, 11, 2830–2839. [Google Scholar] [CrossRef]
- Kato, W.; Nishio, M.; To, Y.; Togashi, H.; Mak, T.W.; Takada, H.; Ohga, S.; Maehama, T.; Suzuki, A. MOB1 regulates thymocyte egress and T-cell survival in mice in a YAP1-independent manner. Genes Cells 2019, 24, 485–495. [Google Scholar] [CrossRef]
- Han, H.; Huang, Z.; Xu, C.; Seo, G.; An, J.; Yang, B.; Liu, Y.; Lan, T.; Yan, J.; Ren, S.; et al. Functional annotation of the Hippo pathway somatic mutations in human cancers. Nat. Commun. 2024, 15, 10106. [Google Scholar] [CrossRef]
- Cunningham, R.; Hansen, C.G. The Hippo pathway in cancer: YAP/TAZ and TEAD as therapeutic targets in cancer. Clin. Sci. 2022, 136, 197–222. [Google Scholar] [CrossRef]
- Wu, Y.; Li, M.; Lin, J.; Hu, C. Hippo/TEAD4 signaling pathway as a potential target for the treatment of breast cancer. Oncol. Lett. 2021, 21, 313. [Google Scholar] [CrossRef] [PubMed]
- Chi, M.; Liu, J.; Mei, C.; Shi, Y.; Liu, N.; Jiang, X.; Liu, C.; Xue, N.; Hong, H.; Xie, J.; et al. TEAD4 functions as a prognostic biomarker and triggers EMT via PI3K/AKT pathway in bladder cancer. J. Exp. Clin. Cancer Res. 2022, 41, 175. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Yan, Y.; Tang, P.; Cai, J.; Cao, X.; Wang, Z.; Zhang, F.; Shen, B. TEAD4 as a Prognostic Marker Promotes Cell Migration and Invasion of Urinary Bladder Cancer via EMT. OncoTargets Ther. 2021, 14, 937–949. [Google Scholar] [CrossRef]
- Park, M.A.; Lee, Y.H.; Gu, M.J. High TEAD4 Expression is Associated With Aggressive Clear Cell Renal Cell Carcinoma, Regardless of YAP1 Expression. Appl. Immunohistochem. Mol. Morphol. 2023, 31, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Li, N.; Sun, C.; Li, Z.; Xie, H. A Four-Gene Prognostic Signature Based on the TEAD4 Differential Expression Predicts Overall Survival and Immune Microenvironment Estimation in Lung Adenocarcinoma. Front. Pharmacol. 2022, 13, 874780. [Google Scholar] [CrossRef]
- Xu, A.; Wang, X.; Zeng, Y.; Zhou, M.; Yi, R.; Wu, Z.; Lin, J.; Song, Y. Overexpression of TEAD4 correlates with poor prognosis of glioma and promotes cell invasion. Int. J. Clin. Exp. Pathol. 2018, 11, 4827–4835. [Google Scholar]
- Oka, T.; Mazack, V.; Sudol, M. Mst2 and Lats Kinases Regulate Apoptotic Function of Yes Kinase-associated Protein (YAP). J. Biol. Chem. 2008, 283, 27534–27546. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Maglic, D.; Dill, M.T.; Mojumdar, K.; Ng, P.K.; Jeong, K.J.; Tsang, Y.H.; Moreno, D.; Bhavana, V.H.; et al. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep. 2018, 25, 1304–1317.e1305. [Google Scholar] [CrossRef]
- Brooks, H.L.; de Castro Brás, L.E.; Brunt, K.R.; Sylvester, M.A.; Parvatiyar, M.S.; Sirish, P.; Bansal, S.S.; Sule, R.; Eadie, A.L.; Knepper, M.A.; et al. Guidelines on antibody use in physiology research. Am. J. Physiol. Ren. Physiol. 2024, 326, F511–F533. [Google Scholar] [CrossRef]
- Whyte, C.E.; Tumes, D.J.; Liston, A.; Burton, O.T. Do more with Less: Improving High Parameter Cytometry Through Overnight Staining. Curr. Protoc. 2022, 2, e589. [Google Scholar] [CrossRef]
- Cacioppo, R.; Akman, H.B.; Tuncer, T.; Erson-Bensan, A.E.; Lindon, C. Differential translation of mRNA isoforms underlies oncogenic activation of cell cycle kinase Aurora A. eLife 2023, 12, RP87253. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Cao, C.; Kumar, M.; Sinha, S.; Chanda, A.; McNeil, R.; Samuel, D.; Arora, R.K.; Matthews, T.W.; Chandarana, S.; et al. Spatial transcriptomics reveals distinct and conserved tumor core and edge architectures that predict survival and targeted therapy response. Nat. Commun. 2023, 14, 5029. [Google Scholar] [CrossRef]
- Jing, S.-y.; Wang, H.-q.; Lin, P.; Yuan, J.; Tang, Z.-x.; Li, H. Quantifying and interpreting biologically meaningful spatial signatures within tumor microenvironments. NPJ Precis. Oncol. 2025, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Borreguero-Muñoz, N.; Fletcher, G.C.; Aguilar-Aragon, M.; Elbediwy, A.; Vincent-Mistiaen, Z.I.; Thompson, B.J. The Hippo pathway integrates PI3K-Akt signals with mechanical and polarity cues to control tissue growth. PLoS Biol. 2019, 17, e3000509. [Google Scholar] [CrossRef]
- Miyanaga, A.; Masuda, M.; Tsuta, K.; Kawasaki, K.; Nakamura, Y.; Sakuma, T.; Asamura, H.; Gemma, A.; Yamada, T. Hippo Pathway Gene Mutations in Malignant Mesothelioma: Revealed by RNA and Targeted Exon Sequencing. J. Thorac. Oncol. 2015, 10, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Tetreault, M.; Bareke, E.; Nadaf, J.; Alirezaie, N.; Majewski, J. Whole-exome sequencing as a diagnostic tool: Current challenges and future opportunities. Expert Rev. Mol. Diagn. 2015, 15, 749–760. [Google Scholar] [CrossRef]
- Vashisht, V.; Vashisht, A.; Mondal, A.K.; Woodall, J.; Kolhe, R. From Genomic Exploration to Personalized Treatment: Next-Generation Sequencing in Oncology. Curr. Issues Mol. Biol. 2024, 46, 12527–12549. [Google Scholar] [CrossRef]
- Lao, Z.; Chen, X.; Pan, B.; Fang, B.; Yang, W.; Qian, Y. Pharmacological regulators of Hippo pathway: Advances and challenges of drug development. FASEB J. 2025, 39, e70438. [Google Scholar] [CrossRef]
- Guo, S.; Huang, C.; Shrishrimal, S.; Cui, J.; Zhang, V.; Deng, N.; Dong, I.; Wang, G.; Begley, C.G.; Luo, S.; et al. Covalent pan-TEAD inhibitors for the treatment of cancers with Hippo pathway alterations. Eur. J. Cancer 2022, 174, S41. [Google Scholar] [CrossRef]
- Hagenbeek, T.J.; Zbieg, J.R.; Hafner, M.; Mroue, R.; Lacap, J.A.; Sodir, N.M.; Noland, C.L.; Afghani, S.; Kishore, A.; Bhat, K.P.; et al. An allosteric pan-TEAD inhibitor blocks oncogenic YAP/TAZ signaling and overcomes KRAS G12C inhibitor resistance. Nat. Cancer 2023, 4, 812–828. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Huang, H.Y.; Lin, Z.; Ranieri, M.; Li, S.; Sahu, S.; Liu, Y.; Ban, Y.; Guidry, K.; Hu, H.; et al. Genome-Wide CRISPR Screens Identify Multiple Synthetic Lethal Targets That Enhance KRASG12C Inhibitor Efficacy. Cancer Res. 2023, 83, 4095–4111. [Google Scholar] [CrossRef] [PubMed]
- Chapeau, E.A.; Sansregret, L.; Galli, G.G.; Chène, P.; Wartmann, M.; Mourikis, T.P.; Jaaks, P.; Baltschukat, S.; Barbosa, I.A.M.; Bauer, D.; et al. Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers. Nat. Cancer 2024, 5, 1102–1120. [Google Scholar] [CrossRef] [PubMed]
- Elm, L.; Levidou, G. Deciphering Molecular Alterations of the Hippo Signaling Pathway in Thymic Epithelial Tumors Using Immunohistochemistry. In Proceedings of the Oral presentation at the the 3rd International Electronic Conference on Biomedicines, Online, 12–15 May 2025; MDPI: Basel, Switzerland, 2025. [Google Scholar]





| Molecule | Immunolocalization | Tumor Size | WHO Histological Types | Masaoka-Koga Stage | Survival Analysis |
|---|---|---|---|---|---|
| MST1 | Cytoplasmic (96.1%) | No significant correlation (Spearman’s correlation coefficient, p = 0.152) | Higher expression in B3/TC (Mann–Whitney U test, p = 0.032) | Higher expression in III/IV (Mann–Whitney U test, p = 0.002) | No significant correlation (log-rank, >10% versus ≥10%, p = 0.99) |
| SAV1 | Cytoplasmic (100%) | No significant correlation (Spearman’s correlation coefficient, p = 0.237) | More uniformly high expression in B3/TC (Fisher’s exact test, p = 0.025) | Higher expression in III/IV (Fisher’s exact test, I/II versus III/IV, p = 0.027) | No significant correlation (log-rank, <100% versus 100%, p = 0.479) |
| LATS1 | Cytoplasmic (100%) | No significant correlation (Mann–Whitney U test, p = 0.566) | Higher expression in B3/TC (Mann–Whitney U test, p < 0.001) | Higher expression in III/IV (Mann–Whitney U test, p = 0.007) | No significant correlation (log-rank, p = 0.838) |
| MOB1A | Cytoplasmic (100%) | No significant correlation (Mann–Whitney U test, p = 0.389) | Higher in B3/TC (Mann–Whitney U test, marginal, p = 0.063) | Higher in III/IV (Mann–Whitney U test, I/II versus III/IV, p = 0.033) | No significant correlation (log-rank, >100% versus 100%, p = 0.592) |
| YAP1 | Nuclear (90.9%), Cytoplasmic (63.4%) | No significant correlation (Mann–Whitney U test, nuclear: p = 0.870; cytoplasmic: p = 0.144) | Lower nuclear expression in B3/TC (Mann–Whitney U test, p = 0.010) | Lower nuclear in III/IV (Mann–Whitney U test, I/II versus III/IV, p = 0.023); higher cytoplasmic in III/IV (Mann-Whitney U test, I versus II/III/IV, p = 0.032) | No significant correlation (nuclear: log-rank, <40% versus ≥40%, p = 0.323; cytoplasmic: <15% versus ≥15%, p = 0.468) |
| AYAP | Nuclear (93.5%), Cytoplasmic (75.3%) | Nuclear and cytoplasmic: no significant correlation (Mann–Whitney U test, p = 0.131) | Lower nuclear expression in TC (Mann–Whitney U test, p < 0.001); highest cytoplasmic expression in A/AB (Kruskal–Wallis ANOVA, p = 0.011) | Lower nuclear in III/IV (Mann–Whitney U test, I/II versus III/IV, p = 0.007) | No significant correlation (nuclear: log-rank, <40% versus ≥40%, p = 0.296; cytoplasmic: <30% versus ≥30%, p = 0.614) |
| TAZ | Nuclear (68.8%), Cytoplasmic (51.9%) | No significant correlation (Mann–Whitney U test, nuclear: p > 0.999; cytoplasmic: p = 0.123) | Nuclear higher in A/AB (Kruskal–Wallis ANOVA, p = 0.004); cytoplasmic lower in B3/TC (Mann–Whitney U test, p = 0.004) | No significant correlation (nuclear: Mann–Whitney U test, I/II versus III/IV, p = 0.182; cytoplasmic: Mann–Whitney U test, I/II versus III/IV, p = 0.281) | No significant correlation ( nuclear: log-rank, <10% versus ≥10%, p = 0.951; cytoplasmic: log-rank, <2% versus ≥2%, p = 0.218) |
| TEAD4 | Nuclear (63.6%), Cytoplasmic (94.8%) | No significant correlation (Mann–Whitney U test, nuclear: p = 0.348; cytoplasmic: p = 0.222) | Higher cytoplasmic in TCs (Mann–Whitney U test, p = 0.002); higher nuclear in A/AB and TCs (Kruskal–Wallis ANOVA, p = 0.005) | Higher cytoplasmic in III/IV (Mann–Whitney U test, I/II versus III/IV, p < 0.001) | Nuclear: no siginifcant correlation (log-rank, <15% versus ≥15%, p = 0.850) Worse OS if cytoplasmic ≥70% (log-rank, p = 0.003) |
| n- MST1 | c-MST1 | c-SAV1 | c-LATS1 | c-MOB1A | n-TAZ | c-TAZ | n-YAP1 | c-YAP1 | n-AYAP | c-AYAP | n-TEAD4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c-MST1 | R = −0.19 p = 0.095 | |||||||||||
| c-SAV1 | R = 0.048 p = 0.68 | R = 0.19 p = 0.096 | ||||||||||
| c-LATS1 | R = −0.14 p = 0.206 | R = 0.45 p < 0.001 | R = 0.33 p = 0.003 | |||||||||
| c-MOB1A | R = 0.09 p = 0.441 | R = 0.24 p = 0.032 | R = 0.05 p = 0.624 | R = 0.34 p = 0.002 | ||||||||
| n-TAZ | R = 0.12 p = 0.275 | R = −0.08 p = 0.459 | R = −0.10 p = 0.366 | R = 0.25 p = 0.025 | R = 0.02 p = 0.814 | |||||||
| c-TAZ | R = 0.13 p = 0.246 | R = 0.24 p = 0.038 | R = 0.01 p = 0.919 | R = 0.22 p = 0.056 | R = 0.23 p = 0.042 | R = 0.19 p = 0.097 | ||||||
| n-YAP1 | R = −0.02 p = 0.816 | R = −0.21 p = 0.070 | R = −0.21 p = 0.064 | R = −0.05 p = 0.631 | R = 0.04 p = 0.729 | R = 0.30 p = 0.007 | R = 0.32 p = 0.004 | |||||
| c-YAP1 | R = −0.13 p = 0.244 | R = 0.11 p = 0.340 | R = −0.02 p = 0.857 | R = 0.35 p = 0.001 | R = 0.23 p = 0.044 | R = 0.13 p = 0.239 | R = 0.27 p = 0.016 | R = 0.36 p = 0.001 | ||||
| n-AYAP | R = 0.16 p = 0.16 | R = −0.20 p = 0.07 | R = −0.20 p = 0.080 | R = −0.02 p = 0.826 | R = 0.05 p = 0.653 | R = 0.37 p = 0.001 | R = 0.44 p < 0.001 | R = 0.80 p < 0.001 | R = 0.30 p = 0.009 | |||
| C-AYAP | R = 0.08 p = 0.497 | R = −0.01 p = 0.872 | R = −0.03 p = 0.788 | R = 0.29 p = 0.010 | R = 0.29 p = 0.009 | R = 0.28 p = 0.013 | R = 0.23 p = 0.047 | R = 0.45 p < 0.001 | R = 0.68 p < 0.001 | R = 0.58 p < 0.001 | ||
| n-TEAD4 | R = −0.01 p = 0.867 | R = 0.13 p = 0.265 | R = −0.01 p = 0.892 | R = 0.36 p = 0.001 | R = 0.01 p = 0.940 | R = 0.34 p = 0.002 | R = 0.06 p = 0.600 | R = 0.07 p = 0.526 | R = 0.24 p = 0.038 | R = 0.07 p = 0.519 | R = 0.13 p = 0.245 | |
| c-TEAD4 | R = 0.08 p = 0.468 | R = 0.31 p = 0.005 | R = 0.13 p = 0.265 | R = 0.62 p < 0.001 | R = 0.29 p = 0.012 | R = 0.25 p = 0.029 | R = 0.11 p = 0.318 | R = −0.04 p = 0.706 | R = 0.27 p = 0.017 | R = −0.07 p = 0.523 | R = 0.24 p = 0.031 | R = 0.31 p = 0.007 |
| Parameter | Median | Min–Max |
|---|---|---|
| Age (years) | 69 | 21–88 |
| Tumor size (cm) | 6.5 | 0.9–14 |
| Number | % | |
| Gender | ||
| Male | 39 | 50.6 |
| Female | 38 | 49.4 |
| WHO subtypes | ||
| Type A | 3 | 3.9 |
| Type AB | 26 | 33.7 |
| Type B1 | 7 | 9 |
| Type B2 | 11 | 14.3 |
| Type B3 | 14 | 18.2 |
| Micronodular with lymphoid stroma (MNT) | 2 | 2.6 |
| Thymic carcinoma (TC) | 14 | 18.2 |
| Masaoka–Koga stage | ||
| I | 22 | 33.8 |
| II | 25 | 38.5 |
| III | 9 | 13.8 |
| IVa | 5 | 7.7 |
| IVb | 4 | 6.1 |
| Presence of myasthenia Gravis | 11 | 14.1 |
| Event | ||
| Cencored-Alive | 51/60, follow-up 0.3–109, 4 months | 85 |
| Dead | 9/60, within 0.1–48.6 months | 15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elm, L.; Gerlitz, N.; Hochholzer, A.; Papadopoulos, T.; Levidou, G. Hippo Pathway Dysregulation in Thymic Epithelial Tumors (TETs): Associations with Clinicopathological Features and Patients’ Prognosis. Int. J. Mol. Sci. 2025, 26, 5938. https://doi.org/10.3390/ijms26135938
Elm L, Gerlitz N, Hochholzer A, Papadopoulos T, Levidou G. Hippo Pathway Dysregulation in Thymic Epithelial Tumors (TETs): Associations with Clinicopathological Features and Patients’ Prognosis. International Journal of Molecular Sciences. 2025; 26(13):5938. https://doi.org/10.3390/ijms26135938
Chicago/Turabian StyleElm, Lisa, Nadja Gerlitz, Anke Hochholzer, Thomas Papadopoulos, and Georgia Levidou. 2025. "Hippo Pathway Dysregulation in Thymic Epithelial Tumors (TETs): Associations with Clinicopathological Features and Patients’ Prognosis" International Journal of Molecular Sciences 26, no. 13: 5938. https://doi.org/10.3390/ijms26135938
APA StyleElm, L., Gerlitz, N., Hochholzer, A., Papadopoulos, T., & Levidou, G. (2025). Hippo Pathway Dysregulation in Thymic Epithelial Tumors (TETs): Associations with Clinicopathological Features and Patients’ Prognosis. International Journal of Molecular Sciences, 26(13), 5938. https://doi.org/10.3390/ijms26135938







