Adapting to UV: Integrative Genomic and Structural Analysis in Bacteria from Chilean Extreme Environments
Abstract
1. Introduction
2. Results and Discussion
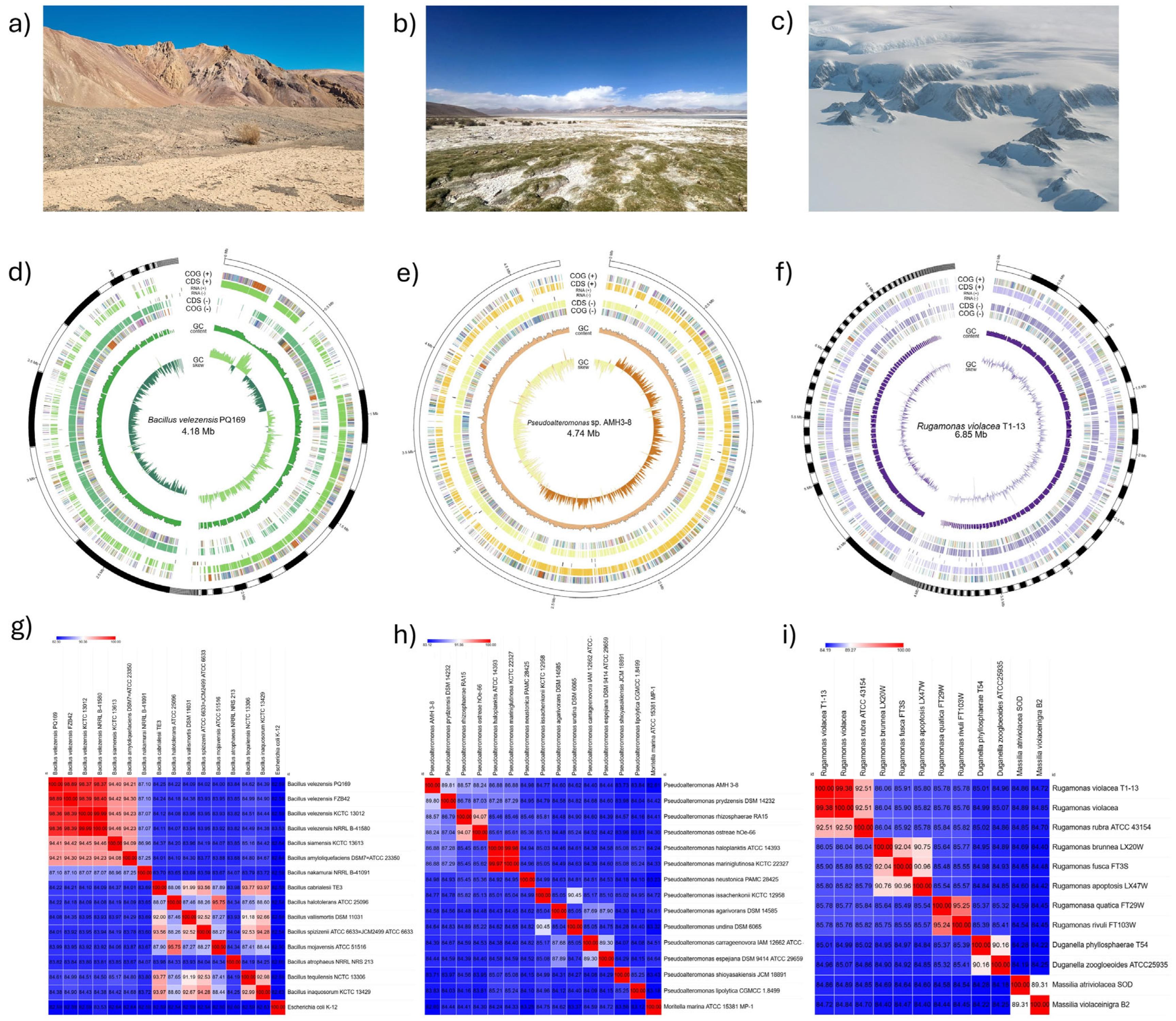
2.1. Genome Assembly and ANI-Based Taxonomic Assignment
2.2. Gene-Content Analysis and DNA Repair-Pathway Components
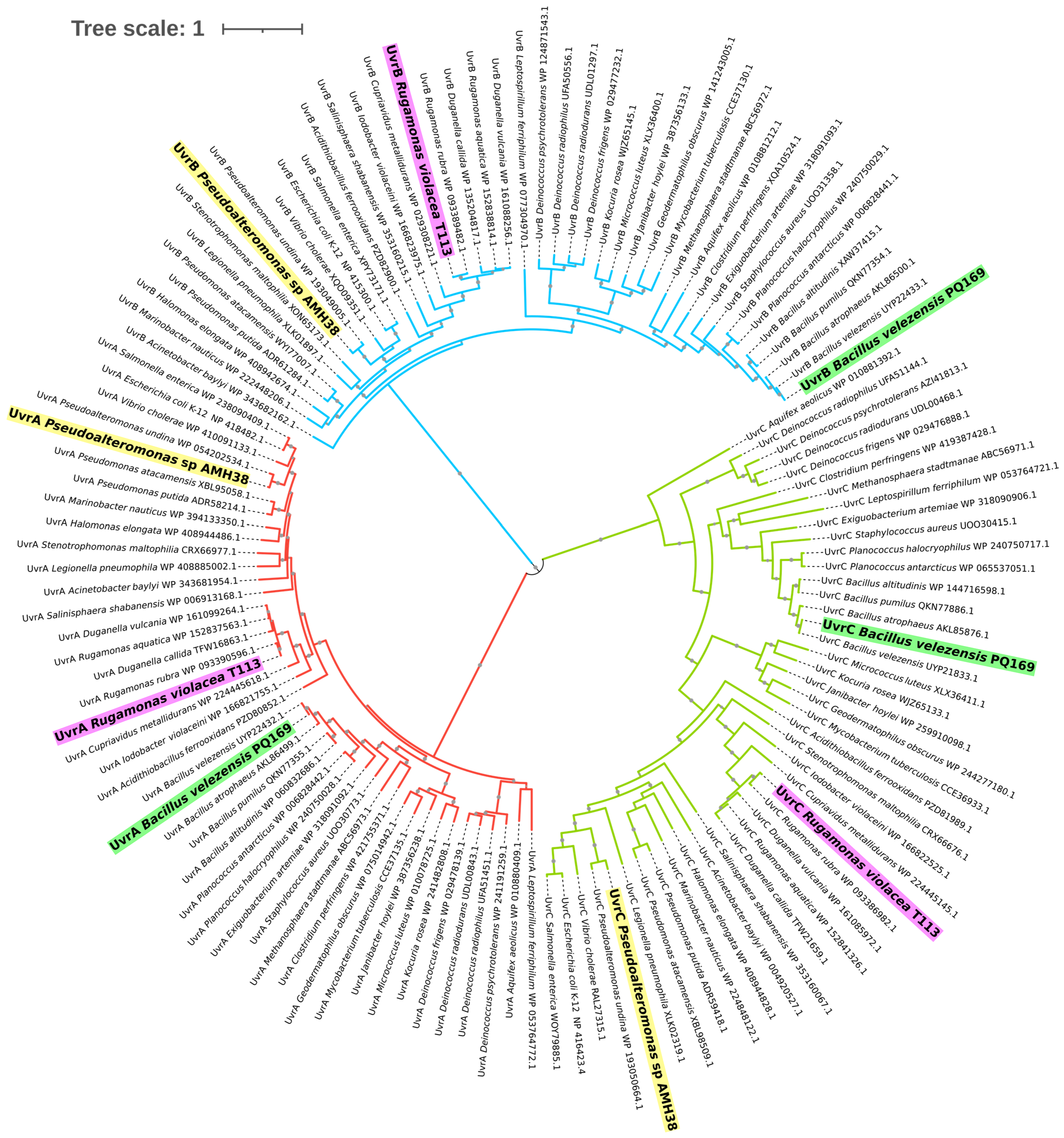
2.2.1. Phylogenetic Analysis of the Nucleotide Excision Repair (NER) System
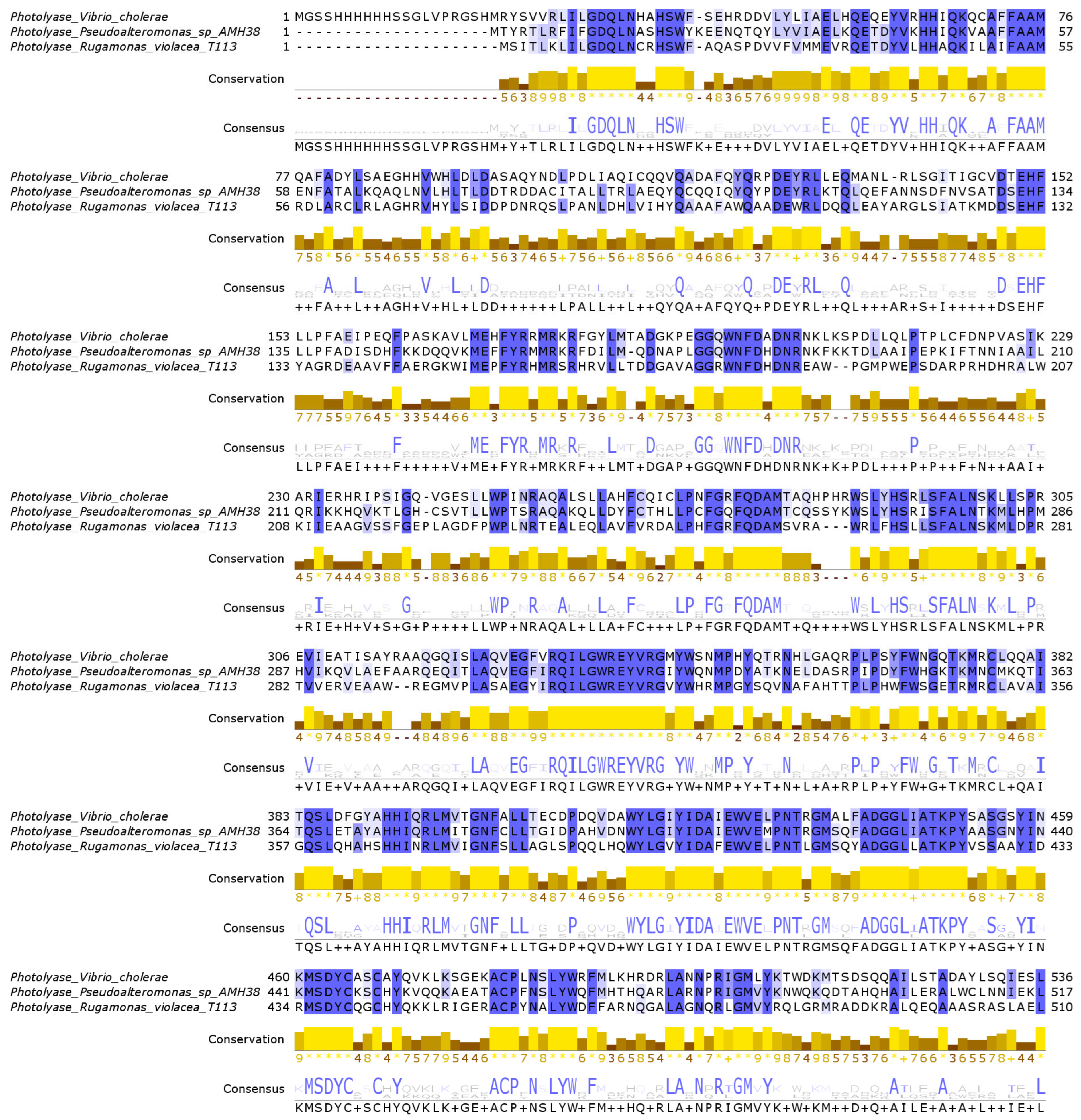
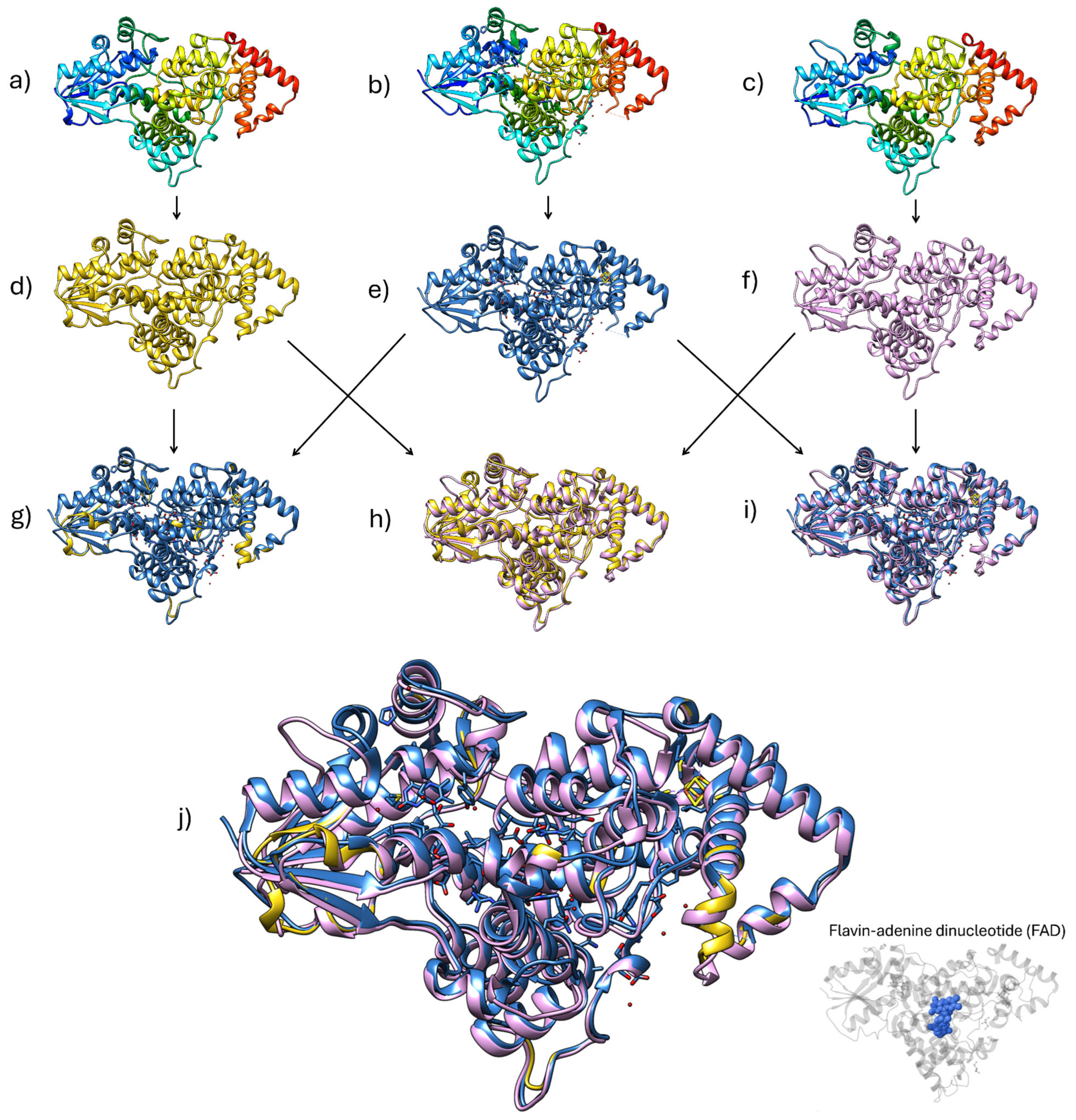
2.2.2. Photolyases and Structural Insights
2.3. Pigment-Based Mechanisms
2.4. Sporulation as an Adaptive Response
3. Materials and Methods
3.1. Bacterial Isolates and Genomic DNA Extraction
3.2. Genome Sequencing, Assembly, Annotation, and ANIm-Based Taxonomic Analysis
3.3. Phylogenetic Analysis of UV Resistance-Related Proteins
3.4. Protein Structure Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rampelotto, P.H. Extremophiles and extreme environments. Life 2013, 3, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Liu, G.; Zhang, G.; Li, X. Radiation-resistant bacteria in desiccated soil and their potentiality in applied sciences. Front. Microbiol. 2024, 15, 1348758. [Google Scholar] [CrossRef] [PubMed]
- Martin-Cuadrado, A.B.; Senel, E.; Martínez-García, M.; Cifuentes, A.; Santos, F.; Almansa, C.; Moreno-Paz, M.; Blanco, Y.; García-Villadangos, M.; García del Cura, M.A.; et al. Prokaryotic and viral community of the sulfate-rich crust from Peñahueca ephemeral lake, an astrobiology analogue. Environ. Microbiol. 2019, 21, 3577–3600. [Google Scholar] [CrossRef] [PubMed]
- Cordero, R.R.; Damiani, A.; Jorquera, J.; Sepúlveda, E.; Caballero, M.; Fernandez, S.; Feron, S.; Llanillo, P.J.; Carrasco, J.; Laroze, D.; et al. Ultraviolet radiation in the Atacama desert. Antonie Leeuwenhoek 2018, 111, 1301–1313. [Google Scholar] [CrossRef]
- Azua-Bustos, A.; González-Silva, C.; Fairén, A.G. The Atacama Desert in northern Chile as an analog model of Mars. Front. Astron. Space Sci. 2022, 8, 810426. [Google Scholar] [CrossRef]
- Azua-Bustos, A.; Urrejola, C.; Vicuña, R. Life at the dry edge: Microorganisms of the Atacama Desert. FEBS Lett. 2012, 586, 2939–2945. [Google Scholar] [CrossRef]
- Castro-Severyn, J.; Remonsellez, F.; Valenzuela, S.L.; Salinas, C.; Fortt, J.; Aguilar, P.; Pardo-Esté, C.; Dorador, C.; Quatrini, R.; Molina, F.; et al. Comparative genomics analysis of a new Exiguobacterium strain from Salar de Huasco reveals a repertoire of stress-related genes and arsenic resistance. Front. Microbiol. 2017, 8, 456. [Google Scholar] [CrossRef]
- Wierzchos, J.; de los Ríos, A.; Ascaso, C. Microorganisms in desert rocks: The edge of life on earth. Int. Microbiol. 2012, 15, 173–183. [Google Scholar] [CrossRef]
- Castro-Severyn, J.; Pardo-Esté, C.; Mendez, K.N.; Fortt, J.; Marquez, S.; Molina, F.; Castro-Nallar, E.; Remonsellez, F.; Saavedra, C.P. Living to the high extreme: Unraveling the composition, structure, and functional insights of bacterial communities thriving in the arsenic-rich Salar de Huasco altiplanic ecosystem. Microbiol. Spectr. 2021, 9, e0044421. [Google Scholar] [CrossRef]
- Hernández, K.L.; Yannicelli, B.; Olsen, L.M.; Dorador, C.; Menschel, E.J.; Molina, V.; Remonsellez, F.; Hengst, M.B.; Jeffrey, W.H. Microbial activity response to solar radiation across contrasting environmental conditions in Salar de Huasco, northern Chilean Altiplano. Front. Microbiol. 2016, 7, 1857. [Google Scholar] [CrossRef]
- Pérez, V.; Dorador, C.; Molina, V.; Yáñez, C.; Hengst, M. Rhodobacter sp. Rb3, an aerobic anoxygenic phototroph which thrives in the polyextreme ecosystem of the Salar de Huasco, in the Chilean Altiplano. Antonie Leeuwenhoek 2018, 111, 1449–1465. [Google Scholar] [CrossRef] [PubMed]
- Salwan, R.; Sharma, V. Genomics of prokaryotic extremophiles to unfold the mystery of survival in extreme environments. Microbiol. Res. 2022, 264, 127156. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, G.N.; Psarologaki, C.; Pirintsos, S.; Kotzabasis, K. Extremophiles and extremophilic behaviour-new insights and perspectives. Life 2024, 14, 1425. [Google Scholar] [CrossRef] [PubMed]
- Cordero, R.R.; Feron, S.; Damiani, A.; Redondas, A.; Carrasco, J.; Sepúlveda, E.; Jorquera, J.; Fernandoy, F.; Llanillo, P.; Rowe, P.M.; et al. Persistent extreme ultraviolet irradiance in Antarctica despite the ozone recovery onset. Sci. Rep. 2022, 12, 1266. [Google Scholar] [CrossRef]
- He, Y.; Qu, C.; Zhang, L.; Miao, J. DNA photolyase from Antarctic marine bacterium Rhodococcus sp. NJ-530 can repair DNA damage caused by ultraviolet. 3 Biotech 2021, 11, 102. [Google Scholar] [CrossRef]
- Marizcurrena, J.J.; Morel, M.A.; Braña, V.; Morales, D.; Martinez-López, W.; Castro-Sowinski, S. Searching for novel photolyases in UVC-resistant Antarctic bacteria. Extremophiles 2017, 21, 409–418. [Google Scholar] [CrossRef]
- Silva, T.R.E.; Silva, L.C.F.J.; de Queiroz, A.C.; Alexandre Moreira, M.S.; de Carvalho Fraga, C.A.; de Menezes, G.C.A.; Rosa, L.H.; Bicas, J.; de Oliveira, V.M.; Duarte, A.W.F. Pigments from Antarctic bacteria and their biotechnological applications. Crit. Rev. Biotechnol. 2021, 41, 809–826. [Google Scholar] [CrossRef]
- Orellana, R.; Macaya, C.; Bravo, G.; Dorochesi, F.; Cumsille, A.; Valencia, R.; Rojas, C.; Seeger, M. Living at the frontiers of life: Extremophiles in Chile and their potential for bioremediation. Front. Microbiol. 2018, 9, 2309. [Google Scholar] [CrossRef]
- Meslier, V.; DiRuggiero, J. Endolithic microbial communities as model systems for ecology and astrobiology. In Model Ecosystems in Extreme Environments; Acdemic Press (Elsevier): Cambridge, MA, USA, 2019; pp. 145–168. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Kim, S.-J.; Kwon, S.-W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar] [CrossRef]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The Gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef]
- Parrilli, E.; Tedesco, P.; Fondi, M.; Tutino, M.L.; Lo Giudice, A.; de Pascale, D.; Fani, R. The art of adapting to extreme environments: The model system Pseudoalteromonas. Phys. Life Rev. 2021, 36, 137–161. [Google Scholar] [CrossRef] [PubMed]
- Sedláček, I.; Holochová, P.; Sobotka, R.; Busse, H.-J.; Švec, P.; Králová, S.; Šedo, O.; Pilný, J.; Staňková, E.; Koublová, V.; et al. Classification of a violacein-producing psychrophilic group of isolates associated with freshwater in Antarctica and description of Rugamonas violacea sp. Nov. Microbiol. Spectr. 2021, 9, e0045221. [Google Scholar] [CrossRef] [PubMed]
- Vechtomova, Y.L.; Telegina, T.A.; Buglak, A.A.; Kritsky, M.S. UV radiation in DNA damage and repair involving DNA-photolyases and cryptochromes. Biomedicines 2021, 9, 1564. [Google Scholar] [CrossRef]
- Goosen, N.; Moolenaar, G.F. Repair of UV damage in bacteria. DNA Repair 2008, 7, 353–379. [Google Scholar] [CrossRef]
- Morita, R.; Nakane, S.; Shimada, A.; Inoue, M.; Iino, H.; Wakamatsu, T.; Fukui, K.; Nakagawa, N.; Masui, R.; Kuramitsu, S. Molecular mechanisms of the whole DNA repair system: A comparison of bacterial and eukaryotic systems. J. Nucleic Acids 2010, 2010, 179594. [Google Scholar] [CrossRef]
- Van Houten, B.; Kad, N. Investigation of bacterial nucleotide excision repair using single-molecule techniques. DNA Repair 2014, 20, 41–48. [Google Scholar] [CrossRef]
- Truglio, J.J.; Croteau, D.L.; Van Houten, B.; Kisker, C. Prokaryotic nucleotide excision repair: The UvrABC system. Chem. Rev. 2006, 106, 233–252. [Google Scholar] [CrossRef]
- Riesenman, P.J.; Nicholson, W.L. Role of the spore coat layers in Bacillus subtilis spore resistance to hydrogen peroxide, artificial UV-C, UV-b, and solar UV radiation. Appl. Environ. Microbiol. 2000, 66, 620–626. [Google Scholar] [CrossRef]
- Davis, G.M.; Greenhill, A.R.; Munro, T.W.; Horner, J.; Marsden, S.C.; Carter, B.D. Partial protection of Bacillus subtilis spores on simulated martian media enhances survival against UV radiation. Discov. Space 2025, 129, 3. [Google Scholar] [CrossRef]
- Mao, Y.; Yang, Y.; Lin, F.; Chu, H.; Zhou, L.; Han, J.; Zhou, J.; Su, X. Functional analysis of stress resistance of Bacillus cereus SCL10 strain based on whole-genome sequencing. Microorganisms 2024, 12, 1168. [Google Scholar] [CrossRef]
- Suárez, V.P.; Martínez, L.E.; Leyva-Sánchez, H.C.; Valenzuela-García, L.I.; Lara-Martínez, R.; Jiménez-García, L.F.; Ramírez-Ramírez, N.; Obregon-Herrera, A.; Cuéllar-Cruz, M.; Robleto, E.A.; et al. Transcriptional coupling and repair of 8-OxoG activate a RecA-dependent checkpoint that controls the onset of sporulation in Bacillus subtilis. Sci. Rep. 2021, 11, 2513. [Google Scholar] [CrossRef] [PubMed]
- Gaasbeek, E.J.; van der Wal, F.J.; van Putten, J.P.M.; de Boer, P.; van der Graaf-van Bloois, L.; de Boer, A.G.; Vermaning, B.J.; Wagenaar, J.A. Functional characterization of excision repair and RecA-dependent recombinational DNA repair in Campylobacter jejuni. J. Bacteriol. 2009, 191, 3785–3793. [Google Scholar] [CrossRef] [PubMed]
- Briaud, P.; Gautier, T.; Rong, V.; Mereghetti, L.; Lanotte, P.; Hiron, A. The Streptococcus agalactiae exonuclease ExoVII is required for resistance to exogenous DNA-damaging agents. J. Bacteriol. 2023, 205, e0002423. [Google Scholar] [CrossRef] [PubMed]
- On, Y.Y.; Welch, M. The methylation-independent mismatch repair machinery in Pseudomonas aeruginosa. Microbiology 2021, 167, 001120. [Google Scholar] [CrossRef]
- Blanchard, L.; de Groot, A. Coexistence of SOS-dependent and SOS-independent regulation of DNA repair genes in radiation-resistant Deinococcus bacteria. Cells 2021, 10, 924. [Google Scholar] [CrossRef]
- de Groot, A.; Blanchard, L. DNA repair and oxidative stress defense systems in radiation-resistant Deinococcus murrayi. Can. J. Microbiol. 2023, 69, 416–431. [Google Scholar] [CrossRef]
- Kim, S.R.; Maenhaut-Michel, G.; Yamada, M.; Yamamoto, Y.; Matsui, K.; Sofuni, T.; Nohmi, T.; Ohmori, H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: An overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. USA 1997, 94, 13792–13797. [Google Scholar] [CrossRef]
- Crowley, D.J.; Boubriak, I.; Berquist, B.R.; Clark, M.; Richard, E.; Sullivan, L.; DasSarma, S.; McCready, S. The uvrA, uvrB and uvrC genes are required for repair of ultraviolet light induced DNA photoproducts in Halobacterium sp. NRC-1. Saline Syst. 2006, 2, 11. [Google Scholar] [CrossRef][Green Version]
- Au, N.; Kuester-Schoeck, E.; Mandava, V.; Bothwell, L.E.; Canny, S.P.; Chachu, K.; Colavito, S.A.; Fuller, S.N.; Groban, E.S.; Hensley, L.A.; et al. Genetic composition of the Bacillus subtilis SOS system. J. Bacteriol. 2005, 187, 7655–7666. [Google Scholar] [CrossRef]
- Subramani, G.; Srinivasan, S. Involvement of nucleotide excision repair and Rec-dependent pathway genes for UV radiation resistance in Deinococcus irradiatisoli 17bor-2. Genes 2023, 14, 1803. [Google Scholar] [CrossRef]
- Kurth, D.; Belfiore, C.; Gorriti, M.F.; Cortez, N.; Farias, M.E.; Albarracín, V.H. Genomic and proteomic evidences unravel the UV-resistome of the poly-extremophile Acinetobacter sp. Ver3. Front. Microbiol. 2015, 6, 328. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.; Schultz, J.; Almeida Trapp, M.; Modolon, F.; Romanenko, A.; Kumar Jaiswal, A.; Gomes, L.; Rodrigues-Filho, E.; Rosado, A.S. Investigating polyextremophilic bacteria in Al Wahbah crater, Saudi Arabia: A terrestrial model for life on Saturn’s moon Enceladus. Astrobiology 2024, 24, 824–838. [Google Scholar] [CrossRef] [PubMed]
- Kraithong, T.; Hartley, S.; Jeruzalmi, D.; Pakotiprapha, D. A peek inside the machines of bacterial nucleotide excision repair. Int. J. Mol. Sci. 2021, 22, 952. [Google Scholar] [CrossRef]
- Ellington, A.J.; Schult, T.J.; Reisch, C.R.; Christner, B.C. The genetic determinants of extreme UV radiation and desiccation tolerance in a bacterium recovered from the stratosphere. Microorganisms 2025, 13, 756. [Google Scholar] [CrossRef]
- Ramírez-Gamboa, D.; Díaz-Zamorano, A.L.; Meléndez-Sánchez, E.R.; Reyes-Pardo, H.; Villaseñor-Zepeda, K.R.; López-Arellanes, M.E.; Sosa-Hernández, J.E.; Coronado-Apodaca, K.G.; Gámez-Méndez, A.; Afewerki, S.; et al. Photolyase production and current applications: A review. Molecules 2022, 27, 5998. [Google Scholar] [CrossRef]
- Sancar, A. Structure and function of DNA photolyase. Biochemistry 1994, 33, 2–9. [Google Scholar] [CrossRef]
- Portero, L.R.; Alonso-Reyes, D.G.; Zannier, F.; Vazquez, M.P.; Farías, M.E.; Gärtner, W.; Albarracín, V.H. Photolyases and cryptochromes in UV-resistant bacteria from high-altitude Andean lakes. Photochem. Photobiol. 2019, 95, 315–330. [Google Scholar] [CrossRef]
- Walker, E.L.; Bose, J.L.; Stabb, E.V. Photolyase confers resistance to UV light but does not contribute to the symbiotic benefit of bioluminescence in Vibrio fischeri ES114. Appl. Environ. Microbiol. 2006, 72, 6600–6606. [Google Scholar] [CrossRef]
- Balaji, S.; Srinivasan, N. Comparison of sequence-based and structure-based phylogenetic trees of homologous proteins: Inferences on protein evolution. J. Biosci. 2007, 32, 83–96. [Google Scholar] [CrossRef]
- Pellegrini, M.; Marcotte, E.M.; Thompson, M.J.; Eisenberg, D.; Yeates, T.O. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA 1999, 96, 4285–4288. [Google Scholar] [CrossRef]
- Damm, K.L.; Carlson, H.A. Gaussian-weighted RMSD superposition of proteins: A structural comparison for flexible proteins and predicted protein structures. Biophys. J. 2006, 90, 4558–4573. [Google Scholar] [CrossRef] [PubMed]
- Magala, P.; Klevit, R.E.; Thomas, W.E.; Sokurenko, E.V.; Stenkamp, R.E. RMSD analysis of structures of the bacterial protein FimH identifies five conformations of its lectin domain. Proteins 2020, 88, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Kothari, V.; Sharma, S.; Padia, D. Recent research advances on Chromobacterium violaceum. Asian Pac. J. Trop. Med. 2017, 10, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Órdenes-Aenishanslins, N.; Anziani-Ostuni, G.; Vargas-Reyes, M.; Alarcón, J.; Tello, A.; Pérez-Donoso, J.M. Pigments from UV-resistant Antarctic bacteria as photosensitizers in dye sensitized solar cells. J. Photochem. Photobiol. B 2016, 162, 707–714. [Google Scholar] [CrossRef]
- Reis-Mansur, M.C.P.P.; Cardoso-Rurr, J.S.; Silva, J.V.M.A.; de Souza, G.R.; Cardoso, V.d.S.; Mansoldo, F.R.P.; Pinheiro, Y.; Schultz, J.; Lopez Balottin, L.B.; da Silva, A.J.R.; et al. Carotenoids from UV-resistant Antarctic Microbacterium sp. LEMMJ01. Sci. Rep. 2019, 9, 9554. [Google Scholar] [CrossRef]
- Guerrero M., G.G. Sporulation, structure assembly, and germination in the soil bacterium Bacillus thuringiensis: Survival and success in the environment and the insect host. Microbiol. Res. 2023, 14, 466–491. [Google Scholar] [CrossRef]
- Al-Hinai, M.A.; Jones, S.W.; Papoutsakis, E.T. The Clostridium sporulation programs: Diversity and preservation of endospore differentiation. Microbiol. Mol. Biol. Rev. 2015, 79, 19–37. [Google Scholar] [CrossRef]
- Fisher, P.R. Microbial development. In Encyclopedia of Molecular Cell Biology and Molecular Medicine; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2006. [Google Scholar] [CrossRef]
- Ramírez-Guadiana, F.H.; Barajas-Ornelas, R.d.C.; Corona-Bautista, S.U.; Setlow, P.; Pedraza-Reyes, M. The RecA-dependent SOS response is active and required for processing of DNA damage during Bacillus subtilis sporulation. PLoS ONE 2016, 11, e0150348. [Google Scholar] [CrossRef]
- Vlašić, I.; Mertens, R.; Seco, E.M.; Carrasco, B.; Ayora, S.; Reitz, G.; Commichau, F.M.; Alonso, J.C.; Moeller, R. Bacillus subtilis RecA and its accessory factors, RecF, RecO, RecR and RecX, are required for spore resistance to DNA double-strand break. Nucleic Acids Res. 2014, 42, 2295–2307. [Google Scholar] [CrossRef]
- Richardson, E.A.; Abruzzo, N.O.; Taylor, C.E.; Stevens, B.R.; Cuda, J.P.; Weeks, E.N.I. Combining DL-methionine and Bacillus thuringiensis subspecies israelensis: Prospects for a mosquito larvicide. Insects 2020, 11, 880. [Google Scholar] [CrossRef]
- Chen, W.P.; Kuo, T.T. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993, 21, 2260. [Google Scholar] [CrossRef]


| DNA-Repair Components | Function | B. velezensis PQ169 | Pseudoalteromonas sp. AMH3-8 | R. violacea T1-13 |
|---|---|---|---|---|
| UvrA | Initiates nucleotide excision repair (NER) by recognizing DNA damage. | ✓ | ✓ | ✓ |
| UvrB | Participates in NER by verifying DNA damage and forming the incision complex. | ✓ | ✓ | ✓ |
| UvrC | Makes incisions on both sides of the DNA lesion in NER. | ✓ | ✓ | ✓ |
| UvrD | DNA helicase that removes the damaged DNA strand during NER. | ✓ | ✓ | ✓ |
| Phr | DNA photolyase that directly reverses UV-induced pyrimidine dimers using light energy. | – | ✓ | ✓ |
| RecA | Promotes homologous recombination and SOS response by facilitating LexA autocleavage. | ✓ | ✓ | ✓ |
| LexA | Repressor of SOS response; inactivated by RecA to induce DNA-repair genes. | ✓ | ✓ | ✓ |
| MutS | Mismatch recognition protein involved in mismatch repair (MMR). | ✓ | ✓ | ✓ |
| MutL | Acts as a matchmaker in MMR by coordinating repair proteins. | ✓ | ✓ | ✓ |
| MutT | Prevents incorporation of oxidized guanine (8-oxo-dGTP) into DNA. | ✓ | – | ✓ |
| MutM | Removes 8-oxoguanine from DNA via base excision repair (BER). | ✓ | ✓ | ✓ |
| MutY | Removes adenines misincorporated opposite 8-oxoguanine (G:A mismatch repair). | ✓ | ✓ | ✓ |
| PolB | DNA polymerase II involved in translesion synthesis and repair. | – | ✓ | ✓ |
| DinB | DNA polymerase IV, error-prone translesion polymerase involved in bypassing UV lesions. | ✓ | ✓ | ✓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Núñez, M.; Naciff, A.; Cuadros, F.; Rojas, C.; Carvallo, G.; Yáñez, C. Adapting to UV: Integrative Genomic and Structural Analysis in Bacteria from Chilean Extreme Environments. Int. J. Mol. Sci. 2025, 26, 5842. https://doi.org/10.3390/ijms26125842
Núñez M, Naciff A, Cuadros F, Rojas C, Carvallo G, Yáñez C. Adapting to UV: Integrative Genomic and Structural Analysis in Bacteria from Chilean Extreme Environments. International Journal of Molecular Sciences. 2025; 26(12):5842. https://doi.org/10.3390/ijms26125842
Chicago/Turabian StyleNúñez, Mauricio, Antonia Naciff, Fabián Cuadros, Constanza Rojas, Gastón Carvallo, and Carolina Yáñez. 2025. "Adapting to UV: Integrative Genomic and Structural Analysis in Bacteria from Chilean Extreme Environments" International Journal of Molecular Sciences 26, no. 12: 5842. https://doi.org/10.3390/ijms26125842
APA StyleNúñez, M., Naciff, A., Cuadros, F., Rojas, C., Carvallo, G., & Yáñez, C. (2025). Adapting to UV: Integrative Genomic and Structural Analysis in Bacteria from Chilean Extreme Environments. International Journal of Molecular Sciences, 26(12), 5842. https://doi.org/10.3390/ijms26125842






