Two Peas in a Pod: Retroviral RNA Dimers Organize Gag–RNA Nanoclusters with Novel Biophysical Properties
Abstract
1. Introduction
2. Results
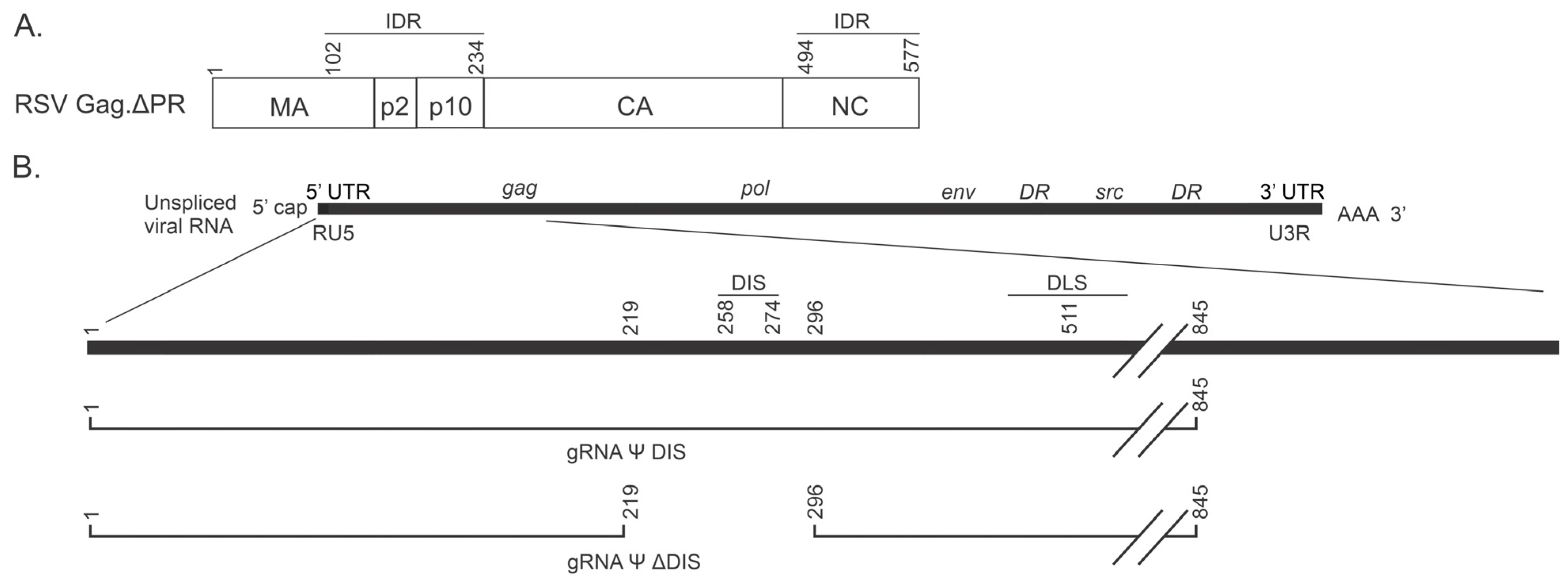
2.1. Biophysical Comparison of Similar Ψ-Containing Viral RNAs That Differ in Their Ability to Dimerize
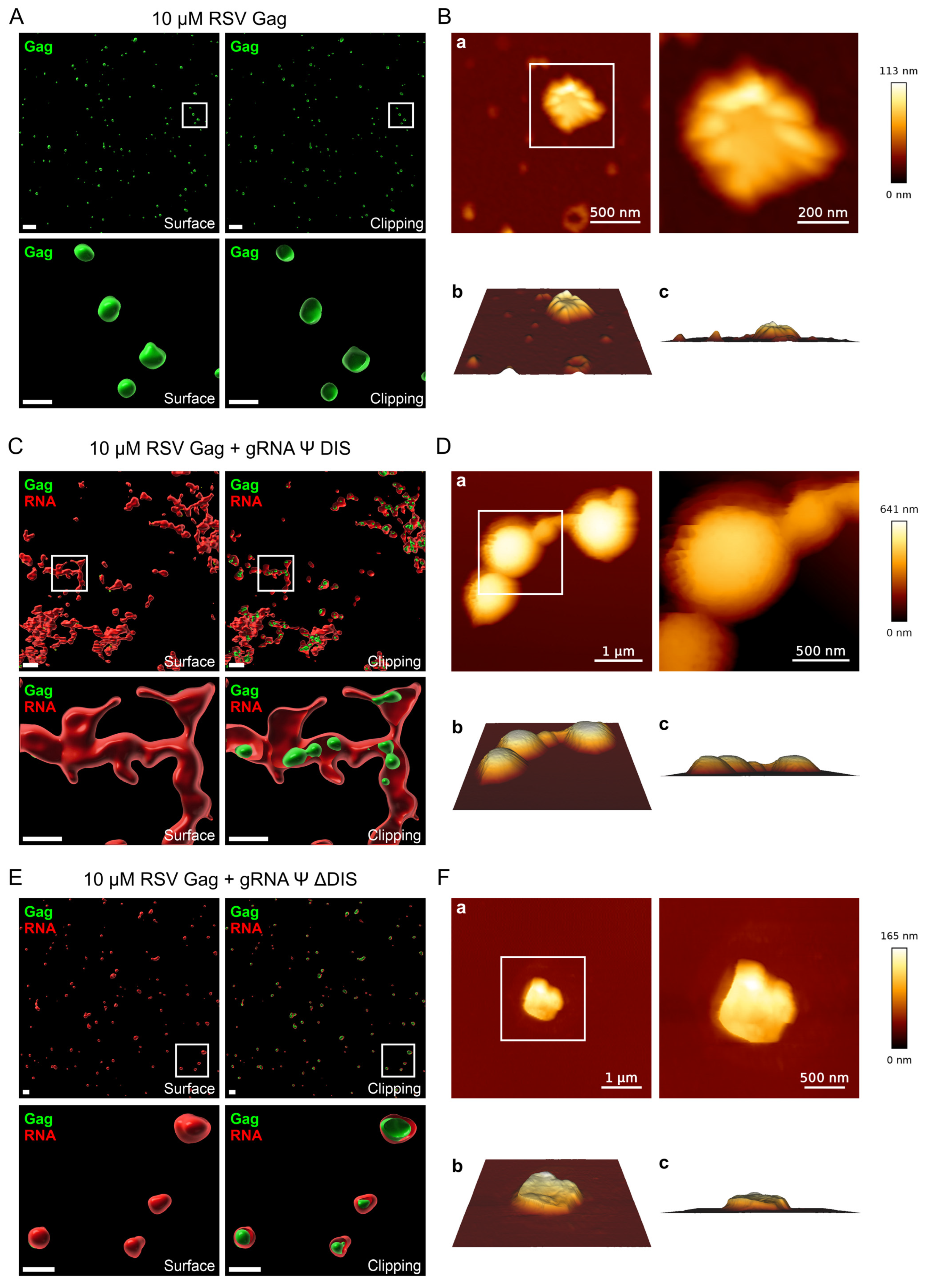
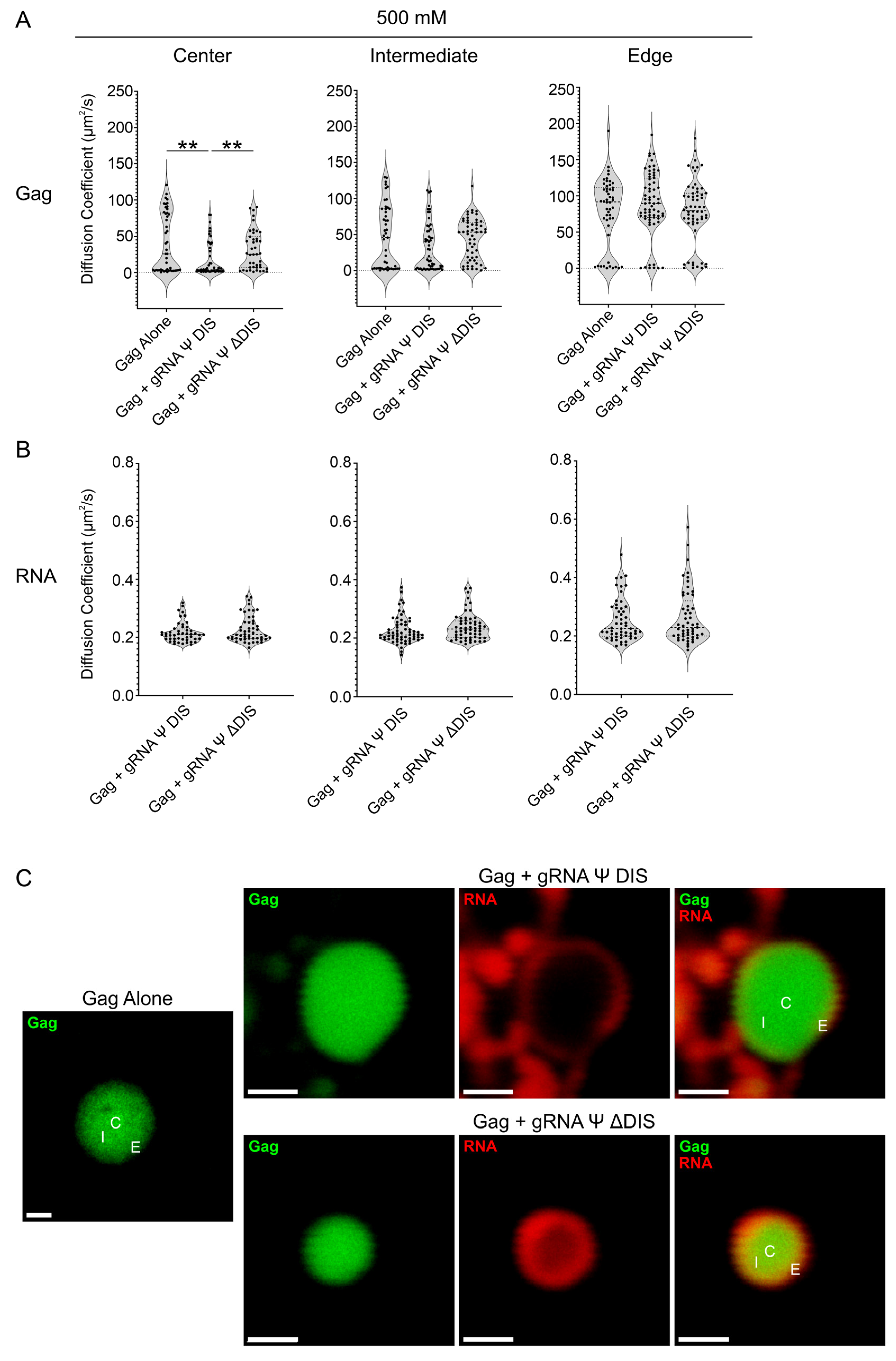
2.2. Assessing the Nanoscale Architecture of RSV Gag–vRNA Co-Condensates
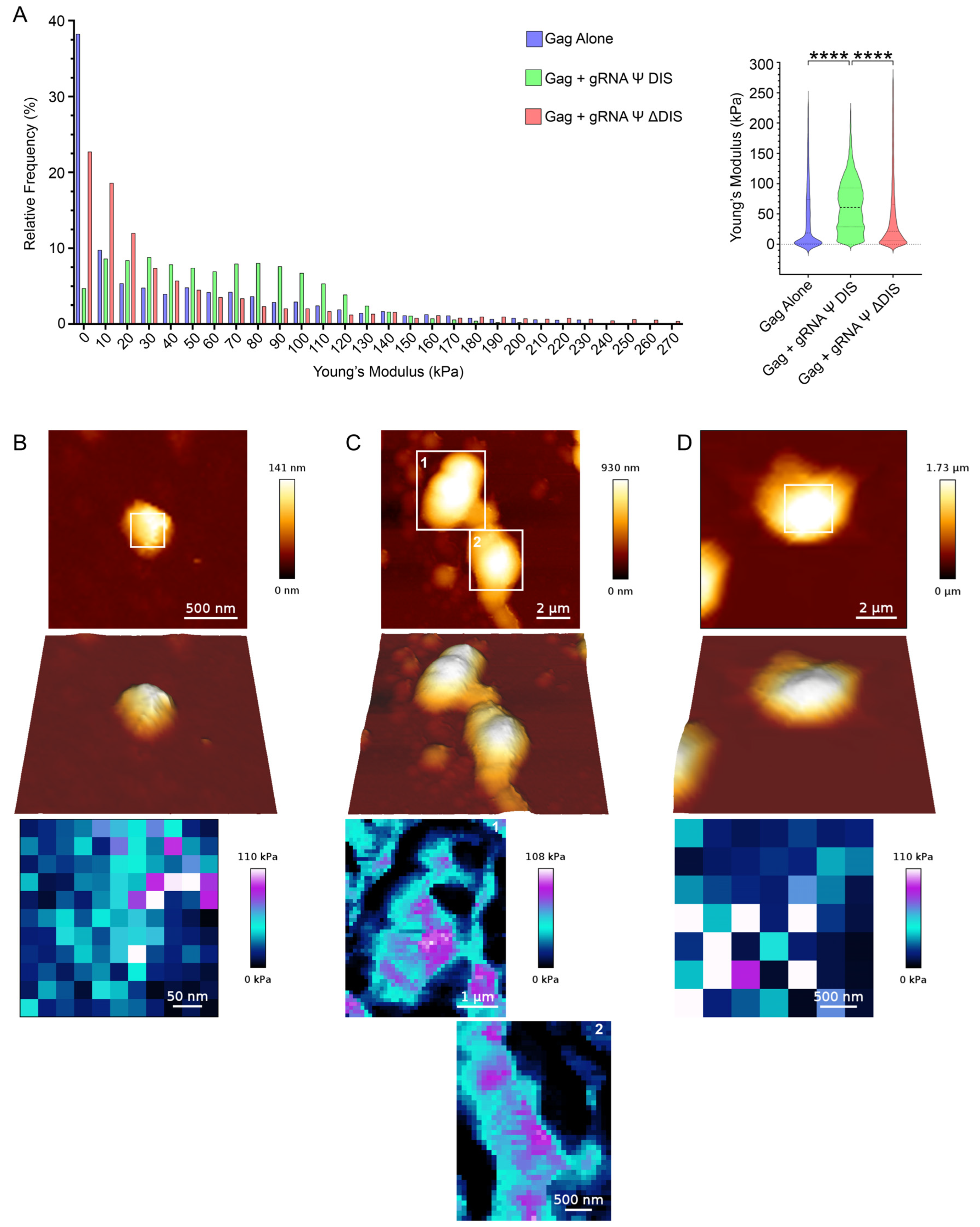
2.3. Viral RNA Dimerization Promotes the Formation of Heterogeneous Condensate Assemblies Displaying Increased Resistance to Mechanical Deformation
2.4. Viral RNA Dimerization Results in the Formation of RNA Scaffolds
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Gag Protein Expression, Purification, and Fluorescent Labeling
4.3. RNA Synthesis by In Vitro Transcription and Fluorescent Labeling
4.4. RNA Sequences
- gRNA Ψ DIS: 5′ GCCAUUUGACCAUUCACCACAUUGGUGUGCACCUGGGUUGAUGGCCGGACCGUUGAUUCCCUGACGACUACGAGCACCUGCAUGAAGCAGAAGGCUUCAUUUGGUGACCCCGACGUGAUAGUUAGGGAAUAGUGGUCGGCCACAGACGGCGUGGCGAUCCUGUCUCCAUCCGUCUCGUCUAUCGGGAGGCGAGUUCGAUGACCCUGGUGGAGGGGGCUGCGGCUUAGGGAGGCAGAAGCUGAGUACCGUCGGAGGGAGCUCUACUGCAGGGAGCCCAGAUACCCUACCGAGAACUCAGAGAGUCGUUGGAAGACGGGAAGGAAGCCCGACGACUGAGCAGUCCACCCCAGGCGUGAUUCUGGUCGCCCGGUGGAUCAAGCAUGGAAGCCGUCAUAAAGGUGAUUUCGUCCGCGUGUAAAACCUAUUGCGGGAAAACCUCUCCUUCUAAGAAGGAAAUAGGGGCCAUGUUGUCCCUCUUACAAAAGGAAGGGUUGCUUAUGUCUCCCUCAGACUUAUAUUCCCCGGGGUCCUGGGAUCCCAUUACCGCGGCGCUAUCCCAGCGGGCUAUGAUACUUGGGAAAUCGGGAGAGUUAAAAACCUGGGGAUUGGUUUUGGGGGCAUUGAAGGCGGCUCGAGAGGAACAGGUUACAUCUGAGCAAGCAAAGUUUUGGUUGGGAUUAGGGGGAGGGAGGGUCUCUCCCCCAGGUCCGGAGUGCAUCGAGAAACCAGCAACGGAGCGGCGAAUCGACAAAGGGGAGGAAGUGGGAGAAACAACUGUGCAGCGAGAUGCGAAGAUGGCGCCGGAGGAAACGGCCACACCUAAAACCGUUGGCACAUCCUGCUAU 3′.
- gRNA Ψ ΔDIS: 5′ GCCAUUUGACCAUUCACCACAUUGGUGUGCACCUGGGUUGAUGGCCGGACCGUUGAUUCCCUGACGACUACGAGCACCUGCAUGAAGCAGAAGGCUUCAUUUGGUGACCCCGACGUGAUAGUUAGGGAAUAGUGGUCGGCCACAGACGGCGUGGCGAUCCUGUCUCCAUCCGUCUCGUCUAUCGGGAGGCGAGUUCGAUGACCCUGGUGGAGGGGGCUAGAGAGUCGUUGGAAGACGGGAAGGAAGCCCGACGACUGAGCAGUCCACCCCAGGCGUGAUUCUGGUCGCCCGGUGGAUCAAGCAUGGAAGCCGUCAUAAAGGUGAUUUCGUCCGCGUGUAAAACCUAUUGCGGGAAAACCUCUCCUUCUAAGAAGGAAAUAGGGGCCAUGUUGUCCCUCUUACAAAAGGAAGGGUUGCUUAUGUCUCCCUCAGACUUAUAUUCCCCGGGGUCCUGGGAUCCCAUUACCGCGGCGCUAUCCCAGCGGGCUAUGAUACUUGGGAAAUCGGGAGAGUUAAAAACCUGGGGAUUGGUUUUGGGGGCAUUGAAGGCGGCUCGAGAGGAACAGGUUACAUCUGAGCAAGCAAAGUUUUGGUUGGGAUUAGGGGGAGGGAGGGUCUCUCCCCCAGGUCCGGAGUGCAUCGAGAAACCAGCAACGGAGCGGCGAAUCGACAAAGGGGAGGAAGUGGGAGAAACAACUGUGCAGCGAGAUGCGAAGAUGGCGCCGGAGGAAACGGCCACACCUAAAACCGUUGGCACAUCCUGCUAU 3′.
4.5. In Vitro Condensate Formation
4.6. Confocal Imaging and Three-Dimensional Reconstructions
4.7. Atomic Force Microscopy (AFM)
4.8. Fluorescence Correlation Spectroscopy (FCS)
4.9. Quantitative Image and Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| gRNA | Genomic RNA |
| vRNA | Viral RNA |
| BMC | Biomolecular condensate/biomolecular condensation |
| Ψ | Psi |
| AFM | Atomic force microscopy |
| FCS | Fluorescence correlation spectroscopy |
| NC | Nucleocapsid |
| MA | Matrix |
| PR | Protease |
| USvRNA | Unspliced viral RNA |
| DIS | Dimerization initiation site |
| DLS | Dimerization linkage structure |
| RSV | Rous sarcoma virus |
| HIV-1 | Human immunodeficiency virus type-1 |
| PEG-8000 | Polyethylene glycol-8000 |
| IDR | Intrinsically disordered region |
| C | Center |
| I | Intermediate |
| FRAP | Fluorescence recovery after photobleaching |
| nM | Nanomolar |
| µM | Micromolar |
| mM | Millimolar |
| M | Molar |
| WLL | White light laser |
| LASX | Leica Application Suite |
| s | Second |
| µm | Micrometer |
References
- Maldonado, R.J.K.; Rice, B.; Chen, E.C.; Tuffy, K.M.; Chiari, E.F.; Fahrbach, K.M.; Hope, T.J.; Parent, L.J. Visualizing Association of the Retroviral Gag Protein with Unspliced Viral RNA in the Nucleus. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Tuffy, K.M.; Maldonado, R.J.K.; Chang, J.; Rosenfeld, P.; Cochrane, A.; Parent, L.J. HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites. Viruses 2020, 12, 1281. [Google Scholar] [CrossRef] [PubMed]
- Kaddis Maldonado, R.J.; Parent, L.J. Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors. Viruses 2016, 8, 257. [Google Scholar] [CrossRef] [PubMed]
- Hanson, H.M.; Willkomm, N.A.; Yang, H.; Mansky, L.M. Human Retrovirus Genomic RNA Packaging. Viruses 2022, 14, 1094. [Google Scholar] [CrossRef]
- Banks, J.D.; Linial, M.L. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J. Virol. 2000, 74, 456–464. [Google Scholar] [CrossRef]
- Zhou, J.; McAllen, J.K.; Tailor, Y.; Summers, M.F. High affinity nucleocapsid protein binding to the muPsi RNA packaging signal of Rous sarcoma virus. J. Mol. Biol. 2005, 349, 976–988. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Bean, R.L.; Vogt, V.M.; Summers, M. Solution structure of the Rous sarcoma virus nucleocapsid protein: MuPsi RNA packaging signal complex. J. Mol. Biol. 2007, 365, 453–467. [Google Scholar] [CrossRef]
- Méric, C.; Spahr, P.F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J. Virol. 1986, 60, 450–459. [Google Scholar] [CrossRef]
- Katz, R.A.; Terry, R.W.; Skalka, A.M. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J. Virol. 1986, 59, 163–167. [Google Scholar] [CrossRef]
- Lear, A.L.; Haddrick, M.; Heaphy, S. A study of the dimerization of Rous sarcoma virus RNA in vitro and in vivo. Virology 1995, 212, 47–57. [Google Scholar] [CrossRef]
- Aronoff, R.; Linial, M. Specificity of retroviral RNA packaging. J. Virol. 1991, 65, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Kaddis Maldonado, R.; Rye-McCurdy, T.; Binkley, C.; Bah, A.; Chen, E.C.; Rice, B.L.; Parent, L.J.; Musier-Forsyth, K. Rous sarcoma virus Genomic RNA Dimerization Capability In Vitro Is Not a Prerequisite for Viral Infectivity. Viruses 2020, 12, 568. [Google Scholar] [CrossRef]
- Chen, E.C.; Maldonado, R.J.K.; Parent, L.J. Visualizing Rous sarcoma virus Genomic RNA Dimerization in the Nucleus, Cytoplasm, and at the Plasma Membrane. Viruses 2021, 13, 903. [Google Scholar] [CrossRef] [PubMed]
- Houzet, L.; Paillart, J.C.; Smagulova, F.; Maurel, S.; Morichaud, Z.; Marquet, R.; Mougel, M. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007, 35, 2695–2704. [Google Scholar] [CrossRef]
- Mangel, W.F.; Delius, H.; Duesberg, P.H. Structure and molecular weight of the 60–70S RNA and the 30–40S RNA of the Rous sarcoma virus. Proc. Natl. Acad. Sci. USA 1974, 71, 4541–4545. [Google Scholar] [CrossRef] [PubMed]
- Murti, K.G.; Bondurant, M.; Tereba, A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J. Virol. 1981, 37, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Garbitt, R.A.; Albert, J.A.; Kessler, M.D.; Parent, L.J. trans-acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J. Virol. 2001, 75, 260–268. [Google Scholar] [CrossRef]
- Dubois, N.; Marquet, R.; Paillart, J.C.; Bernacchi, S. Retroviral RNA Dimerization: From Structure to Functions. Front. Microbiol. 2018, 9, 527. [Google Scholar] [CrossRef]
- Paillart, J.C.; Berthoux, L.; Ottmann, M.; Darlix, J.L.; Marquet, R.; Ehresmann, B.; Ehresmann, C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 1996, 70, 8348–8354. [Google Scholar] [CrossRef]
- Mikkelsen, J.G.; Lund, A.H.; Duch, M.; Pedersen, F.S. Mutations of the kissing-loop dimerization sequence influence the site specificity of murine leukemia virus recombination in vivo. J. Virol. 2000, 74, 600–610. [Google Scholar] [CrossRef]
- Mikkelsen, J.G.; Pedersen, F.S. Genetic reassortment and patch repair by recombination in retroviruses. J. Biomed. Sci. 2000, 7, 77–99. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Vogt, V.M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 1995, 69, 6487–6497. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.M.; Vogt, V.M. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 2002, 76, 5452–5462. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.M.; Vogt, V.M. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 2004, 78, 52–60. [Google Scholar] [CrossRef]
- Alberti, S.; Hyman, A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 196–213. [Google Scholar] [CrossRef]
- Kaddis Maldonado, R.; Lambert, G.S.; Rice, B.L.; Sudol, M.; Flanagan, J.M.; Parent, L.J. The Rous sarcoma virus Gag Polyprotein Forms Biomolecular Condensates Driven by Intrinsically-disordered Regions. J. Mol. Biol. 2023, 435, 168182. [Google Scholar] [CrossRef]
- Monette, A.; Niu, M.; Maldonado, R.K.; Chang, J.; Lambert, G.S.; Flanagan, J.M.; Cochrane, A.; Parent, L.J.; Mouland, A.J. Influence of HIV-1 Genomic RNA on the Formation of Gag Biomolecular Condensates. J. Mol. Biol. 2023, 435, 168190. [Google Scholar] [CrossRef]
- Uversky, V.N. Proteins without unique 3D structures: Biotechnological applications of intrinsically unstable/disordered proteins. Biotechnol. J. 2015, 10, 356–366. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Fonin, A.V.; Darling, A.L.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Intrinsically disordered proteins in crowded milieu: When chaos prevails within the cellular gumbo. Cell Mol. Life Sci. 2018, 75, 3907–3929. [Google Scholar] [CrossRef]
- Lin, Y.; Protter, D.S.; Rosen, M.K.; Parker, R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 2015, 60, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Tauber, D.; Tauber, G.; Parker, R. Mechanisms and Regulation of RNA Condensation in RNP Granule Formation. Trends Biochem. Sci. 2020, 45, 764–778. [Google Scholar] [CrossRef]
- Lambert, G.S.; Rice, B.L.; Maldonado, R.J.K.; Chang, J.; Parent, L.J. Comparative analysis of retroviral Gag-host cell interactions: Focus on the nuclear interactome. Retrovirology 2024, 21, 13. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Lyons, H.; Veettil, R.T.; Pradhan, P.; Fornero, C.; De La Cruz, N.; Ito, K.; Eppert, M.; Roeder, R.G.; Sabari, B.R. Functional partitioning of transcriptional regulators by patterned charge blocks. Cell 2023, 186, 327–345.e328. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef]
- Alberti, S. Phase separation in biology. Curr. Biol. 2017, 27, R1097–R1102. [Google Scholar] [CrossRef] [PubMed]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Wheeler, R.J.; Hyman, A.A. Controlling compartmentalization by non-membrane-bound organelles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170193. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Jülicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef] [PubMed]
- Lochmann, T.L.; Bann, D.V.; Ryan, E.P.; Beyer, A.R.; Mao, A.; Cochrane, A.; Parent, L.J. NC-mediated nucleolar localization of retroviral gag proteins. Virus Res. 2013, 171, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Rice, B.L.; Kaddis, R.J.; Stake, M.S.; Lochmann, T.L.; Parent, L.J. Interplay between the alpharetroviral Gag protein and SR proteins SF2 and SC35 in the nucleus. Front. Microbiol. 2015, 6, 925. [Google Scholar] [CrossRef]
- Mouland, A.J.; Parent, L.; Weber, S.C.; Holehouse, A.S. Virus Induced Membraneless Organelles and Biomolecular Condensates. J. Mol. Biol. 2023, 435, 168213. [Google Scholar] [CrossRef]
- Sagan, S.M.; Weber, S.C. Let’s phase it: Viruses are master architects of biomolecular condensates. Trends Biochem. Sci. 2023, 48, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Asimi, V.; Sampath Kumar, A.; Niskanen, H.; Riemenschneider, C.; Hetzel, S.; Naderi, J.; Fasching, N.; Popitsch, N.; Du, M.; Kretzmer, H.; et al. Hijacking of transcriptional condensates by endogenous retroviruses. Nat. Genet. 2022, 54, 1238–1247. [Google Scholar] [CrossRef]
- Borodavka, A.; Acker, J. Seeing Biomolecular Condensates Through the Lens of Viruses. Annu. Rev. Virol. 2023, 10, 163–182. [Google Scholar] [CrossRef]
- Alston, J.J.; Soranno, A. Condensation Goes Viral: A Polymer Physics Perspective. J. Mol. Biol. 2023, 435, 167988. [Google Scholar] [CrossRef]
- Martin, E.W.; Iserman, C.; Olety, B.; Mitrea, D.M.; Klein, I.A. Biomolecular Condensates as Novel Antiviral Targets. J. Mol. Biol. 2024, 436, 168380. [Google Scholar] [CrossRef]
- Charman, M.; Grams, N.; Kumar, N.; Halko, E.; Dybas, J.M.; Abbott, A.; Lum, K.K.; Blumenthal, D.; Tsopurashvili, E.; Weitzman, M.D. A viral biomolecular condensate coordinates assembly of progeny particles. Nature 2023, 616, 332–338. [Google Scholar] [CrossRef]
- Pei, J.; Beri, N.R.; Zou, A.J.; Hubel, P.; Dorando, H.K.; Bergant, V.; Andrews, R.D.; Pan, J.; Andrews, J.M.; Sheehan, K.C.F.; et al. Nuclear-localized human respiratory syncytial virus NS1 protein modulates host gene transcription. Cell Rep. 2021, 37, 109803. [Google Scholar] [CrossRef] [PubMed]
- Rovnak, J.; Quackenbush, S.L. Exploitation of the Mediator complex by viruses. PLoS Pathog. 2022, 18, e1010422. [Google Scholar] [CrossRef] [PubMed]
- Vijayalingam, S.; Chinnadurai, G. Adenovirus L-E1A activates transcription through mediator complex-dependent recruitment of the super elongation complex. J. Virol. 2013, 87, 3425–3434. [Google Scholar] [CrossRef]
- Lambert, G.S.; Maldonado, R.J.K.; Parent, L.J. Role of the Psi Packaging Signal and Dimerization Initiation Sequence in the Organization of Rous sarcoma virus Gag-gRNA Co-Condensates. Viruses 2025, 17, 97. [Google Scholar] [CrossRef]
- Webb, J.A.; Jones, C.P.; Parent, L.J.; Rouzina, I.; Musier-Forsyth, K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: Implications for viral genomic RNA packaging. RNA 2013, 19, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Rye-McCurdy, T.; Olson, E.D.; Liu, S.; Binkley, C.; Reyes, J.P.; Thompson, B.R.; Flanagan, J.M.; Parent, L.J.; Musier-Forsyth, K. Functional Equivalence of Retroviral MA Domains in Facilitating Psi RNA Binding Specificity by Gag. Viruses 2016, 8, 256. [Google Scholar] [CrossRef]
- Ibrahim, K.A.; Naidu, A.S.; Miljkovic, H.; Radenovic, A.; Yang, W. Label-Free Techniques for Probing Biomolecular Condensates. ACS Nano 2024, 18, 10738–10757. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Šneideris, T.; Vendruscolo, M.; Knowles, T.P.J. Atomic force microscopy for single molecule characterisation of protein aggregation. Arch. Biochem. Biophys. 2019, 664, 134–148. [Google Scholar] [CrossRef]
- Alshareedah, I.; Banerjee, P.R. Measurement of Protein and Nucleic Acid Diffusion Coefficients Within Biomolecular Condensates Using In-Droplet Fluorescence Correlation Spectroscopy. Methods Mol. Biol. 2023, 2563, 199–213. [Google Scholar] [CrossRef]
- Incicco, J.J.; Roy, D.; Stuchell-Brereton, M.D.; Soranno, A. Fluorescence Correlation Spectroscopy and Phase Separation. Methods Mol. Biol. 2023, 2563, 161–198. [Google Scholar] [CrossRef]
- Li, C.; Bian, Y.; Tang, Y.; Meng, L.; Yin, P.; Hong, Y.; Cheng, J.; Li, Y.; Lin, J.; Tang, C.; et al. Deciphering the molecular mechanism underlying morphology transition in two-component DNA-protein cophase separation. Structure 2025, 33, 62–77.e68. [Google Scholar] [CrossRef] [PubMed]
- Qamar, S.; Wang, G.; Randle, S.J.; Ruggeri, F.S.; Varela, J.A.; Lin, J.Q.; Phillips, E.C.; Miyashita, A.; Williams, D.; Ströhl, F.; et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell 2018, 173, 720–734.e715. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, A.; Alam, J.M.; Noshiro, D.; Hirata, E.; Fujioka, Y.; Suzuki, K.; Ohsumi, Y.; Noda, N.N. Liquidity Is a Critical Determinant for Selective Autophagy of Protein Condensates. Mol. Cell 2020, 77, 1163–1175.e1169. [Google Scholar] [CrossRef] [PubMed]
- Heath, G.R.; Kots, E.; Robertson, J.L.; Lansky, S.; Khelashvili, G.; Weinstein, H.; Scheuring, S. Localization atomic force microscopy. Nature 2021, 594, 385–390. [Google Scholar] [CrossRef]
- Ramalho, R.; Rankovic, S.; Zhou, J.; Aiken, C.; Rousso, I. Analysis of the mechanical properties of wild type and hyperstable mutants of the HIV-1 capsid. Retrovirology 2016, 13, 17. [Google Scholar] [CrossRef]
- Deshpande, A.; Bryer, A.J.; Andino-Moncada, J.R.; Shi, J.; Hong, J.; Torres, C.; Harel, S.; Francis, A.C.; Perilla, J.R.; Aiken, C.; et al. Elasticity of the HIV-1 core facilitates nuclear entry and infection. PLoS Pathog. 2024, 20, e1012537. [Google Scholar] [CrossRef]
- Wang, Z.; Lou, J.; Zhang, H. Essence determines phenomenon: Assaying the material properties of biological condensates. J. Biol. Chem. 2022, 298, 101782. [Google Scholar] [CrossRef]
- Vinckier, A.; Semenza, G. Measuring elasticity of biological materials by atomic force microscopy. FEBS Lett. 1998, 430, 12–16. [Google Scholar] [CrossRef]
- Kontomaris, S.V.; Stylianou, A.; Chliveros, G.; Malamou, A. Determining Spatial Variability of Elastic Properties for Biological Samples Using AFM. Micromachines 2023, 14, 182. [Google Scholar] [CrossRef]
- Zeng, M.; Chen, X.; Guan, D.; Xu, J.; Wu, H.; Tong, P.; Zhang, M. Reconstituted Postsynaptic Density as a Molecular Platform for Understanding Synapse Formation and Plasticity. Cell 2018, 174, 1172–1187.e1116. [Google Scholar] [CrossRef]
- Yu, W.; Liu, J.; Huang, X.; Ren, J. Study on Phase Separation of Fused in Sarcoma by Fluorescence Correlation Spectroscopy. Langmuir 2024, 40, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Giles, K.E.; Caputi, M.; Beemon, K.L. Packaging and reverse transcription of snRNAs by retroviruses may generate pseudogenes. RNA 2004, 10, 299–307. [Google Scholar] [CrossRef]
- Eckwahl, M.J.; Telesnitsky, A.; Wolin, S.L. Host RNA Packaging by Retroviruses: A Newly Synthesized Story. mBio 2016, 7, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Cen, S.; Javanbakht, H.; Kim, S.; Shiba, K.; Craven, R.; Rein, A.; Ewalt, K.; Schimmel, P.; Musier-Forsyth, K.; Kleiman, L. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J. Virol. 2002, 76, 13111–13115. [Google Scholar] [CrossRef] [PubMed]
- Nadaraia-Hoke, S.; Bann, D.V.; Lochmann, T.L.; Gudleski-O’Regan, N.; Parent, L.J. Alterations in the MA and NC domains modulate phosphoinositide-dependent plasma membrane localization of the Rous sarcoma virus Gag protein. J. Virol. 2013, 87, 3609–3615. [Google Scholar] [CrossRef]
- Sawyer, R.C.; Dahlberg, J.E. Small RNAs of Rous sarcoma virus: Characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J. Virol. 1973, 12, 1226–1237. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.M.; Levinson, W.E.; Sullivan, D.; Fanshier, L.; Quintrell, N.; Jackson, J. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology 1970, 42, 927–937. [Google Scholar] [CrossRef]
- Erikson, E.; Erikson, R.L.; Henry, B.; Pace, N.R. Comparison of oligonucleotides produced by RNase T1 digestion of 7 S RNA from avian and murine oncornaviruses and from uninfected cells. Virology 1973, 53, 40–46. [Google Scholar] [CrossRef]
- Fossé, P.; Motté, N.; Roumier, A.; Gabus, C.; Muriaux, D.; Darlix, J.L.; Paoletti, J. A short autocomplementary sequence plays an essential role in avian sarcoma-leukosis virus RNA dimerization. Biochemistry 1996, 35, 16601–16609. [Google Scholar] [CrossRef]
- Schwartz, D.E.; Tizard, R.; Gilbert, W. Nucleotide sequence of Rous sarcoma virus. Cell 1983, 32, 853–869. [Google Scholar] [CrossRef]
- Butterfield-Gerson, K.L.; Scheifele, L.Z.; Ryan, E.P.; Hopper, A.K.; Parent, L.J. Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J. Virol. 2006, 80, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Scheifele, L.Z.; Ryan, E.P.; Parent, L.J. Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J. Virol. 2005, 79, 8732–8741. [Google Scholar] [CrossRef]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar] [CrossRef]
- Jäger, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global landscape of HIV–human protein complexes. Nature 2011, 481, 365. [Google Scholar] [CrossRef] [PubMed]
- Engeland, C.E.; Oberwinkler, H.; Schümann, M.; Krause, E.; Müller, G.A.; Kräusslich, H.-G. The Cellular Protein Lyric Interacts with HIV-1 Gag. J. Virol. 2011, 85, 13322–13332. [Google Scholar] [CrossRef] [PubMed]
- Engeland, C.E.; Brown, N.P.; Börner, K.; Schümann, M.; Krause, E.; Kaderali, L.; Müller, G.A.; Kräusslich, H.-G. Proteome analysis of the HIV-1 Gag interactome. Virology 2014, 460, 194–206. [Google Scholar] [CrossRef]
- Ritchie, C.; Cylinder, I.; Platt, E.J.; Barklis, E. Analysis of HIV-1 Gag Protein Interactions via Biotin Ligase Tagging. J. Virol. 2015, 89, 3988–4001. [Google Scholar] [CrossRef]
- Le Sage, V.; Cinti, A.; Valiente-Echeverría, F.; Mouland, A.J. Proteomic analysis of HIV-1 Gag interacting partners using proximity-dependent biotinylation. Virol. J. 2015, 12, 138. [Google Scholar] [CrossRef]
- Li, Y.; Frederick, K.M.; Haverland, N.A.; Ciborowski, P.; Belshan, M. Investigation of the HIV-1 matrix interactome during virus replication. Proteom.–Clin. Appl. 2016, 10, 156–163. [Google Scholar] [CrossRef]
- Bose, M.; Rankovic, B.; Mahamid, J.; Ephrussi, A. An architectural role of specific RNA-RNA interactions in oskar granules. Nat. Cell Biol. 2024, 26, 1934–1942. [Google Scholar] [CrossRef]
- Ferrandon, D.; Koch, I.; Westhof, E.; Nüsslein-Volhard, C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997, 16, 1751–1758. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.; Palacios, I.; Jaeger, L.; St Johnston, D.; Ehresmann, B.; Ehresmann, C.; Brunel, C. Dimerization of the 3′ UTR of bicoid mRNA involves a two-step mechanism. J. Mol. Biol. 2001, 313, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Little, S.C.; Sinsimer, K.S.; Lee, J.J.; Wieschaus, E.F.; Gavis, E.R. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 2015, 17, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.Z.; Ruff, K.M.; Dar, F.; Jalihal, A.; King, M.R.; Lalmansingh, J.M.; Posey, A.E.; Erkamp, N.A.; Seim, I.; Gladfelter, A.S.; et al. Dynamical control enables the formation of demixed biomolecular condensates. Nat. Commun. 2023, 14, 7678. [Google Scholar] [CrossRef]
- Langdon, E.M.; Qiu, Y.; Ghanbari Niaki, A.; McLaughlin, G.A.; Weidmann, C.A.; Gerbich, T.M.; Smith, J.A.; Crutchley, J.M.; Termini, C.M.; Weeks, K.M.; et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 2018, 360, 922–927. [Google Scholar] [CrossRef]
- Conti, B.A.; Oppikofer, M. Biomolecular condensates: New opportunities for drug discovery and RNA therapeutics. Trends Pharmacol. Sci. 2022, 43, 820–837. [Google Scholar] [CrossRef]
- Rice, B.L.; Stake, M.S.; Parent, L.J. TNPO3-Mediated Nuclear Entry of the Rous Sarcoma virus Gag Protein Is Independent of the Cargo-Binding Domain. J. Virol. 2020, 94, 17. [Google Scholar] [CrossRef]
- Gudleski, N.; Flanagan, J.M.; Ryan, E.P.; Bewley, M.C.; Parent, L.J. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 9358–9363. [Google Scholar] [CrossRef]
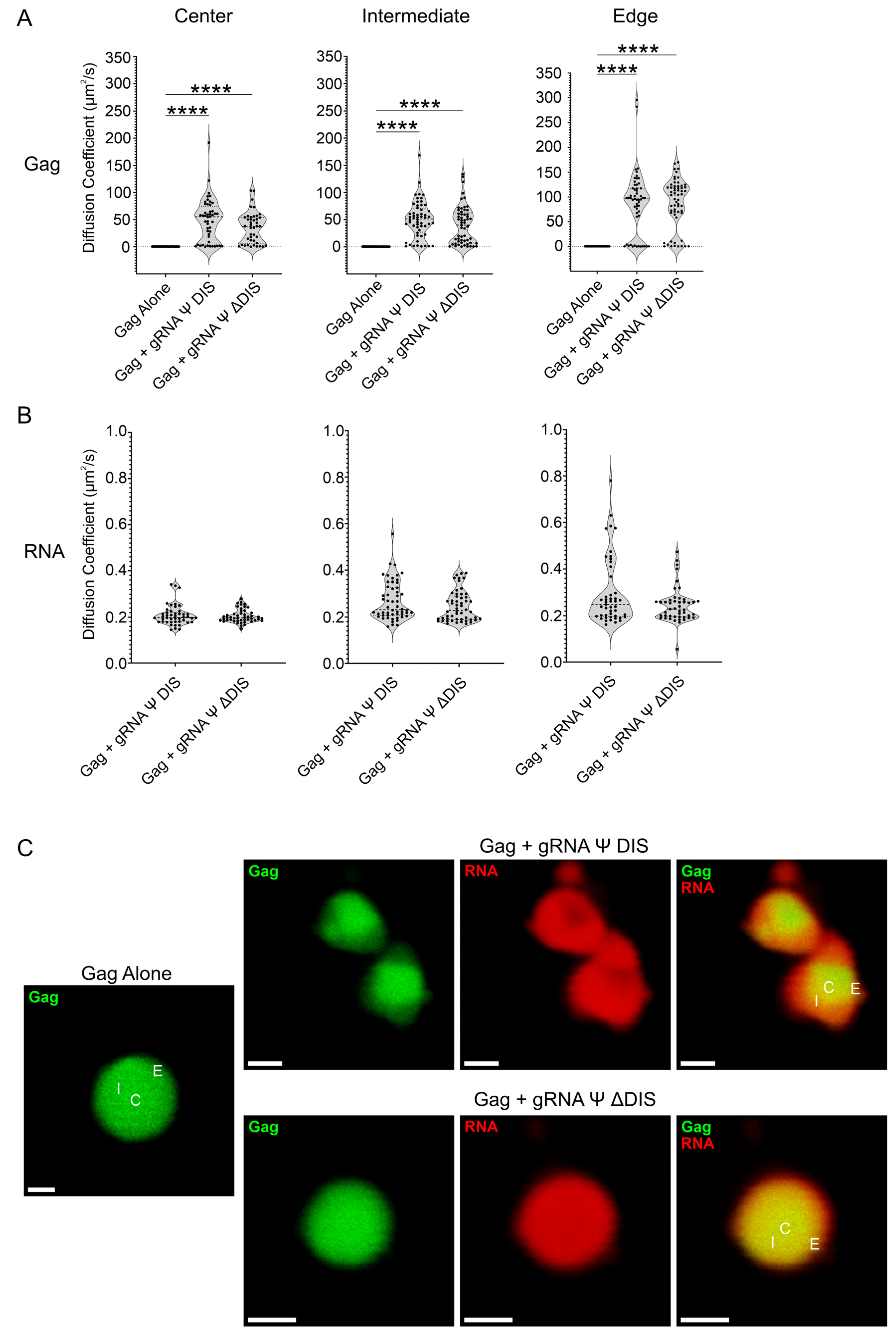

| Condition | Gag Diffusion Coefficient (µm2/s) | RNA Diffusion Coefficient (µm2/s) |
|---|---|---|
| Gag Alone: Center, 150 mM | 0.423 | N/A |
| Gag Alone: Intermediate, 150 mM | 0.429 | N/A |
| Gag Alone: Edge, 150 mM | 0.363 | N/A |
| gRNA Ψ DIS: Center, 150 mM | 48.3 | 0.209 |
| gRNA Ψ DIS: Intermediate, 150 mM | 50.5 | 0.268 |
| gRNA Ψ DIS: Edge, 150 mM | 87.2 | 0.299 |
| gRNA Ψ ΔDIS: Center, 150 mM | 35.4 | 0.205 |
| gRNA Ψ ΔDIS: Intermediate, 150 mM | 40.2 | 0.247 |
| gRNA Ψ ΔDIS: Edge, 150 mM | 86.3 | 0.244 |
| Gag Alone: Center, 500 mM | 40.4 | N/A |
| Gag Alone: Intermediate, 500 mM | 47.1 | N/A |
| Gag Alone: Edge, 500 mM | 78.5 | N/A |
| gRNA Ψ DIS: Center 500 mM | 17.1 | 0.217 |
| gRNA Ψ DIS: Intermediate, 500 mM | 32.7 | 0.226 |
| gRNA Ψ DIS: Edge, 500 M | 89.0 | 0.252 |
| gRNA Ψ ΔDIS: Center 500 mM | 30.8 | 0.228 |
| gRNA Ψ ΔDIS: Intermediate, 500 mM | 43.9 | 0.239 |
| gRNA Ψ ΔDIS: Edge, 500 mM | 78.5 | 0.265 |
| Gag Alone: Center, 1 M | 39.9 | N/A |
| Gag Alone: Intermediate, 1 M | 27.1 | N/A |
| Gag Alone: Edge, 1 M | 68.3 | N/A |
| gRNA Ψ DIS: Center 1 M | 30.6 | 0.260 |
| gRNA Ψ DIS: Intermediate, 1 M | 32.0 | 0.249 |
| gRNA Ψ DIS: Edge, 1 M | 71.7 | 0.215 |
| gRNA Ψ ΔDIS: Center 1 M | 16.8 | 0.249 |
| gRNA Ψ ΔDIS: Intermediate, 1 M | 20.7 | 0.266 |
| gRNA Ψ ΔDIS: Edge, 1 M | 73.1 | 0.244 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lambert, G.S.; Siedlecki, C.A.; Parent, L.J. Two Peas in a Pod: Retroviral RNA Dimers Organize Gag–RNA Nanoclusters with Novel Biophysical Properties. Int. J. Mol. Sci. 2025, 26, 5679. https://doi.org/10.3390/ijms26125679
Lambert GS, Siedlecki CA, Parent LJ. Two Peas in a Pod: Retroviral RNA Dimers Organize Gag–RNA Nanoclusters with Novel Biophysical Properties. International Journal of Molecular Sciences. 2025; 26(12):5679. https://doi.org/10.3390/ijms26125679
Chicago/Turabian StyleLambert, Gregory S., Christopher A. Siedlecki, and Leslie J. Parent. 2025. "Two Peas in a Pod: Retroviral RNA Dimers Organize Gag–RNA Nanoclusters with Novel Biophysical Properties" International Journal of Molecular Sciences 26, no. 12: 5679. https://doi.org/10.3390/ijms26125679
APA StyleLambert, G. S., Siedlecki, C. A., & Parent, L. J. (2025). Two Peas in a Pod: Retroviral RNA Dimers Organize Gag–RNA Nanoclusters with Novel Biophysical Properties. International Journal of Molecular Sciences, 26(12), 5679. https://doi.org/10.3390/ijms26125679






