Emerging Concepts of Targeted Protein Degrader Technologies via Lysosomal Pathways
Abstract
1. Introduction
2. An Overview of the History of TPD
3. New Frontiers in Lysosome-Mediated TPD
4. Emerging Lysosome-Based Targeted Protein Degradation
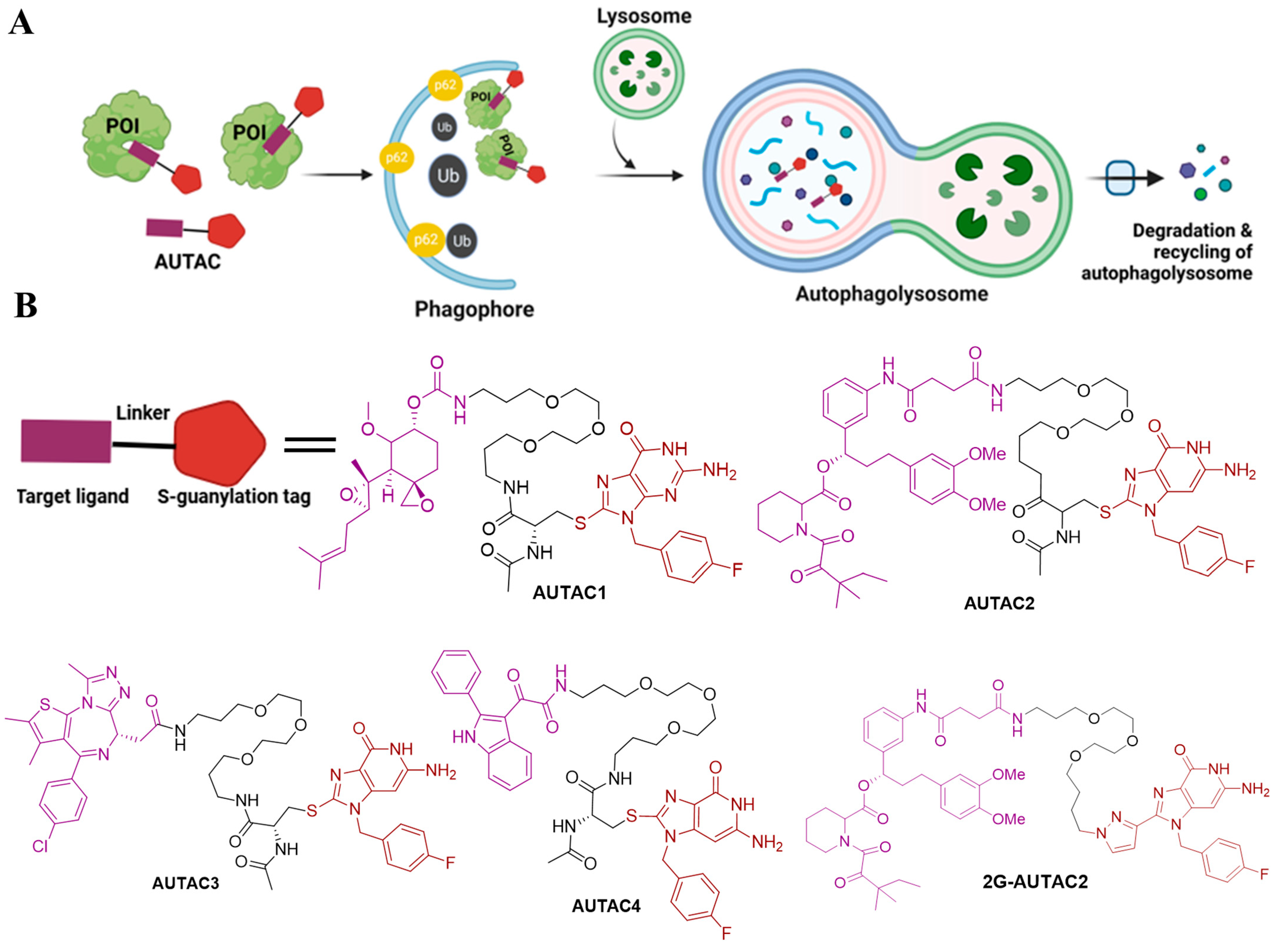
4.1. AUTACs
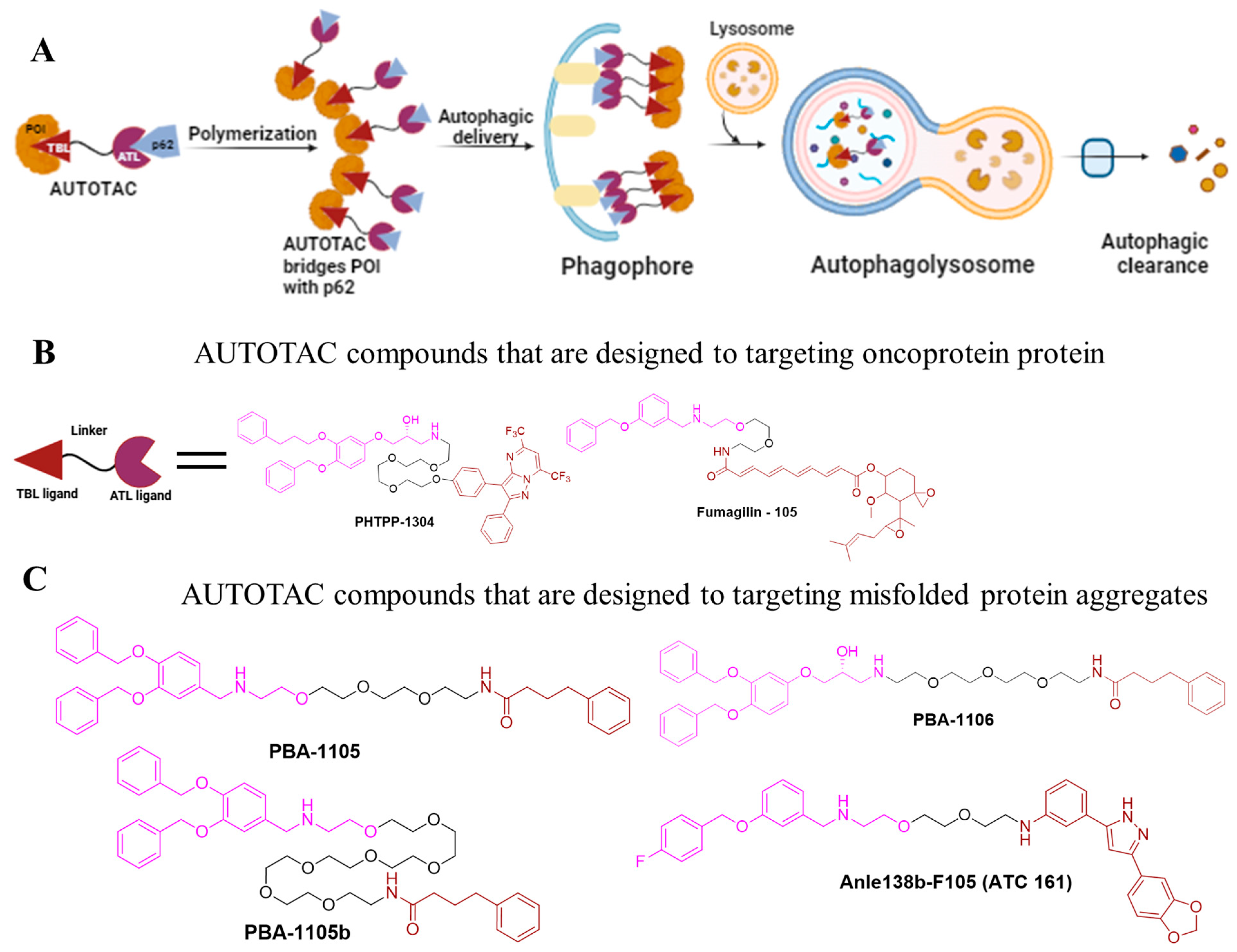
4.2. AUTOTACs
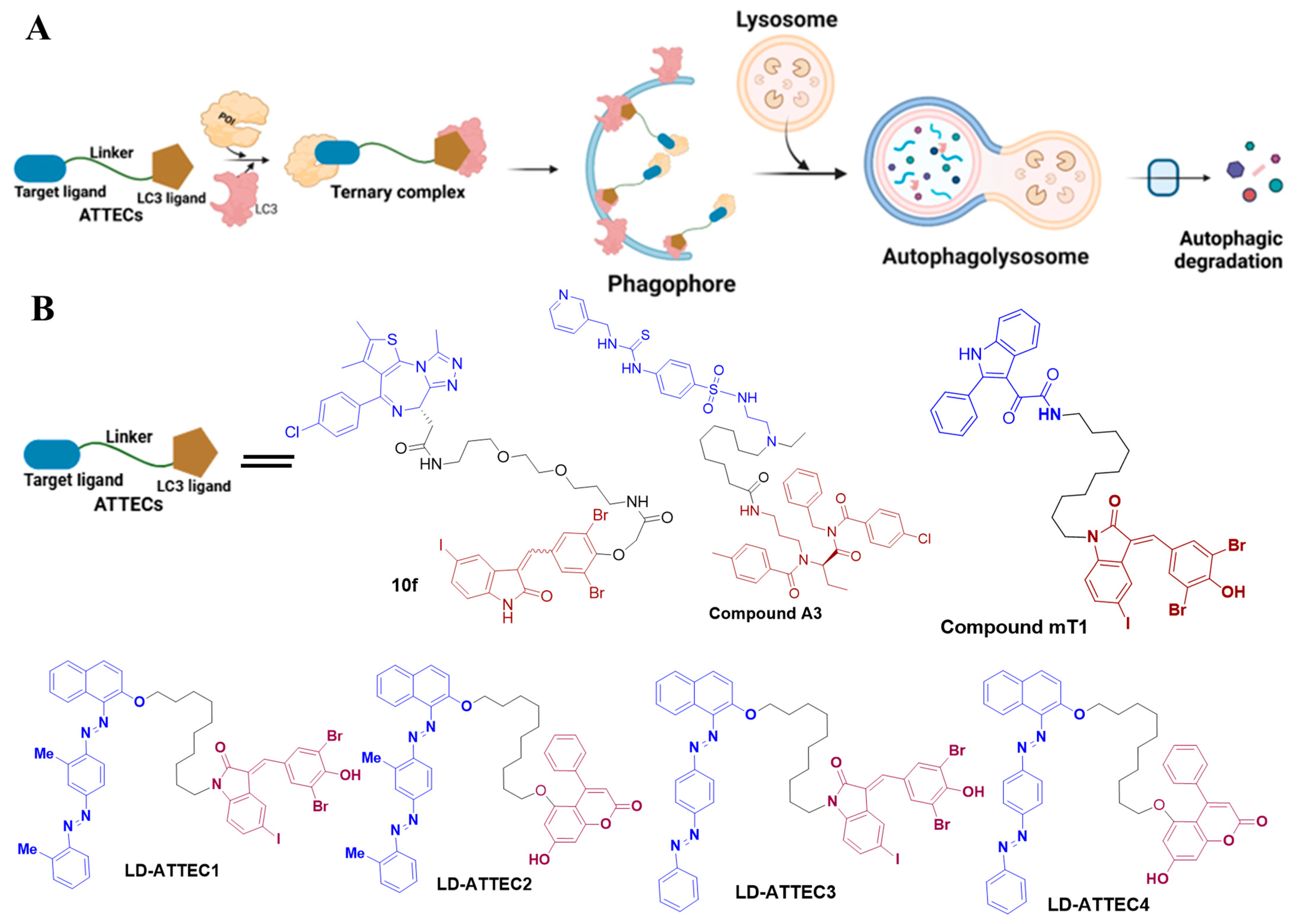
4.3. ATTECs
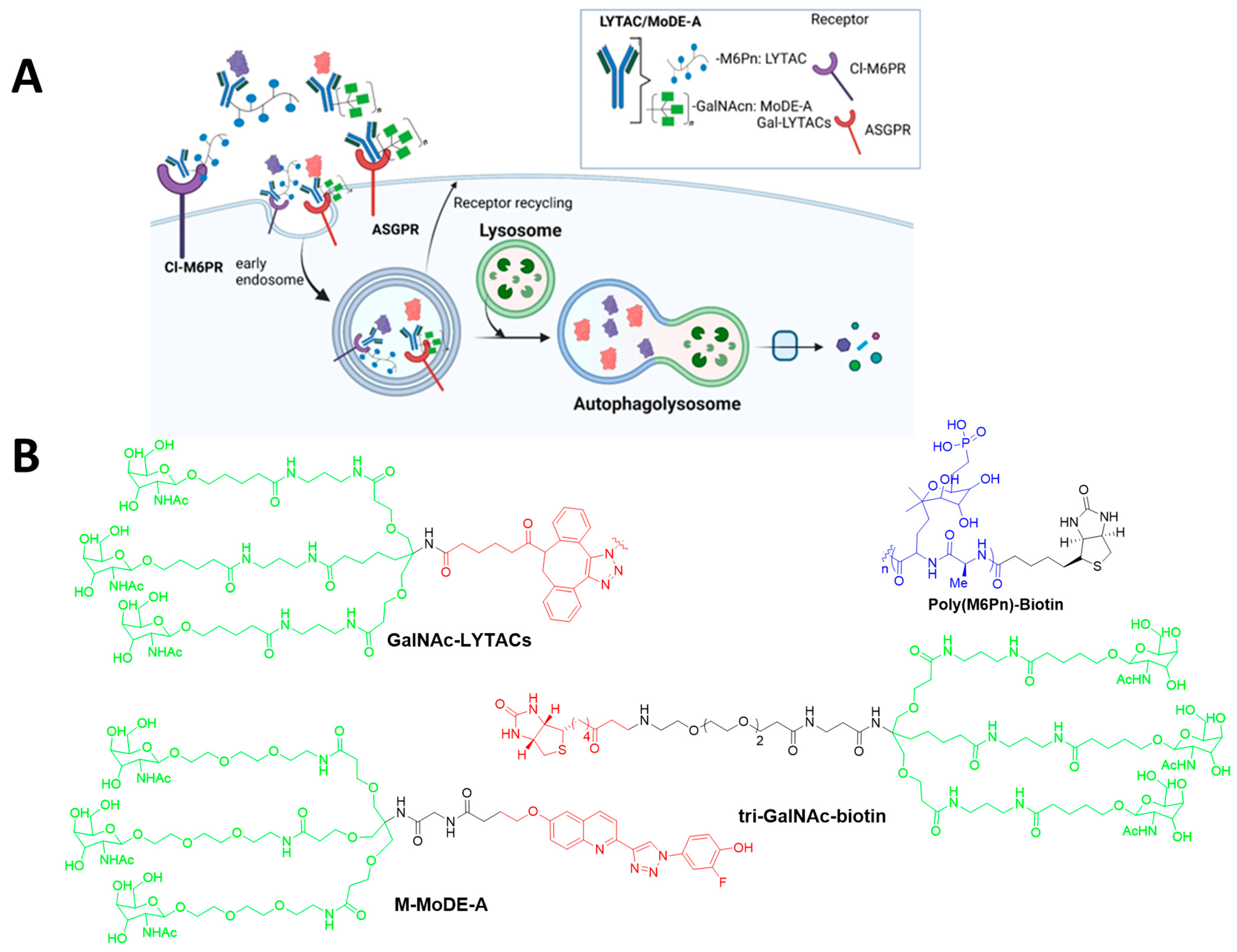
4.4. LYTACs
4.5. MoDE-As, TransMoDEs, and IFLDs
4.6. AbTACs and PROTABs
4.7. GlueTACs
4.8. KineTACs
4.9. TransTACs
4.10. AceTACs
4.11. FRTACs
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef]
- Samarasinghe, K.T.G.; Crews, C.M. Targeted protein degradation: A promise for undruggable proteins. Cell Chem. Biol. 2021, 28, 934–951. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef]
- Rousseau, A.; Bertolotti, A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 697–712. [Google Scholar] [CrossRef]
- Varshavsky, A. Regulated protein degradation. Trends Biochem. Sci. 2005, 30, 283–286. [Google Scholar] [CrossRef]
- Lilienbaum, A. Relationship between the proteasomal system and autophagy. Int. J. Biochem. Mol. Biol. 2013, 4, 1–26. [Google Scholar] [PubMed]
- Ding, Y.; Xing, D.; Fei, Y.; Lu, B. Emerging degrader technologies engaging lysosomal pathways. Chem. Soc. Rev. 2022, 51, 8832–8876. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Byun, I.; Kim, D.Y.; Joh, H.; Kim, H.J.; Lee, M.J. Targeted protein degradation directly engaging lysosomes or proteasomes. Chem. Soc. Rev. 2024, 53, 3253–3272. [Google Scholar] [CrossRef]
- Bond, M.J.; Crews, C.M. Proteolysis targeting chimeras (PROTACs) come of age: Entering the third decade of targeted protein degradation. RSC Chem. Biol. 2021, 2, 725–742. [Google Scholar] [CrossRef]
- Burslem, G.M.; Crews, C.M. Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell 2020, 181, 102–114. [Google Scholar] [CrossRef]
- Qi, S.M.; Dong, J.; Xu, Z.Y.; Cheng, X.D.; Zhang, W.D.; Qin, J.J. PROTAC: An Effective Targeted Protein Degradation Strategy for Cancer Therapy. Front. Pharmacol. 2021, 12, 692574. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef]
- Zou, Y.; Ma, D.; Wang, Y. The PROTAC technology in drug development. Cell Biochem. Funct. 2019, 37, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Bekes, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef]
- Takahashi, D.; Moriyama, J.; Nakamura, T.; Miki, E.; Takahashi, E.; Sato, A.; Akaike, T.; Itto-Nakama, K.; Arimoto, H. AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol. Cell 2019, 76, 797–810.e710. [Google Scholar] [CrossRef]
- Ji, C.H.; Kim, H.Y.; Lee, M.J.; Heo, A.J.; Park, D.Y.; Lim, S.; Shin, S.; Ganipisetti, S.; Yang, W.S.; Jung, C.A.; et al. The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat. Commun. 2022, 13, 904. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, C.; Ding, Y.; Fei, Y.; Lu, B. ATTEC: A potential new approach to target proteinopathies. Autophagy 2020, 16, 185–187. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, N.; Wang, Z.; Luo, S.; Ding, Y.; Lu, B. Degradation of lipid droplets by chimeric autophagy-tethering compounds. Cell Res. 2021, 31, 965–979. [Google Scholar] [CrossRef]
- Banik, S.M.; Pedram, K.; Wisnovsky, S.; Ahn, G.; Riley, N.M.; Bertozzi, C.R. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 2020, 584, 291–297. [Google Scholar] [CrossRef]
- Ahn, G.; Banik, S.M.; Miller, C.L.; Riley, N.M.; Cochran, J.R.; Bertozzi, C.R. LYTACs that engage the asialoglycoprotein receptor for targeted protein degradation. Nat. Chem. Biol. 2021, 17, 937–946. [Google Scholar] [CrossRef]
- Caianiello, D.F.; Zhang, M.; Ray, J.D.; Howell, R.A.; Swartzel, J.C.; Branham, E.M.J.; Chirkin, E.; Sabbasani, V.R.; Gong, A.Z.; McDonald, D.M.; et al. Bifunctional small molecules that mediate the degradation of extracellular proteins. Nat. Chem. Biol. 2021, 17, 947–953. [Google Scholar] [CrossRef]
- Howell, R.A.; Wang, S.; Khambete, M.; McDonald, D.M.; Spiegel, D.A. Bifunctional Molecules That Induce Both Targeted Degradation and Transcytosis of Extracellular Proteins in Brain Cells. J. Am. Chem. Soc. 2024. [Google Scholar] [CrossRef]
- Zheng, J.; He, W.; Li, J.; Feng, X.; Li, Y.; Cheng, B.; Zhou, Y.; Li, M.; Liu, K.; Shao, X.; et al. Bifunctional Compounds as Molecular Degraders for Integrin-Facilitated Targeted Protein Degradation. J. Am. Chem. Soc. 2022, 144, 21831–21836. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhao, J.; Zhong, K.; Tong, A.; Jia, D. Targeted protein degradation: Mechanisms, strategies and application. Signal Transduct. Target. Ther. 2022, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Cotton, A.D.; Nguyen, D.P.; Gramespacher, J.A.; Seiple, I.B.; Wells, J.A. Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1. J. Am. Chem. Soc. 2021, 143, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Han, Y.; Yang, Y.; Lin, F.; Li, K.; Kong, L.; Liu, H.; Dang, Y.; Lin, J.; Chen, P.R. Covalently Engineered Nanobody Chimeras for Targeted Membrane Protein Degradation. J. Am. Chem. Soc. 2021, 143, 16377–16382. [Google Scholar] [CrossRef]
- Pance, K.; Gramespacher, J.A.; Byrnes, J.R.; Salangsang, F.; Serrano, J.C.; Cotton, A.D.; Steri, V.; Wells, J.A. Modular cytokine receptor-targeting chimeras for targeted degradation of cell surface and extracellular proteins. Nat. Biotechnol. 2023, 41, 273–281. [Google Scholar] [CrossRef]
- Rhee, K.; Zhou, X. Two in one: The emerging concept of bifunctional antibodies. Curr. Opin. Biotechnol. 2024, 85, 103050. [Google Scholar] [CrossRef]
- Jiang, Z.; Kuo, Y.H.; Arkin, M.R. Autophagy Receptor-Inspired Antibody-Fusion Proteins for Targeted Intracellular Degradation. J. Am. Chem. Soc. 2023, 145, 23939–23947. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, C.; Chen, X.; Zhao, Y.; Liao, Y.; Huang, P.; Wu, W.; Nieto, N.S.; Li, L.; Tang, W. Development of folate receptor targeting chimeras for cancer selective degradation of extracellular proteins. Nat. Commun. 2024, 15, 8695. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Gao, Q.; Mao, M.; Zhang, C.; Yang, L.; Yang, Y.; Han, D. Bispecific Aptamer Chimeras Enable Targeted Protein Degradation on Cell Membranes. Angew. Chem. Int. Ed. Engl. 2021, 60, 11267–11271. [Google Scholar] [CrossRef] [PubMed]
- Paudel, R.R.; Lu, D.; Roy Chowdhury, S.; Monroy, E.Y.; Wang, J. Targeted Protein Degradation via Lysosomes. Biochemistry 2023, 62, 564–579. [Google Scholar] [CrossRef] [PubMed]
- Noblejas-Lopez, M.D.M.; Tebar-Garcia, D.; Lopez-Rosa, R.; Alcaraz-Sanabria, A.; Cristobal-Cueto, P.; Pinedo-Serrano, A.; Rivas-Garcia, L.; Galan-Moya, E.M. TACkling Cancer by Targeting Selective Protein Degradation. Pharmaceutics 2023, 15, 2442. [Google Scholar] [CrossRef]
- Haridas, V.; Dutta, S.; Munjal, A.; Singh, S. Inhibitors to degraders: Changing paradigm in drug discovery. iScience 2024, 27, 109574. [Google Scholar] [CrossRef]
- Ding, Y.; Fei, Y.; Lu, B. Emerging New Concepts of Degrader Technologies. Trends Pharmacol. Sci. 2020, 41, 464–474. [Google Scholar] [CrossRef]
- Takahashi, D.; Ora, T.; Sasaki, S.; Ishii, N.; Tanaka, T.; Matsuda, T.; Ikeda, M.; Moriyama, J.; Cho, N.; Nara, H.; et al. Second-Generation AUTACs for Targeted Autophagic Degradation. J. Med. Chem. 2023, 66, 12342–12372. [Google Scholar] [CrossRef]
- Pei, J.; Pan, X.; Wang, A.; Shuai, W.; Bu, F.; Tang, P.; Zhang, S.; Zhang, Y.; Wang, G.; Ouyang, L. Developing potent LC3-targeting AUTAC tools for protein degradation with selective autophagy. Chem. Commun. 2021, 57, 13194–13197. [Google Scholar] [CrossRef]
- Dong, G.; Wu, Y.; Cheng, J.; Chen, L.; Liu, R.; Ding, Y.; Wu, S.; Ma, J.; Sheng, C. Ispinesib as an Effective Warhead for the Design of Autophagosome-Tethering Chimeras: Discovery of Potent Degraders of Nicotinamide Phosphoribosyltransferase (NAMPT). J. Med. Chem. 2022, 65, 7619–7628. [Google Scholar] [CrossRef]
- Tan, S.; Wang, D.; Fu, Y.; Zheng, H.; Liu, Y.; Lu, B. Targeted clearance of mitochondria by an autophagy-tethering compound (ATTEC) and its potential therapeutic effects. Sci. Bull. 2023, 68, 3013–3026. [Google Scholar] [CrossRef]
- Zhou, Y.; Teng, P.; Montgomery, N.T.; Li, X.; Tang, W. Development of Triantennary N-Acetylgalactosamine Conjugates as Degraders for Extracellular Proteins. ACS Cent. Sci. 2021, 7, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Arimoto, H. Targeting selective autophagy by AUTAC degraders. Autophagy 2020, 16, 765–766. [Google Scholar] [CrossRef]
- Takahashi, D.; Arimoto, H. Selective autophagy as the basis of autophagy-based degraders. Cell Chem. Biol. 2021, 28, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Kim, S.B.; Kim, H.Y.; Lee, S.J.; Lee, J.S.; Kwon, Y.T.; Ji, C.H. Methods to detect AUTOphagy-Targeting Chimera (AUTOTAC)-mediated Targeted Protein Degradation in Tauopathies. Bio Protoc. 2023, 13, e4594. [Google Scholar] [CrossRef]
- Lee, J.; Sung, K.W.; Bae, E.J.; Yoon, D.; Kim, D.; Lee, J.S.; Park, D.H.; Park, D.Y.; Mun, S.R.; Kwon, S.C.; et al. Targeted degradation of ⍺-synuclein aggregates in Parkinson’s disease using the AUTOTAC technology. Mol. Neurodegener. 2023, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M. Protacs for treatment of cancer. Pediatr. Res. 2010, 67, 505–508. [Google Scholar] [CrossRef][Green Version]
- Li, Z.; Wang, C.; Wang, Z.; Zhu, C.; Li, J.; Sha, T.; Ma, L.; Gao, C.; Yang, Y.; Sun, Y.; et al. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature 2019, 575, 203–209. [Google Scholar] [CrossRef]
- Alabi, S.B.; Crews, C.M. Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J. Biol. Chem. 2021, 296, 100647. [Google Scholar] [CrossRef]
- Yim, W.W.; Mizushima, N. Lysosome biology in autophagy. Cell Discov. 2020, 6, 6. [Google Scholar] [CrossRef]
- Ghosh, P.; Dahms, N.M.; Kornfeld, S. Mannose 6-phosphate receptors: New twists in the tale. Nat. Rev. Mol. Cell Biol. 2003, 4, 202–212. [Google Scholar] [CrossRef]
- Wu, Y.; Lin, B.; Lu, Y.; Li, L.; Deng, K.; Zhang, S.; Zhang, H.; Yang, C.; Zhu, Z. Aptamer-LYTACs for Targeted Degradation of Extracellular and Membrane Proteins. Angew. Chem. Int. Ed. Engl. 2023, 62, e202218106. [Google Scholar] [CrossRef] [PubMed]
- Gramespacher, J.A.; Cotton, A.D.; Burroughs, P.W.W.; Seiple, I.B.; Wells, J.A. Roadmap for Optimizing and Broadening Antibody-Based PROTACs for Degradation of Cell Surface Proteins. ACS Chem. Biol. 2022, 17, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Marei, H.; Tsai, W.K.; Kee, Y.S.; Ruiz, K.; He, J.; Cox, C.; Sun, T.; Penikalapati, S.; Dwivedi, P.; Choi, M.; et al. Antibody targeting of E3 ubiquitin ligases for receptor degradation. Nature 2022, 610, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.K.; Spit, M.; Jordens, I.; Low, T.Y.; Stange, D.E.; van de Wetering, M.; van Es, J.H.; Mohammed, S.; Heck, A.J.; Maurice, M.M.; et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012, 488, 665–669. [Google Scholar] [CrossRef]
- Guo, Y.; Che, J.; Dong, X. TransTACs: Transforming antibodies into targeted protein degraders. Acta Pharm. Sin. B 2025, 15, 1186–1188. [Google Scholar] [CrossRef]
- Zhang, D.; Duque-Jimenez, J.; Facchinetti, F.; Brixi, G.; Rhee, K.; Feng, W.W.; Janne, P.A.; Zhou, X. Transferrin receptor targeting chimeras for membrane protein degradation. Nature 2025, 638, 787–795. [Google Scholar] [CrossRef]
- Wang, L.; Liu, H.; Li, L. Autophagy receptor-inspired chimeras: A novel approach to facilitate the removal of protein aggregates and organelle by autophagy degradation. J. Zhejiang Univ. Sci. B 2024, 25, 1115–1119. [Google Scholar] [CrossRef]
- Hu, X.; Kabir, M.; Lin, Y.; Xiong, Y.; Parsons, R.E.; Gu, W.; Jin, J. Design, Synthesis, and Evaluation of p53Y220C Acetylation Targeting Chimeras (AceTACs). J. Med. Chem. 2024, 67, 14633–14648. [Google Scholar] [CrossRef]

| TPD Technology | Compounds | Binding Protein(s) | Target | Activity (DC50, or Else Mentioned) | References |
|---|---|---|---|---|---|
| AUTACs | AUTAC1 | p62 LC3 | MetAP2 | 1 µM in Hela cells | [16] |
| AUTAC2 | p62 LC3 | FKB12 | 10 µM in Hela cells | [16] | |
| AUTAC3 | p62 LC3 | BRD4 | - | [16] | |
| AUTAC4 | p62 LC3 | TSPO | - | [16] | |
| 2G-AUTAC2 | p62 LC3 | FKB12 | 10 µM in Hela cells | [37] | |
| AUTOTACs | PHTPP-1304 | p62 LC3 | ER | ~2 nM | [17] |
| VinclozolinM2-2204 | p62 LC3 | AR | ~200 nM | [17] | |
| Fumagilin-105 | p62 LC3 | MetAP2 | 0.7 µM | [17] | |
| PBA-1105 | p62 LC3 | tauP301L | 1–10 nM | [17] | |
| PBA-1106 | p62 LC3 | tauP301L | 1–10 nM | [17] | |
| Anle138b-F105 (ATC 161) | p62 LC3 | tauP301L | ~3.0 nM | [17] | |
| ATTECs | LD-ATTEC1 | LC3 | LC3B | Kd~4.2 µM | [19] |
| LD-ATTEC2 | LC3 | LC3B | Kd~4.9 µM | [19] | |
| LD-ATTEC3 | LC3 | LC3B | Kd~1.9 µM | [19] | |
| LD-ATTEC4 | LC3 | LC3B | Kd~1.3 µM | [19] | |
| Compound 10f | LC3 | BRD4 | 0.5 µM in MDA-MD-231 and MDA-MB-468 cells | [38] | |
| Compound A3 | LC3 | NMPT | - | [39] | |
| mT1 | LC3 | TSPO | - | [40] | |
| LYTACs | Poly(M6Pn)-biotein | CI-M6PR | NA-647 | - | [20,21] |
| GalNAc-LYTACs antibody (Ctx-GN) | ASGPR | EGFR | 40% downregulation | [21] | |
| Tri-GaINAc-biotein | ASGPR | NA-650 | - | [41] | |
| MoDE-A | M-MoDE-A | ASGPR | MIF | - | [22] |
| D-MoDE-A | a-DNP anti-bodies | NA | [22] | ||
| TransMoDEs | TransMoDEs | ASGPR | LRP1 | NA | [23] |
| IFLD | BMS-L1-RGD | αvβ3 integrin | Neutravidin PD-L1 degrader | NA | [24] |
| AbTAC | R0/Atz | RNF43 or ZNRF3 | PD-L1 | 0.8 nM in HCC2935 | [26] |
| R3/Atz | RNF43 or ZNRF3 | PD-L1 | 1.2 nM | [26] | |
| R0/Ctx | RNF43 or ZNRF3 | EGFR | 5.1 nM | [26] | |
| R3/Ctx | RNF43 or ZNRF3 | EGFR | 6.6 nM | [26] | |
| Z18/Atz | RNF43 or ZNRF3 | PD-L1 | 3.4 nM | [26] | |
| Z18/Ctx | RNF43 or ZNRF3 | EGFR | 0.6 nM | [26] | |
| AC-1 | PD-L1 | PD-L1 | 3.4 nM in MDA-MB-231 | [26] | |
| PROTAB | PROTAB | RNF43 or ZNRF3 | IGF1R/HER2/PD-L1 | NA | [26] |
| GlueTAC | GlueTAC | CPP-LSS | PD-L1 | NA | [27] |
| KineTAC | CXCL12-Atz | CXCR7 | PD-L1 | NA | [28] |
| CXCL12–Tras | ‘’ | HER2 | NA | [28] | |
| CXCL12–Ctx | ‘’ | EGFR | NA | [28] | |
| TransTAC | TransTAC | TfR | EGFR, CD20, PD-L1, and a CD19-targeted CAR | NA | [29] |
| AceTACs | MS182 | p62 LC3 | mHTT, TDP-43, and Tau | NA | [30] |
| FRTAC | FRTAC | Folate receptor (FR) | EGFR PD-L1 CD47 | NA | [31] |
| Degrader Properties | Potential Targets | Type of Proteins Degraded | Degradation System/Pathway | Advantages | Limitations |
|---|---|---|---|---|---|
| PROTAC | Tethers to E3 ubiquitin ligase and triggers K48 polyubiquitination | Short-lived and soluble, misfolded | Ubiquitin–proteasomal system (UPS)/proteasome | Selective protein degradation; clear mechanisms of action; relatively high selectivity; catalytic mode of action | E3-ligase- and ubiquitination-dependent; undesirable PK/PD profile; low BBB penetration capability |
| AUTAC | Triggers K63 polyubiquitin and autophagosome | Long-lived; insoluble protein aggregates | Selective macroautophagy–lysosomal | Potential broad target; independent proteasome; ability to degrade mitochondria | Unclear mechanism of action; dependent on K63 ubiquitination; possible selective autophagy |
| AUTOTAC | p62/SQSRM1, one of the selective autophagy receptors | Long-lived; insoluble protein aggregates | Selective macroautophagy–lysosomal | Broad target scope leverages autophagy pathway; selective degradation; reduced off-target effects | Dependency on autophagy pathway; limited understanding of mechanisms; off-target effects of AUTOTACs |
| ATTEC | LC-3, a key protein of autophagosome | Long-lived; insoluble protein aggregates | Macroautophagy–lysosomal | Potentially a broad target spectrum; direct targeting to the degradation machinery; potentially effective in all cell types; low molecular weight | The LC3-bound chemical moieties need to be solved; lack of studies on designed chimeras |
| LYTAC | Lysosome-targeting receptors (LTRs) | Extracellular; membrane; cell-surface protein | Endosome–lysosomal pathway for degradation of glycosylated proteins | Applicable to extracellular and transmembrane proteins; UPS/proteosomal-independent degradation | Not applicable to cytosolic proteins; large molecular weight and poor permeability; possible immunogenicity as it uses peptide-like structures |
| GalNAc-LYTAC | LTR (ASGPR) | Extracellular; membrane; cell-surface protein | LTR (ASGPR)-mediated internalization and lysosomal delivery | Selective membrane protein degradation; preserves lysosomal integrity | Modulate PK properties to control off-target clearance |
| MoDE-As | ASGPR | Extracellular; membrane; cell-surface proteins | Endosome–lysosomal pathway | Selective targeting of extracellular proteins; proteasome independent degradation; relatively small in size | Limited intracellular targets; challenges of specific cellular targets; short half-life in vivo |
| TransMoDEs | ASGPR | Extracellular; membrane | lysosomal | Hepatocyte-specific degradation; ubiquitin–proteasome independent | Limited to receptor-expressing tissues and extracellular targets; early-stage development |
| IFLD | αvβ3 integrin | Extracellular and membrane proteins | RGD peptide: integrin-mediated endocytosis | Efficient degradation of extracellular and membrane proteins | Dependence on integrin expression; risk of off-target internalization; early-stage research |
| AbTAC/PROTAB | E3-ligase | Extracellular and membrane proteins | RNF43- or ZNRF3-mediated lysosomal degradation | Targets membrane proteins; high specificity vs. antibody | High cost; potential immune response in vivo; unclear endocytosis mechanism; Hook effect |
| GlueTAC | CPP-LSS | Extracellular and membrane proteins | CPP-LSS-mediated internalization and lysosomal degradation | Targets membrane proteins; independent of specific receptors or E3 ligase; irreversible attachment of covalent nanobodies to surface POI | Safety profiles of GlueTACs containing unnatural amino acid half-life needs to be determined |
| KineTAC | CXCL7 | CXCL12: CXCR7-mediated endocytosis | Lysosome | Degrades membrane proteins with intracellular domains and small molecules | Targets few membrane proteins; binds intracellular site of membrane protein and access; UPS in cytosol; importance of linker on binding |
| TransTAC | TfR protein | Transmembrane proteins | Lysosome | Proteasome-independent Target degradation of transmembrane proteins | Poor drug-like properties; Early-stage research; delivery and design complexity |
| IFLD | αvβ3 integrin | Extracellular and membrane proteins | RGD peptide: integrin-mediated endocytosis | Efficient degradation of extracellular and membrane proteins | Dependence on integrin expression; risk of off-target internalization; early-stage research |
| AceTACs | LC3 on the autophagosome membrane | Extracellular and membrane proteins | Lysosome | Proteasome-independent; selective degradation of the target-specific substrates; lower off-target degradation | Poor oral bioavailability and permeability; early-stage research |
| FRTAC | Folate receptor | Extracellular and membrane proteins | Lysosome | Cancer cell selectivity improved precision and efficiency | Off-target effects; unclear mechanism; early-stage research; hook effects |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alam, M.M.; Wasim, S.; Lee, S.-Y. Emerging Concepts of Targeted Protein Degrader Technologies via Lysosomal Pathways. Int. J. Mol. Sci. 2025, 26, 5582. https://doi.org/10.3390/ijms26125582
Alam MM, Wasim S, Lee S-Y. Emerging Concepts of Targeted Protein Degrader Technologies via Lysosomal Pathways. International Journal of Molecular Sciences. 2025; 26(12):5582. https://doi.org/10.3390/ijms26125582
Chicago/Turabian StyleAlam, Mohammad Maqusood, Sobia Wasim, and Sang-Yoon Lee. 2025. "Emerging Concepts of Targeted Protein Degrader Technologies via Lysosomal Pathways" International Journal of Molecular Sciences 26, no. 12: 5582. https://doi.org/10.3390/ijms26125582
APA StyleAlam, M. M., Wasim, S., & Lee, S.-Y. (2025). Emerging Concepts of Targeted Protein Degrader Technologies via Lysosomal Pathways. International Journal of Molecular Sciences, 26(12), 5582. https://doi.org/10.3390/ijms26125582






