Sponge bHLH Gene Expression in Xenopus laevis Disrupts Inner Ear and Lateral Line Neurosensory Development and Otic Afferent Pathfinding
Abstract
1. Introduction
2. Results
2.1. AmqbHLH1 Effects on the Long-Term Development of Ectopic Neurons in the Skin
2.2. Effect on Lateral Line Neurosensory Development
2.3. Effect on Inner Ear Development
2.4. Effect on Inner Ear Afferent Central Pathfinding
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animals
4.3. AmqbHLH1 and Mouse Neurod1 mRNA Injections
4.4. In Situ Hybridization
4.5. Ear Transplantations
4.6. Immunohistochemistry
4.7. Quantification of Ectodermal Neurons
4.8. Lipophilic Dye Tracing
4.9. Statistical Tests
4.10. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Imayoshi, I.; Kageyama, R. bHLH Factors in Self-Renewal, Multipotency, and Fate Choice of Neural Progenitor Cells. Neuron 2014, 82, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, R.; Shimojo, H.; Ohtsuka, T. Dynamic control of neural stem cells by bHLH factors. Neurosci. Res. 2018, 138, 12–18. [Google Scholar] [CrossRef]
- Elliott, K.L.; Pavlínková, G.; Chizhikov, V.V.; Yamoah, E.N.; Fritzsch, B. Development in the Mammalian Auditory System Depends on Transcription Factors. Int. J. Mol. Sci. 2021, 22, 4189. [Google Scholar] [CrossRef]
- Zine, A.; Fritzsch, B. Early Steps towards Hearing: Placodes and Sensory Development. Int. J. Mol. Sci. 2023, 24, 6994. [Google Scholar] [CrossRef] [PubMed]
- Butts, J.C.; Wu, S.R.; Durham, M.A.; Dhindsa, R.S.; Revelli, J.P.; Ljungberg, M.C.; Saulnier, O.; McLaren, M.E.; Taylor, M.D.; Zoghbi, H.Y. A single-cell transcriptomic map of the developing Atoh1 lineage identifies neural fate decisions and neuronal diversity in the hindbrain. Dev. Cell 2024, 59, 2171–2188.e2177. [Google Scholar] [CrossRef]
- Pyott, S.J.; Pavlinkova, G.; Yamoah, E.N.; Fritzsch, B. Harmony in the Molecular Orchestra of Hearing: Developmental Mechanisms from the Ear to the Brain. Annu. Rev. Neurosci. 2024, 47, 1–20. [Google Scholar] [CrossRef]
- Fortunato, S.A.; Vervoort, M.; Adamski, M.; Adamska, M. Conservation and divergence of bHLH genes in the calcisponge Sycon ciliatum. EvoDevo 2016, 7, 23. [Google Scholar] [CrossRef]
- Simionato, E.; Ledent, V.; Richards, G.; Thomas-Chollier, M.; Kerner, P.; Coornaert, D.; Degnan, B.M.; Vervoort, M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: Insights from comparative genomics. BMC Evol. Biol. 2007, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Marletaz, F.; Timoshevskaya, N.; Timoshevskiy, V.; Simakov, O.; Parey, E.; Gavriouchkina, D.; Suzuki, M.; Kubokawa, K.; Brenner, S.; Smith, J. The hagfish genome and the evolution of vertebrates. Nature 2024, 627, 811–820. [Google Scholar] [CrossRef]
- Simakov, O.; Bredeson, J.; Berkoff, K.; Marletaz, F.; Mitros, T.; Schultz, D.T.; O’Connell, B.L.; Dear, P.; Martinez, D.E.; Steele, R.E. Deeply conserved synteny and the evolution of metazoan chromosomes. Sci. Adv. 2022, 8, eabi5884. [Google Scholar] [CrossRef]
- Fritzsch, B.; Elliott, K.L. Gene, cell, and organ multiplication drives inner ear evolution. Dev. Biol. 2017, 431, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Kopecky, B.J.; Jahan, I.; Fritzsch, B. Correct timing of proliferation and differentiation is necessary for normal inner ear development and auditory hair cell viability. Dev. Dyn. 2013, 242, 132–147. [Google Scholar] [CrossRef]
- Sebé-Pedrós, A.; Chomsky, E.; Pang, K.; Lara-Astiaso, D.; Gaiti, F.; Mukamel, Z.; Amit, I.; Hejnol, A.; Degnan, B.M.; Tanay, A. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat. Ecol. Evol. 2018, 2, 1176. [Google Scholar] [CrossRef]
- Mah, J.L.; Leys, S.P. Think like a sponge: The genetic signal of sensory cells in sponges. Dev. Biol. 2017, 431, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Leys, S.P.; Grombacher, L.; Field, D.; Elliott, G.R.; Ho, V.R.; Kahn, A.S.; Reid, P.J.; Riesgo, A.; Lanna, E.; Bobkov, Y. A morphological cell atlas of the freshwater sponge Ephydatia muelleri with key insights from targeted single-cell transcriptomes. EvoDevo 2025, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Fritzsch, B.; Straka, H. Evolution of vertebrate mechanosensory hair cells and inner ears: Toward identifying stimuli that select mutation driven altered morphologies. J. Comp. Physiol. A 2014, 200, 5–18. [Google Scholar] [CrossRef]
- Coyle, M.; King, N. The Evolutionary Foundations of Animal Transcriptional Regulatory Mechanisms. Preprints 2024. [Google Scholar]
- Erives, A.; Fritzsch, B. A screen for gene paralogies delineating evolutionary branching order of early metazoa. G3 Genes Genomes Genet. 2020, 10, 811–826. [Google Scholar] [CrossRef]
- Richards, G.S.; Simionato, E.; Perron, M.; Adamska, M.; Vervoort, M.; Degnan, B.M. Sponge Genes Provide New Insight into the Evolutionary Origin of the Neurogenic Circuit. Curr. Biol. 2008, 18, 1156–1161. [Google Scholar] [CrossRef]
- Kim, P.; Helms, A.W.; Johnson, J.E.; Zimmerman, K. XATH-1,a Vertebrate Homolog ofDrosophila atonal, Induces Neuronal Differentiation within Ectodermal Progenitors. Dev. Biol. 1997, 187, 1–12. [Google Scholar] [CrossRef]
- Lee, J.; Hollenberg, S.; Snider, L.; Turner, D.; Lipnick, N.; Weintraub, H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science 1995, 268, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Sommer, L.; Cserjesi, P.; Anderson, D.J. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J. Neurosci. 1997, 17, 3644–3652. [Google Scholar] [CrossRef]
- Ranganathan, R.; Sari, F.; Wang, S.X.; Thiery, A.; Buzzi, A.L.; Guerra, R.; Moody, S.A.; Streit, A. Targets of the transcription factor Six1 identify previously unreported candidate deafness genes. Development 2025, 152, dev204533. [Google Scholar] [CrossRef] [PubMed]
- Nieuwkoop, P.; Faber, J. Normal Table of Xenopus Laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis; Garland Publishing, Inc.: New York, NY, USA, 1994. [Google Scholar]
- Chagnaud, B.P.; Engelmann, J.; Fritzsch, B.; Glover, J.C.; Straka, H. Sensing External and Self-Motion with Hair Cells: A Comparison of the Lateral Line and Vestibular Systems from a Developmental and Evolutionary Perspective. Brain Behav. Evol. 2017, 90, 98–116. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, G.; Ahrens, K. Molecular anatomy of placode development in Xenopus laevis. Dev. Biol. 2004, 271, 439–466. [Google Scholar] [CrossRef] [PubMed]
- Jahan, I.; Elliott, K.L.; Fritzsch, B. Understanding molecular evolution and development of the organ of Corti can provide clues for hearing restoration. Integr. Comp. Biol. 2018, 58, 351–365. [Google Scholar] [CrossRef]
- Gálvez, H.; Abelló, G.; Giraldez, F. Signaling and transcription factors during inner ear development: The generation of hair cells and otic neurons. Front. Cell Dev. Biol. 2017, 5, 21. [Google Scholar] [CrossRef]
- Filova, I.; Dvorakova, M.; Bohuslavova, R.; Pavlinek, A.; Elliott, K.L.; Vochyanova, S.; Fritzsch, B.; Pavlinkova, G. Combined Atoh1 and Neurod1 deletion reveals autonomous growth of auditory nerve fibers. Mol. Neurobiol. 2020, 57, 5307–5323. [Google Scholar] [CrossRef]
- Schlosser, G. Chapter Four—Making Senses: Development of Vertebrate Cranial Placodes. In International Review of Cell and Molecular Biology; Kwang, J., Ed.; Academic Press: Cambridge, MA, USA, 2010; Volume 283, pp. 129–234. [Google Scholar]
- Elliott, K.L.; Fritzsch, B. Evolution and Development of Lateral Line and Electroreception: An Integrated Perception of Neurons, Hair Cells and Brainstem Nuclei. In The Senses; Fritzsch, B., Bleckmann, H., Eds.; Mechanosensory Lateral Line, electroreception, magnetoreception; Elsevier: Amsterdam, The Netherlands, 2020; Volume 7, pp. 95–115. [Google Scholar]
- Ma, N.X.; Puls, B.; Chen, G. Transcriptomic analyses of NeuroD1-mediated astrocyte-to-neuron conversion. Dev. Neurobiol. 2022, 82, 375–391. [Google Scholar] [CrossRef]
- Pataskar, A.; Jung, J.; Smialowski, P.; Noack, F.; Calegari, F.; Straub, T.; Tiwari, V.K. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J. 2016, 35, 24–45. [Google Scholar] [CrossRef]
- Dvorakova, M.; Macova, I.; Bohuslavova, R.; Anderova, M.; Fritzsch, B.; Pavlinkova, G. Early ear neuronal development, but not olfactory or lens development, can proceed without SOX2. Dev. Biol. 2020, 457, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Macova, I.; Pysanenko, K.; Chumak, T.; Dvorakova, M.; Bohuslavova, R.; Syka, J.; Fritzsch, B.; Pavlinkova, G. Neurod1 Is Essential for the Primary Tonotopic Organization and Related Auditory Information Processing in the Midbrain. J. Neurosci. 2019, 39, 984–1004. [Google Scholar] [CrossRef] [PubMed]
- Filova, I.; Bohuslavova, R.; Tavakoli, M.; Yamoah, E.N.; Fritzsch, B.; Pavlinkova, G. Early Deletion of Neurod1 Alters Neuronal Lineage Potential and Diminishes Neurogenesis in the Inner Ear. Front. Cell Dev. Biol. 2022, 10, 845461. [Google Scholar] [CrossRef]
- Fritzsch, B. Sensing Sound: Evolutionary Neurobiology of a Novel Sense of Hearing; CRC Press: Boca Raton, FL, USA, 2023. [Google Scholar]
- Elliott, K.L.; Fritzsch, B. Transplantation of Xenopus laevis ears reveals the ability to form afferent and efferent connections with the spinal cord. Int. J. Dev. Biol. 2010, 54, 1443–1451. [Google Scholar] [CrossRef]
- Gordy, C.; Straka, H.; Houston, D.W.; Fritzsch, B.; Elliott, K.L. Caudal transplantation of ears provides insights into inner ear afferent pathfinding properties. Dev. Neurobiol. 2018, 78, 1064–1080. [Google Scholar] [CrossRef]
- Jahan, I.; Kersigo, J.; Pan, N.; Fritzsch, B. Neurod1 regulates survival and formation of connections in mouse ear and brain. Cell Tissue Res. 2010, 341, 95–110. [Google Scholar] [CrossRef]
- Elliott, K.L.; Kersigo, J.; Pan, N.; Jahan, I.; Fritzsch, B. Spiral ganglion neuron projection development to the hindbrain in mice lacking peripheral and/or central target differentiation. Front. Neural Circuits 2017, 11, 25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fritzsch, B.; Glover, J.C. Gene networks and the evolution of olfactory organs, eyes, hair cells and motoneurons: A view encompassing lancelets, tunicates and vertebrates. Front. Cell Dev. Biol. 2024, 12, 1340157. [Google Scholar] [CrossRef]
- Duncan, J.S.; Fritzsch, B.; Houston, D.W.; Ketchum, E.M.; Kersigo, J.; Deans, M.R.; Elliott, K.L. Topologically correct central projections of tetrapod inner ear afferents require Fzd3. Sci. Rep. 2019, 9, 10298. [Google Scholar] [CrossRef]
- Jahan, I.; Pan, N.; Kersigo, J.; Fritzsch, B. Neurog1 can partially substitute for Atoh1 function in hair cell differentiation and maintenance during organ of Corti development. Development 2015, 142, 2810–2821. [Google Scholar]
- Maier, E.C.; Saxena, A.; Alsina, B.; Bronner, M.E.; Whitfield, T.T. Sensational placodes: Neurogenesis in the otic and olfactory systems. Dev. Biol. 2014, 389, 50–67. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, G. From so simple a beginning—What amphioxus can teach us about placode evolution. Int. J. Dev. Biol. 2017, 61, 633–648. [Google Scholar] [CrossRef]
- Schlosser, G. Evolutionary origins of vertebrate placodes: Insights from developmental studies and from comparisons with other deuterostomes. J. Exp. Zool. 2005, 304B, 347–399. [Google Scholar] [CrossRef] [PubMed]
- Maharana, S.K.; Schlosser, G. A gene regulatory network underlying the formation of pre-placodal ectoderm in Xenopus laevis. Bmc Biol. 2018, 16, 79. [Google Scholar] [CrossRef]
- Li, J.; Cheng, C.; Xu, J.; Zhang, T.; Tokat, B.; Dolios, G.; Ramakrishnan, A.; Shen, L.; Wang, R.; Xu, P.-X. The transcriptional coactivator Eya1 exerts transcriptional repressive activity by interacting with REST corepressors and REST-binding sequences to maintain nephron progenitor identity. Nucleic Acids Res. 2022, 50, 10343–10359. [Google Scholar] [CrossRef]
- Xu, J.; Li, J.; Zhang, T.; Jiang, H.; Ramakrishnan, A.; Fritzsch, B.; Shen, L.; Xu, P.X. Chromatin remodelers and lineage-specific factors interact to target enhancers to establish proneurosensory fate within otic ectoderm. Proc. Natl. Acad. Sci. USA 2021, 118, e2025196118. [Google Scholar] [CrossRef]
- Fritzsch, B.; Erives, A.; Eberl, D.F.; Yamoah, E.N. 2.16-Genetics of Mechanoreceptor Evolution and Development. In The Senses; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 277–301. [Google Scholar]
- Shimojo, H.; Masaki, T.; Kageyama, R. The Neurog2-Tbr2 axis forms a continuous transition to the neurogenic gene expression state in neural stem cells. Dev. Cell 2024, 59, 1913–1923.e1916. [Google Scholar] [CrossRef] [PubMed]
- Le Dréau, G.; Escalona, R.; Fueyo, R.; Herrera, A.; Martínez, J.D.; Usieto, S.; Menendez, A.; Pons, S.; Martinez-Balbas, M.A.; Marti, E. E proteins sharpen neurogenesis by modulating proneural bHLH transcription factors’ activity in an E-box-dependent manner. eLife 2018, 7, e37267. [Google Scholar] [CrossRef] [PubMed]
- Kopecky, B.; Fritzsch, B. The Myc road to hearing restoration. Cells 2012, 1, 667–698. [Google Scholar] [CrossRef]
- Mann, Z.F.; Galvez, H.; Pedreno, D.; Chen, Z.; Chrysostomou, E.; Żak, M.; Kang, M.; Canden, E.; Daudet, N. Shaping of inner ear sensory organs through antagonistic interactions between Notch signalling and Lmx1a. eLife 2017, 6, e33323. [Google Scholar] [CrossRef]
- Chizhikov, V.V.; Iskusnykh, I.Y.; Fattakhov, N.; Fritzsch, B. Lmx1a and Lmx1b are redundantly required for the development of multiple components of the mammalian auditory system. Neuroscience 2021, 452, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.H.; Pauley, S.; Jahan, I.; Beisel, K.W.; Millen, K.J.; Fritzsch, B. Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res. 2008, 334, 339–358. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Bouma, J.; Kopecky, B.; Jahan, I.; Beisel, K.W.; He, D.; Liu, D.; Fritzsch, B. Interaction with ectopic cochlear crista sensory epithelium disrupts basal cochlear sensory epithelium development in Lmx1a mutant mice. Cell Tissue Res. 2020, 380, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.K.; Frantz, G.D.; Carpenter, D.A.; de Sauvage, F.J.; Gao, W.Q. Distinct expression patterns of notch family receptors and ligands during development of the mammalian inner ear. Mech. Dev. 1998, 78, 159–163. [Google Scholar] [CrossRef]
- Haddon, C.; Mowbray, C.; Whitfield, T.; Jones, D.; Gschmeissner, S.; Lewis, J. Hair cells without supporting cells: Further studies in the ear of the zebrafish mind bomb mutant. J. Neurocytol. 1999, 28, 837–850. [Google Scholar] [CrossRef]
- Matei, V.; Pauley, S.; Kaing, S.; Rowitch, D.; Beisel, K.W.; Morris, K.; Feng, F.; Jones, K.; Lee, J.; Fritzsch, B. Smaller inner ear sensory epithelia in Neurog1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 2005, 234, 633–650. [Google Scholar] [CrossRef]
- Doetzlhofer, A.; Basch, M.L.; Ohyama, T.; Gessler, M.; Groves, A.K.; Segil, N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell 2009, 16, 58–69. [Google Scholar] [CrossRef]
- Elliott, K.L.; Fritzsch, B. Ear transplantations reveal conservation of inner ear afferent pathfinding cues. Sci. Rep. 2018, 8, 13819. [Google Scholar] [CrossRef]
- Glover, J.C.; Elliott, K.L.; Erives, A.; Chizhikov, V.V.; Fritzsch, B. Wilhelm His’ lasting insights into hindbrain and cranial ganglia development and evolution. Dev. Biol. 2018, 444, S14–S24. [Google Scholar] [CrossRef]
- Weintraub, H.; Tapscott, S.J.; Davis, R.L.; Thayer, M.J.; Adam, M.A.; Lassar, A.B.; Miller, A.D. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. USA 1989, 86, 5434–5438. [Google Scholar] [CrossRef]
- Zine, A.; Messat, Y.; Fritzsch, B. A human induced pluripotent stem cell-based modular platform to challenge sensorineural hearing loss. Stem Cells 2021, 39, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Fritzsch, B.; Pan, N.; Jahan, I.; Elliott, K.L. Inner ear development: Building a spiral ganglion and an organ of Corti out of unspecified ectoderm. Cell Tissue Res. 2015, 361, 7–24. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Driver, E.C.; Sillers, L.; Coate, T.M.; Rose, M.F.; Kelley, M.W. The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev. Biol. 2013, 376, 86–98. [Google Scholar] [CrossRef]
- McGovern, M.M.; Zhou, L.; Randle, M.R.; Cox, B.C. Spontaneous hair cell regeneration is prevented by increased notch signaling in supporting cells. Front. Cell. Neurosci. 2018, 12, 120. [Google Scholar] [CrossRef]
- Liu, Z.; Dearman, J.A.; Cox, B.C.; Walters, B.J.; Zhang, L.; Ayrault, O.; Zindy, F.; Gan, L.; Roussel, M.F.; Zuo, J. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J. Neurosci. 2012, 32, 6600–6610. [Google Scholar] [CrossRef]
- Kelly, M.C.; Chang, Q.; Pan, A.; Lin, X.; Chen, P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J. Neurosci. 2012, 32, 6699–6710. [Google Scholar] [CrossRef]
- McGovern, M.M.; Hosamani, I.V.; Niu, Y.; Nguyen, K.Y.; Zong, C.; Groves, A.K. Expression of Atoh1, Gfi1, and Pou4f3 in the mature cochlea reprograms nonsensory cells into hair cells. Proc. Natl. Acad. Sci. USA 2024, 121, e2304680121. [Google Scholar] [CrossRef]
- Yamoah, E.N.; Li, M.; Shah, A.; Elliott, K.L.; Cheah, K.; Xu, P.X.; Phillips, S.; Young, S.M., Jr.; Eberl, D.F.; Fritzsch, B. Using Sox2 to alleviate the hallmarks of age-related hearing loss. Ageing Res. Rev. 2020, 59, 101042. [Google Scholar] [CrossRef] [PubMed]
- Crook, A.C.; Whiteman, H.H. An Evaluation of MS-222 and Benzocaine as Anesthetics for Metamorphic and Paedomorphic Tiger Salamanders (Ambystoma tigrinum nebulosum). Am. Midl. Nat. 2006, 155, 417–421. [Google Scholar] [CrossRef]
- Oschwald, R.; Richter, K.; Grunz, H. Localization of a nervous system-specific class II beta-tubulin gene in Xenopus laevis embryos by whole-mount in situ hybridization. Int. J. Dev. Biol. 2004, 35, 399–405. [Google Scholar]
- Kerr, T.C.; Cuykendall, T.N.; Luettjohann, L.C.; Houston, D.W. Maternal Tgif1 regulates nodal gene expression in Xenopus. Dev. Dyn. 2008, 237, 2862–2873. [Google Scholar] [CrossRef] [PubMed]
- Sive, H.; Grainger, R.; Harland, R. Whole-mount in situ hybridization. In Early Development of Xenopus Laevis. A Laboratory Manual; Cold Spring Habor Laboratory Press: New York, NY, USA, 2000; pp. 249–274. [Google Scholar]
- Elliott, K.L.; Houston, D.W.; DeCook, R.; Fritzsch, B. Ear manipulations reveal a critical period for survival and dendritic development at the single-cell level in Mauthner neurons. Dev. Neurobiol. 2015, 75, 1139–1351. [Google Scholar] [CrossRef] [PubMed]
- Farinas, I.; Jones, K.R.; Tessarollo, L.; Vigers, A.J.; Huang, E.; Kirstein, M.; de Caprona, D.C.; Coppola, V.; Backus, C.; Reichardt, L.F.; et al. Spatial Shaping of Cochlear Innervation by Temporally Regulated Neurotrophin Expression. J. Neurosci. 2001, 21, 6170–6180. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, C.L.; Chapman, B.J.; Guidi, J.L.; Terry, C.E.; Mangiardi, D.A.; Cotanche, D.A. Comparison of activated caspase detection methods in the gentamicin-treated chick cochlea. Hear. Res. 2008, 240, 1–11. [Google Scholar] [CrossRef]
- Fritzsch, B.; Duncan, J.S.; Kersigo, J.; Gray, B.; Elliott, K.L. Neuroanatomical Tracing Techniques in the Ear: History, State of the Art, and Future Developments. In Auditory and Vestibular Research: Methods and Protocols; Sokolowski, B., Ed.; Springer Science+Business Media: New York, NY, USA, 2016; Volume 1427, pp. 243–262. [Google Scholar]

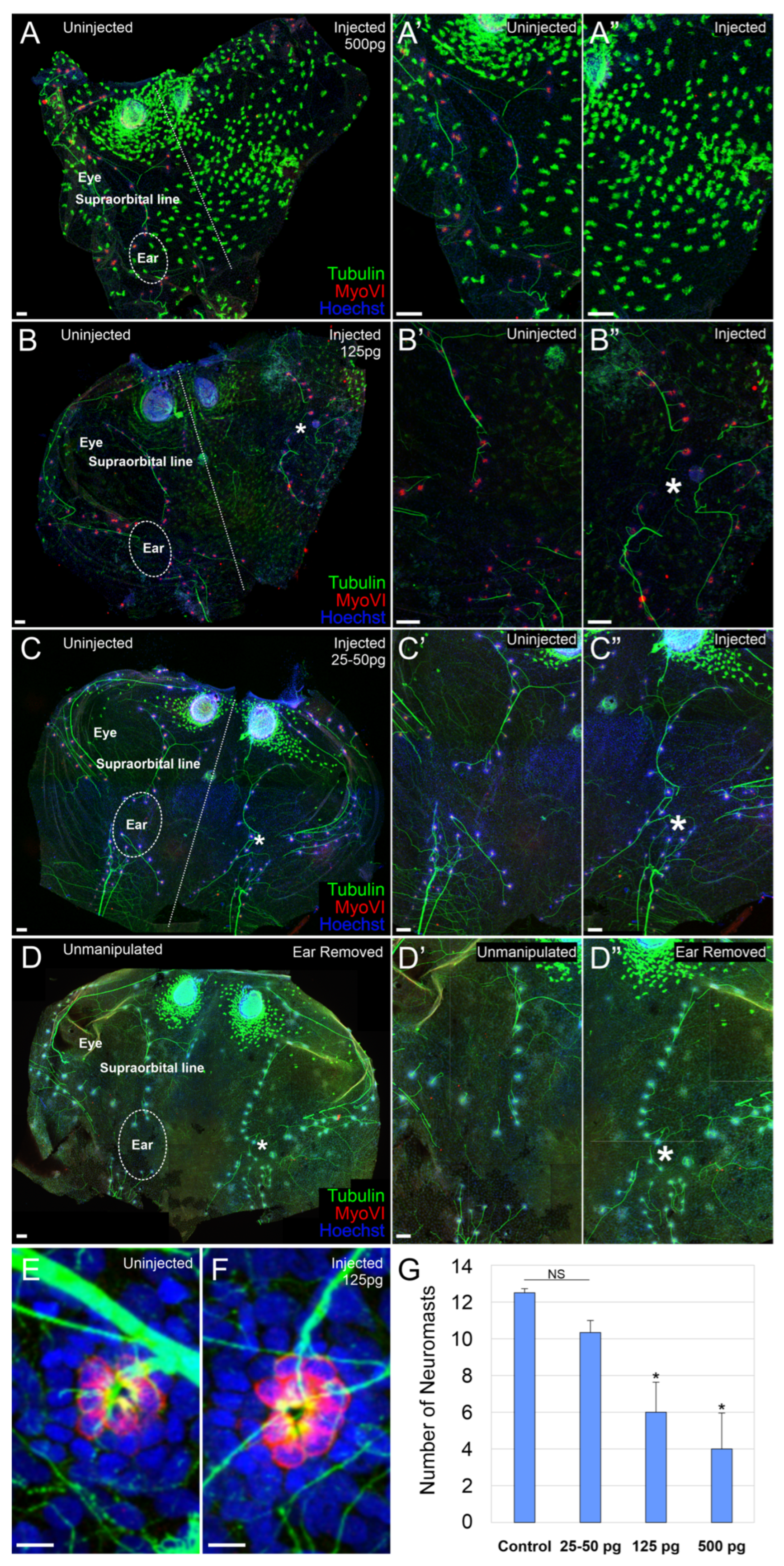
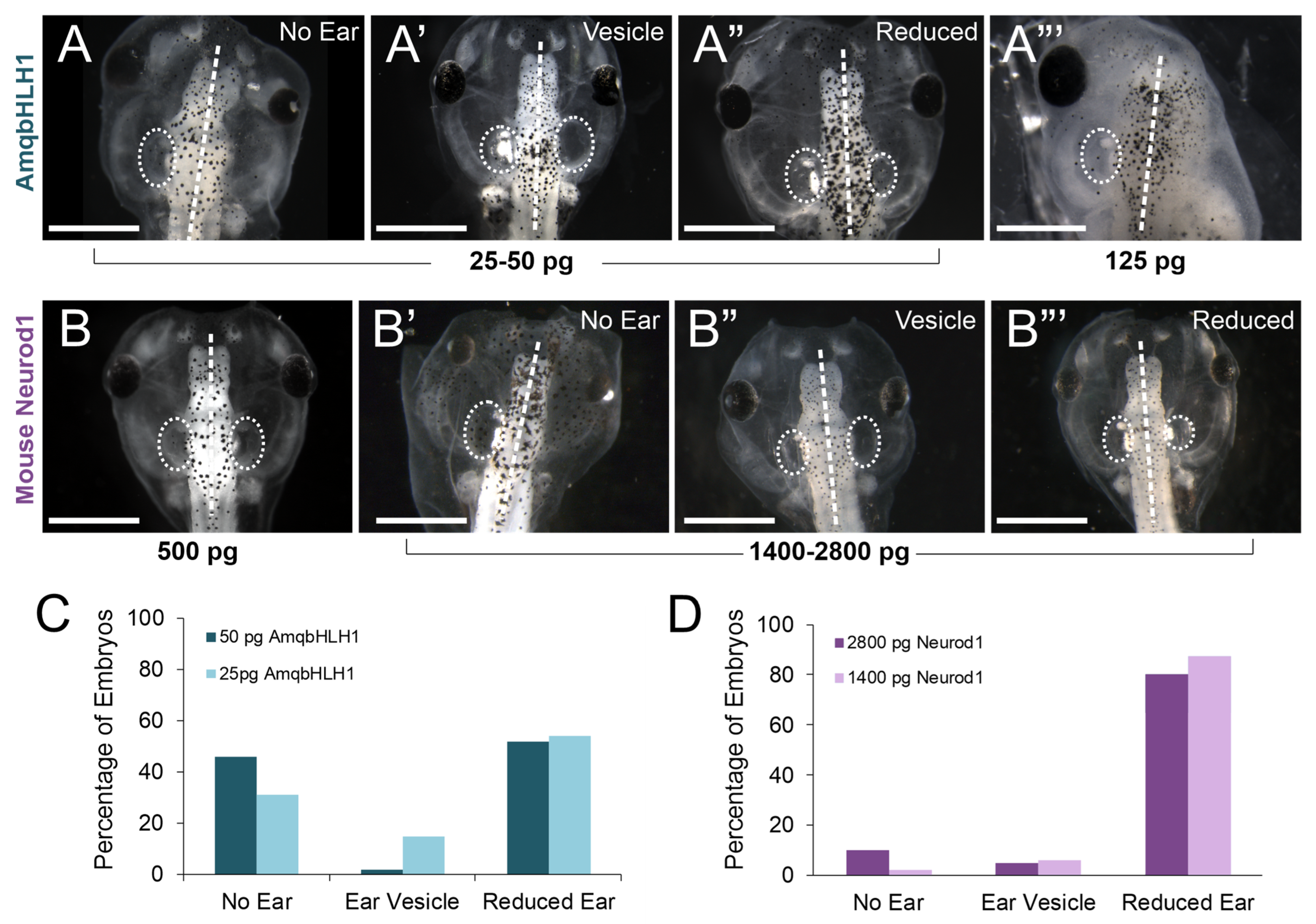

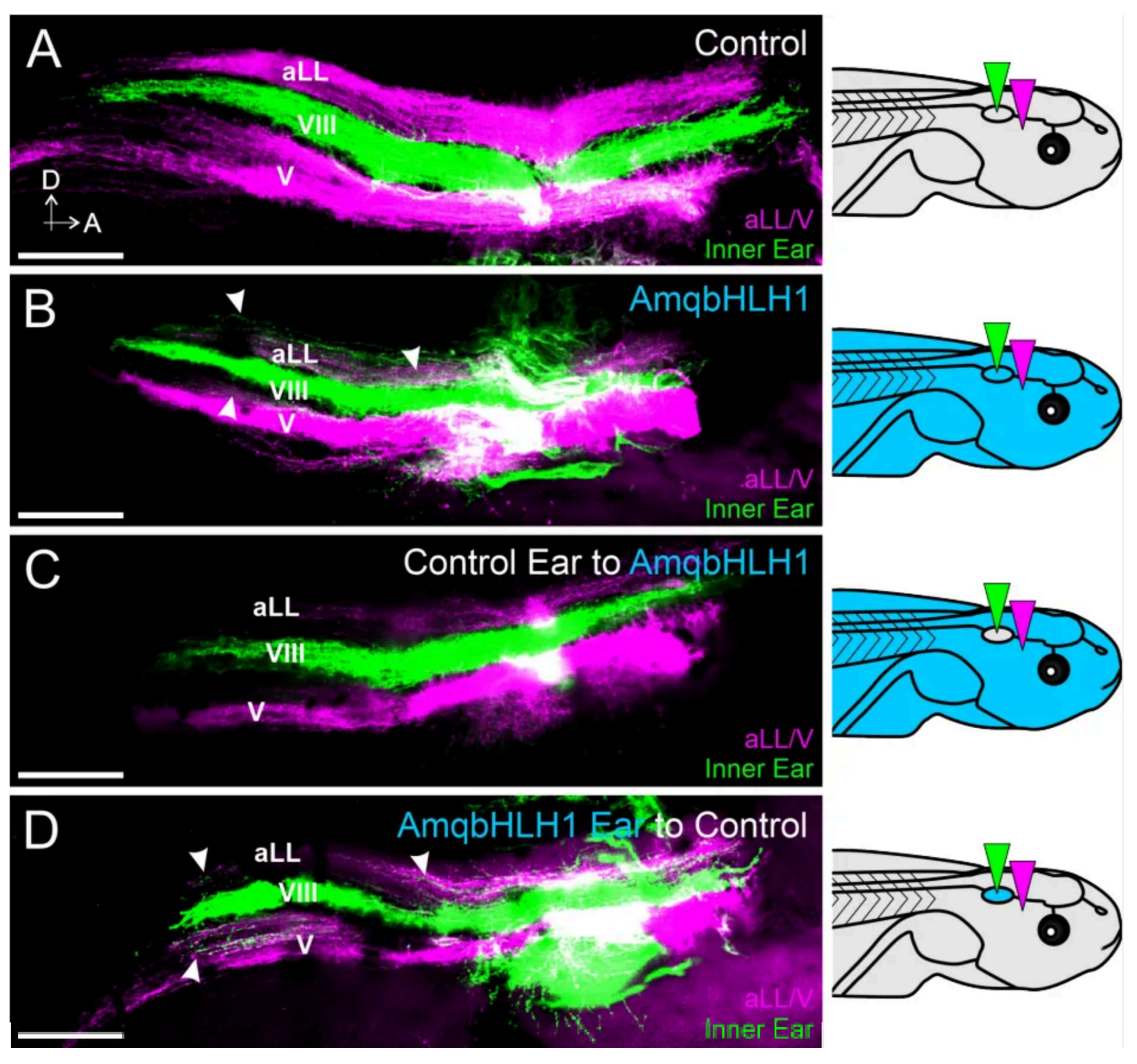
| Dose of mRNA | Total | No Ear | Ear Vesicle | Reduced Ear | No Effect |
|---|---|---|---|---|---|
| 25 pg AmqbHLH1 | 74 | 23 | 11 | 40 | 0 |
| 50 pg AmqbHLH1 | 116 | 53 | 3 | 60 | 0 |
| 125 pg AmqbHLH1 | 35 | 34 | 1 | 0 | 0 |
| 500 pg AmqbHLH1 | 12 | 12 | 0 | 0 | 0 |
| 25 pg Neurod1 | 10 | 0 | 0 | 0 | 10 |
| 500 pg Neurod1 | 14 | 0 | 0 | 0 | 14 |
| 1400 pg Neurod1 | 90 | 2 | 2 | 37 | 49 |
| 2800 pg Neurod1 | 93 | 9 | 3 | 52 | 29 |
| Phenotype | U, Fused S-L, 3 Canals | Fused U-S-L, 4 Canals | Fused U-S-L, 3 Canals | Fused U-S-L 2 Canals | Fused U-S-L 1 Canal | Fused U-S-L No Canals | No HCs |
|---|---|---|---|---|---|---|---|
| Ear Vesicle | 0 | 0 | 0 | 1 | 1 | 3 | 0 |
| Reduced Ear | 2 | 1 | 2 | 1 | 3 | 6 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elliott, K.L.; Gordy, C.; Ingvalson, H.; Holliday, C.; Halyko, J.; Houston, D.W.; Degnan, B.M.; Fritzsch, B. Sponge bHLH Gene Expression in Xenopus laevis Disrupts Inner Ear and Lateral Line Neurosensory Development and Otic Afferent Pathfinding. Int. J. Mol. Sci. 2025, 26, 5487. https://doi.org/10.3390/ijms26125487
Elliott KL, Gordy C, Ingvalson H, Holliday C, Halyko J, Houston DW, Degnan BM, Fritzsch B. Sponge bHLH Gene Expression in Xenopus laevis Disrupts Inner Ear and Lateral Line Neurosensory Development and Otic Afferent Pathfinding. International Journal of Molecular Sciences. 2025; 26(12):5487. https://doi.org/10.3390/ijms26125487
Chicago/Turabian StyleElliott, Karen L., Clayton Gordy, Hannah Ingvalson, Charles Holliday, Jessica Halyko, Douglas W. Houston, Bernard M. Degnan, and Bernd Fritzsch. 2025. "Sponge bHLH Gene Expression in Xenopus laevis Disrupts Inner Ear and Lateral Line Neurosensory Development and Otic Afferent Pathfinding" International Journal of Molecular Sciences 26, no. 12: 5487. https://doi.org/10.3390/ijms26125487
APA StyleElliott, K. L., Gordy, C., Ingvalson, H., Holliday, C., Halyko, J., Houston, D. W., Degnan, B. M., & Fritzsch, B. (2025). Sponge bHLH Gene Expression in Xenopus laevis Disrupts Inner Ear and Lateral Line Neurosensory Development and Otic Afferent Pathfinding. International Journal of Molecular Sciences, 26(12), 5487. https://doi.org/10.3390/ijms26125487






