Genome-Wide Analysis and Expression Profiling of the JAZ Gene Family in Response to Abiotic Stress in Alfalfa
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of the MsJAZs Family of Alfalfa
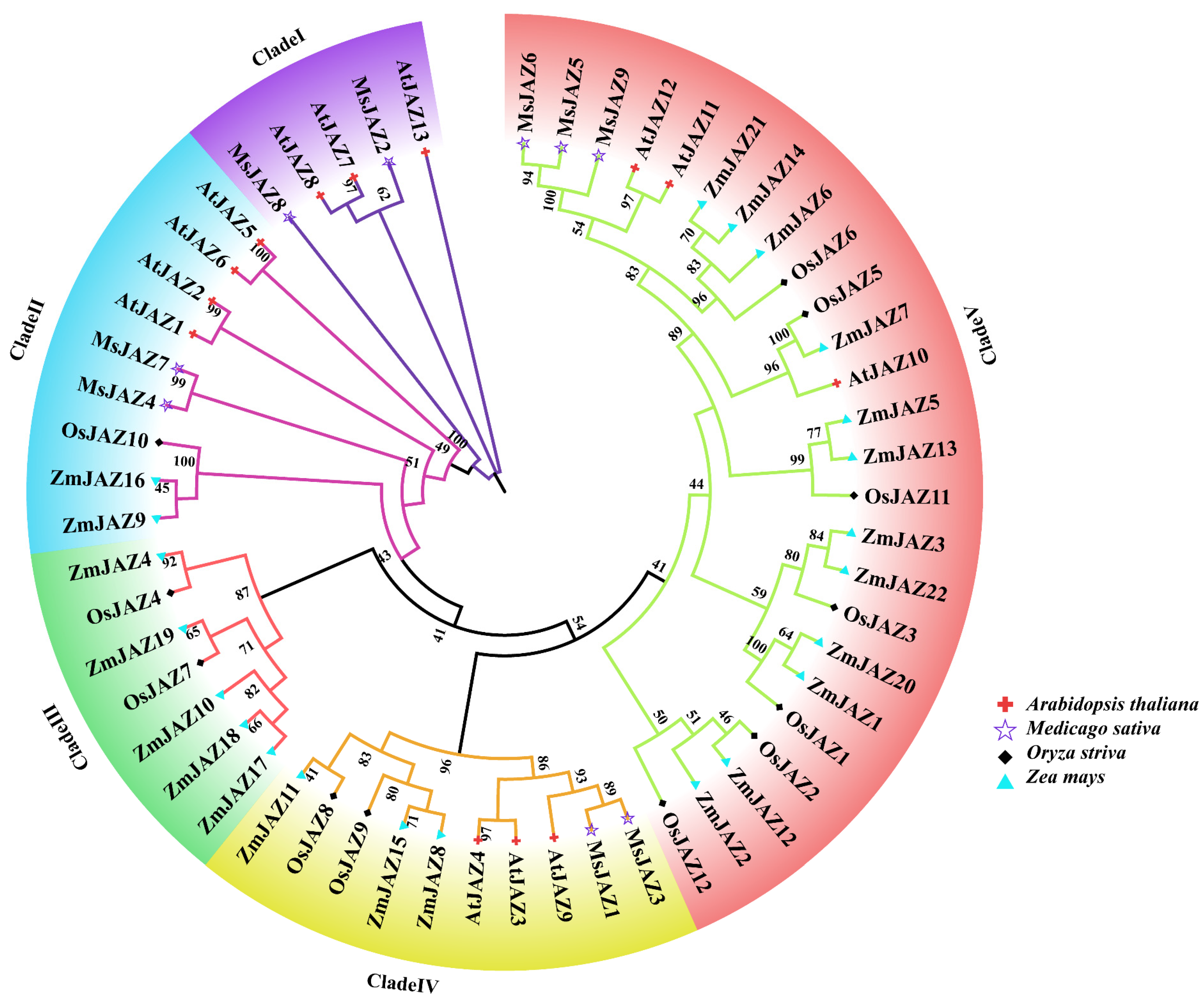
2.2. Phylogenetic Analysis of MsJAZ Proteins in Alfalfa
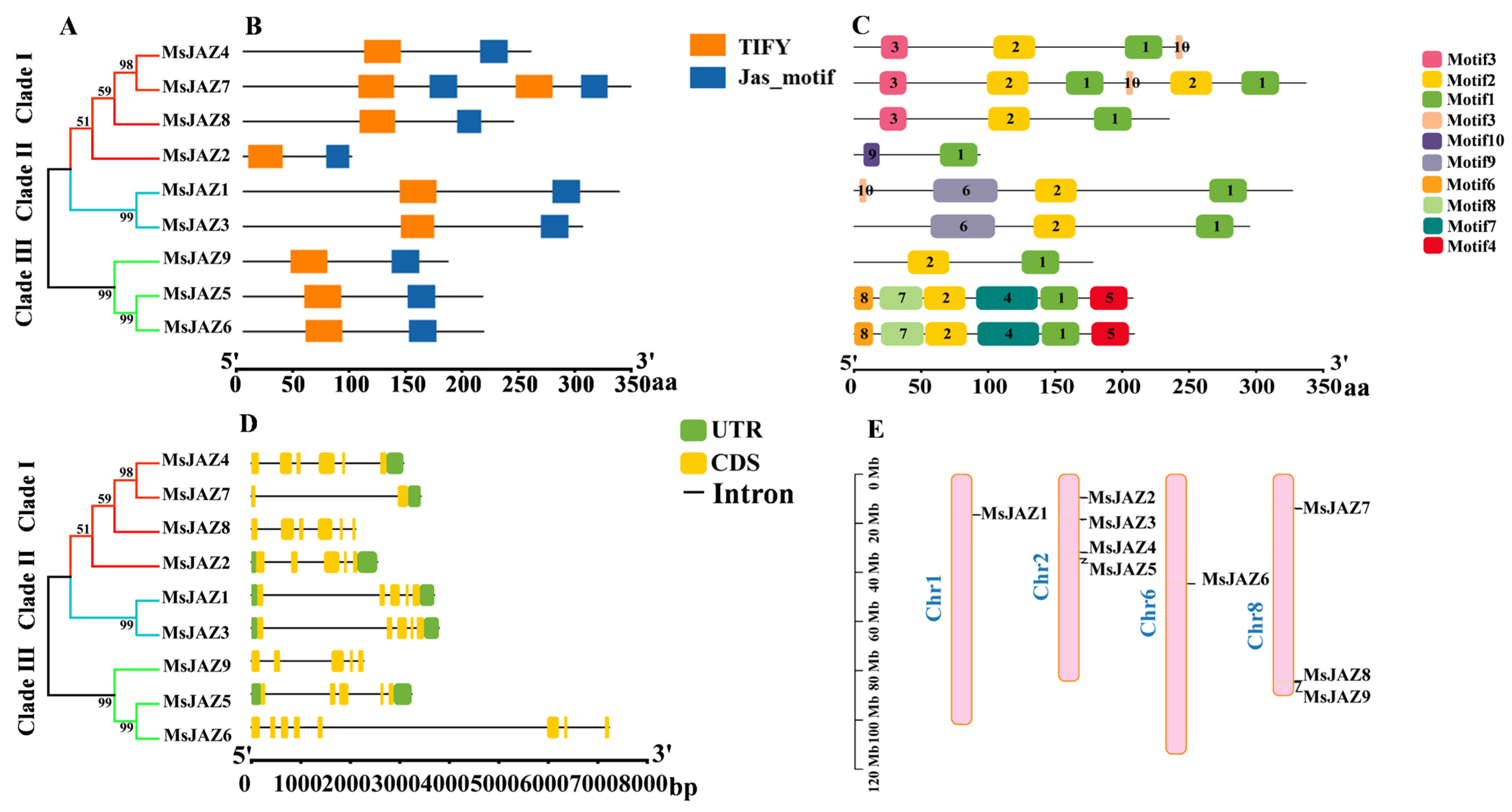
2.3. Functional Domains, Conserved Motifs, and Gene Structure Analysis of MsJAZ Genes
2.4. Analysis of Cis-Acting Elements in the MsJAZ Gene Promoters
2.5. Analysis of the Chromosomal Localization and Collinearity of MsJAZ Genes
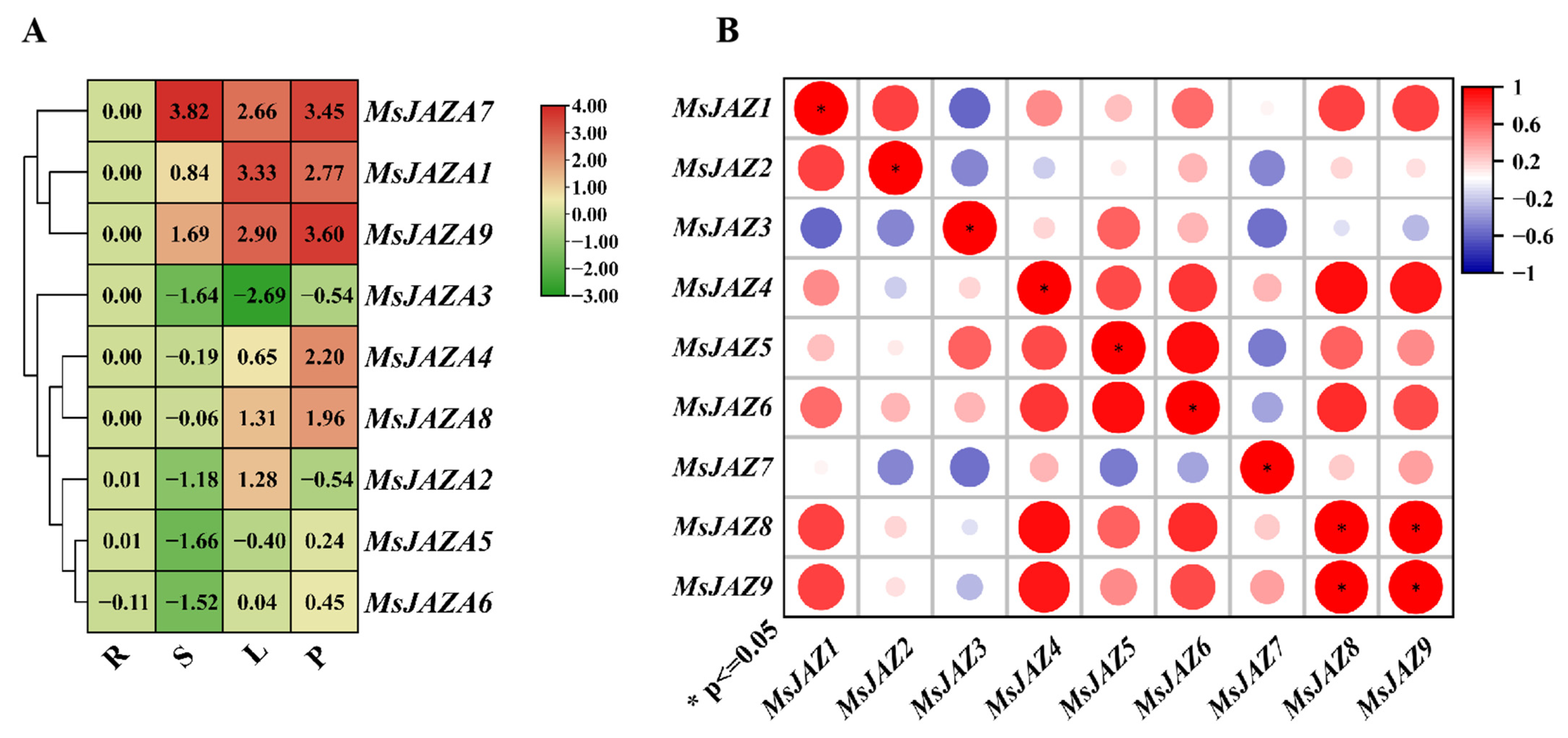
2.6. Expression Patterns of MsJAZ Genes in Different Tissues
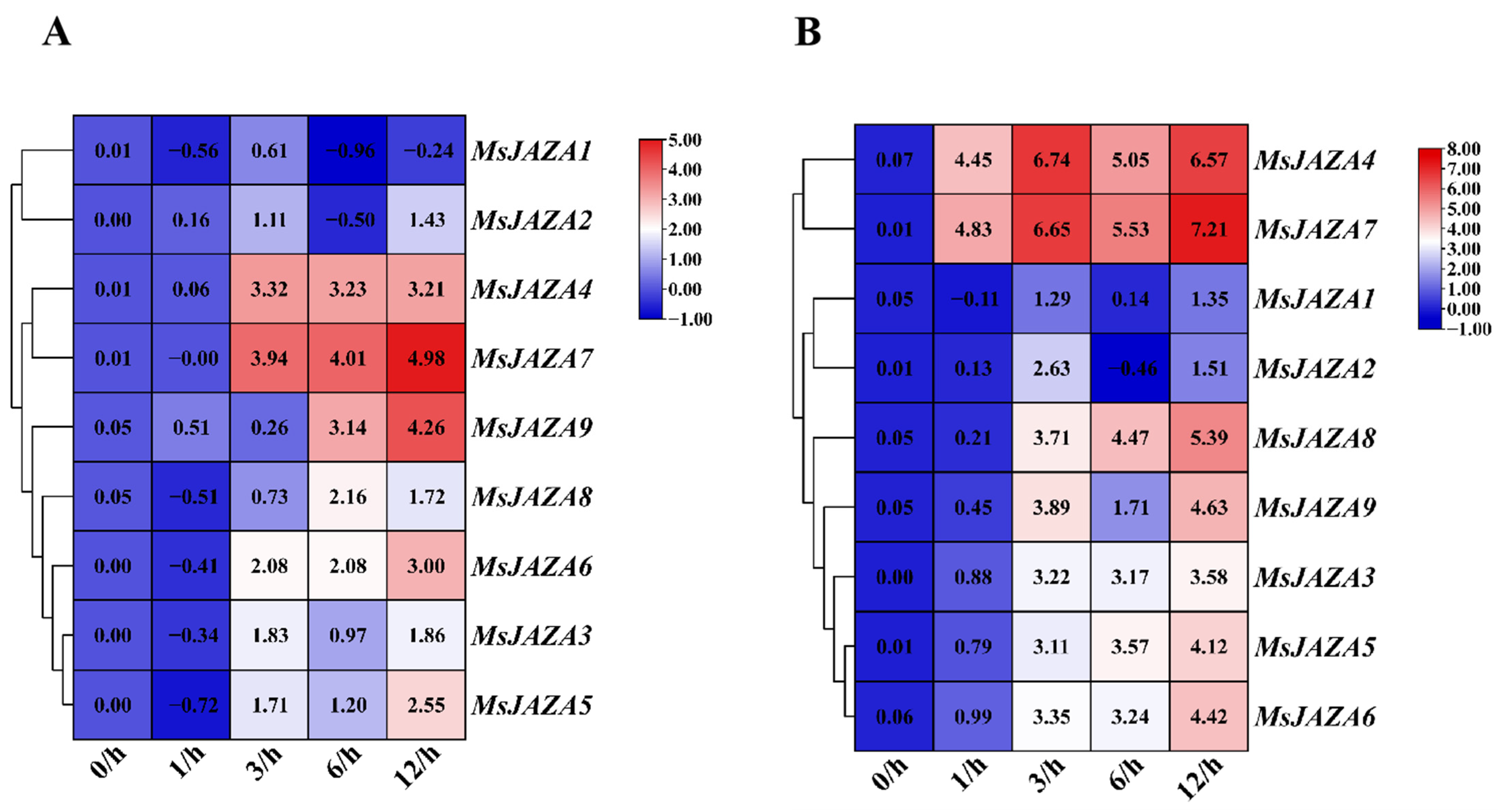
2.7. Expression Patterns of MsJAZ Genes in Response to Abiotic Stress
2.8. Expression Profile of MsJAZ Genes in Response to Chromium and MeJA
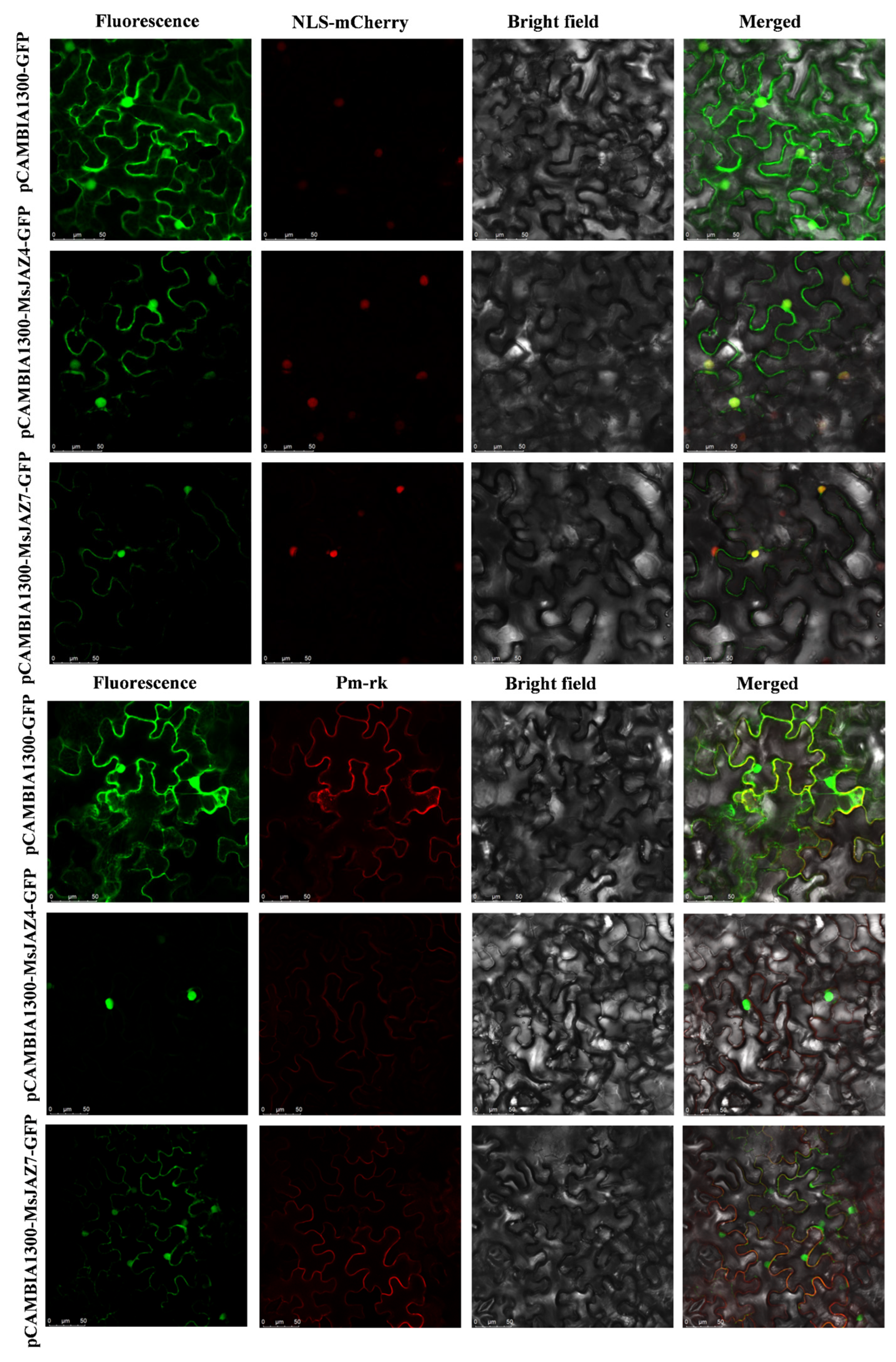
2.9. MsJAZ Protein Interaction Network Prediction and Subcellular Localization
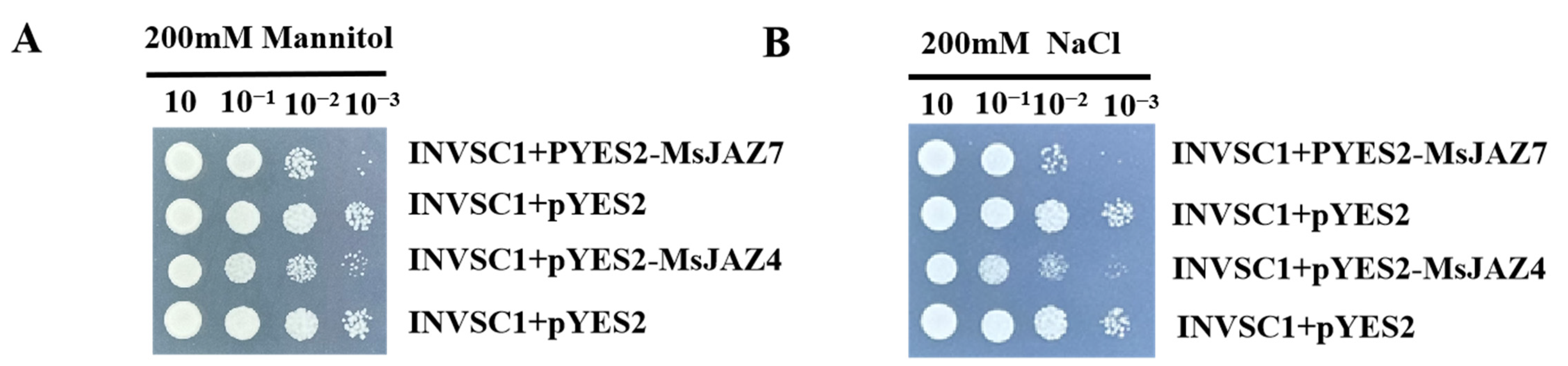
2.10. Functional Validation of the MsJAZ Gene in Yeast
3. Discussion
4. Materials and Methods
4.1. JAZ Gene Identification in Alfalfa
4.2. Basic Analysis of MsJAZ Proteins
4.3. Phylogenetic Analysis
4.4. Functional Domain, Conserved Motifs and Gene Structure Analysis
4.5. Promoter Cis-Acting Element Prediction
4.6. Chromosome Location, Gene Duplication, and Collinearity Analysis
4.7. Plant Materials and Stress Treatments
4.8. RNA Extraction and Quantitative Real Time PCR (RT-qPCR) Analysis
4.9. Subcellular Localization of MsJAZ Proteins
4.10. Functional Validation of the MsJAZ Protein
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Z.; Şakiroğlu, M. The Origin, Evolution, and Genetic Diversity of Alfalfa. In The Alfalfa Genome. Compendium of Plant Genomes; Yu, L.X., Kole, C., Eds.; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, W.H. Priorities for the development of alfalfa pasture in northern China. Fundam. Res. 2022, 3, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.J.; Bi, Y.L.; Geng, M.M.; An, N. Pull-out characteristics of herbaceous roots of alfalfa on the loess in different growth stages and their impacts on slope stability. Soil Tillage Res. 2023, 225, 105542. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, C.; Liu, P.; Jiang, J. The role of root morphology and pulling direction in pullout resistance of alfalfa roots. Front. Plant Sci. 2021, 12, 580825. [Google Scholar] [CrossRef]
- Eiji, N.; Van Wees, S.C.M. Plant hormone functions and interactions in biological systems. Plant J. 2021, 105, 287–289. [Google Scholar] [CrossRef]
- Liu, B.; Seong, K.; Pang, S.; Song, J.; Gao, H.; Wang, C.; Zhai, J.; Zhang, Y.; Gao, S.; Li, X.; et al. Functional specificity, diversity, and redundancy of Arabidopsis JAZ family repressors in jasmonate and COI1-regulated growth, development, and defense. New Phytol. 2021, 231, 1525–1545. [Google Scholar] [CrossRef]
- Ziosi, V.; Bonghi, C.; Bregoli, A.M.; Trainotti, L.; Biondi, S.; Sutthiwal, S.; Kondo, S.; Costa, G.; Torrigiani, P. Jasmonate-induced transcriptional changes suggest a negative interference with the ripening syndrome in peach fruit. J. Exp. Bot. 2008, 59, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Boter, M.; Solano, R. Novel players fine-tune plant trade-offs. Essays Biochem. 2015, 58, 83–100. [Google Scholar] [CrossRef]
- Caarls, L.; Pieterse CM, J.; Van Wees, S.C.M. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Li, M.; Yu, G.; Cao, C.; Liu, P. Metabolism, signaling, and transport of jasmonates. Plant Commun. 2021, 2, 100231. [Google Scholar] [CrossRef]
- Gfeller, A.; Dubugnon, L.; Liechti, R.; Farmer, E.E. Jasmonate biochemical pathway. Sci. Signal 2010, 3, cm3. [Google Scholar] [CrossRef]
- Song, H.Y.; Duan, Z.H.; Wang, Z.; Li, Y.; Wang, Y.Y.; Li, C.M.; Mao, W.M.; Que, Q.M.; Chen, X.Y.; Li, P. Genome-wide identification, expression pattern and subcellular localization analysis of the JAZ gene family in Toona ciliata. Ind. Crops Prod. 2022, 178, 114582. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signaling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Zhao, X.Q.; He, Y.Q.; Liu, Y.X.; Wang, Z.F.; Zhao, J. JAZ proteins: Key regulators of plant growth and stress response. Crop J. 2024, 12, 1505–1516. [Google Scholar] [CrossRef]
- Suza, W.P.; Rowe, M.L.; Hamberg, M.; Staswick, P.E. A tomato enzyme synthesizes (+)-7-iso-jasmonoyl-L-isoleucine in wounded leaves. Planta 2010, 231, 717–728. [Google Scholar] [CrossRef]
- Chung, H.S.; Koo, A.J.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef]
- Oblessuc, P.R.; Obulareddy, N.; DeMott, L.; Matiolli, C.C.; Thompson, B.K.; Melotto, M. JAZ4 is involved in plant defense, growth, and development in Arabidopsis. Plant J. 2020, 101, 371–383. [Google Scholar] [CrossRef]
- Zhu, D.; Cai, H.; Luo, X.; Bai, X.; Deyholos, M.K.; Chen, Q.; Chen, C.; Ji, W.; Zhu, Y. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ding, L.; Jiang, J.; Shentu, Y.; Zhao, W.; Zhao, K.; Zhang, X.; Song, A.; Chen, S.; Chen, F. Overexpression of the CmJAZ1-like gene delays flowering in Chrysanthemum morifolium. Hortic. Res. 2021, 8, 87. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef]
- Zhao, C.; Pan, X.; Yu, Y.; Zhu, Y.; Kong, F.J.; Sun, X.; Wang, F.F. Overexpression of a TIFY family gene, GsJAZ2, exhibits enhanced tolerance to alkaline stress in soybean. Mol. Breed. 2020, 40, 33. [Google Scholar] [CrossRef]
- He, K.; Du, J.; Han, X.; Li, H.; Kui, M.; Zhang, J.; Huang, Z.; Fu, Q.; Jiang, Y.; Hu, Y. PHOSPHATE STARVATION RESPONSE1 (PHR1) interacts with JASMONATE ZIM-DOMAIN (JAZ) and MYC2 to modulate phosphate deficiency-induced jasmonate signaling in Arabidopsis. Plant Cell 2023, 35, 2132–2156. [Google Scholar] [CrossRef]
- Major, I.T.; Guo, Q.; Zhai, J.; Kapali, G.; Kramer, D.M.; Howe, G.A. A Phytochrome B-Independent Pathway Restricts Growth at High Levels of Jasmonate Defense. Plant Physiol. 2020, 183, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Hanan, A.; Basit, A.; Nazir, T.; Majeed, M.Z.; Qiu, D. Anti-insect activity of a partially purified protein derived from the entomopathogenic fungus Lecanicillium lecanii (Zimmermann) and its putative role in a tomato defense mechanism against green peach aphid. J. Invertebr. Pathol. 2020, 170, 107282. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; You, C.X.; Hao, Y.J. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2021, 229, 2707–2729. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Liu, Z.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Genome-Wide Identification, Characterization and Expression Analysis of the JAZ Gene Family in Resistance to Gray Leaf Spots in Tomato. Int. J. Mol. Sci. 2021, 22, 9974. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Luthe, D. Identification and evolution analysis of the JAZ gene family in maize. BMC Genom. 2021, 22, 256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zheng, H.; Wu, H.; Wang, C.; Liang, Z. Recent genome-wide replication promoted expansion and functional differentiation of the JAZs in soybeans. Int. J. Biol. Macromol. 2023, 238, 124064. [Google Scholar] [CrossRef]
- Zhai, Z.; Che, Y.; Geng, S.; Liu, S.; Zhang, S.; Cui, D.; Deng, Z.; Fu, M.; Li, Y.; Zou, X.; et al. Comprehensive comparative analysis of the JAZ gene family in common wheat (Triticum aestivum) and its d-subgenome donor aegilops tauschii. Plants 2024, 13, 1259. [Google Scholar] [CrossRef]
- Bisht, N.; Anshu, A.; Singh, P.C.; Chauhan, P.S. Comprehensive analysis of OsJAZ gene family deciphers rhizobacteria-mediated nutrient stress modulation in rice. Int. J. Biol. Macromol. 2023, 253, 126832. [Google Scholar] [CrossRef]
- Shen, J.; Zou, Z.; Xing, H.; Duan, Y.; Zhu, X.; Ma, Y.; Wang, Y.; Fang, W. Genome-Wide Analysis Reveals Stress and Hormone Responsive Patterns of JAZ Family Genes in Camellia Sinensis. Int. J. Mol. Sci. 2020, 31, 2433. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.F.; Caetano-Anollés, G. Evolution of networks of protein domain organization. Sci. Rep. 2021, 11, 12075. [Google Scholar] [CrossRef]
- Thireault, C.; Shyu, C.; Yoshida, Y.; St Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, X.; Wang, P.; Sun, Y.; Yue, C.; Ye, N. Genome-wide and expression pattern analysis of JAZ family involved in stress responses and postharvest processing treatments in Camellia sinensis. Sci. Rep. 2020, 10, 2792. [Google Scholar] [CrossRef]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). BMC Genom. 2017, 18, 152. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhu, C.; Chen, J.; Zhao, J.; Hu, Z.; Liu, S.; Zhou, Y. Identification and Expression Profile Analysis of the OSCA Gene Family Related to Abiotic and Biotic Stress Response in Cucumber. Biology 2022, 11, 1134. [Google Scholar] [CrossRef]
- Qu, J.J.; Liu, L.L.; Guo, Z.X.; Li, X.D.; Pan, F.Y.; Sun, D.Y.; Ling, Y. The ubiquitous position effect, synergistic effect of recent generated tandem duplicated genes in grapevine, and their co-response and overactivity to biotic stress. Fruit Res. 2023, 3, 16. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Lawton-Rauh, A. Evolutionary dynamics of duplicated genes in plants. Mol. Phylogenetics Evol. 2003, 29, 396–409. [Google Scholar] [CrossRef]
- Hwang, P.I.; Wu, H.B.; Wang, C.D.; Lin, B.L.; Chen, C.T.; Yuan, S.; Wu, G.; Li, K.C. Tissue-specific gene expression templates for accurate molecular characterization of the normal physiological states of multiple human tissues with implication in development and cancer studies. BMC Genom. 2011, 12, 439. [Google Scholar] [CrossRef]
- Xu, D.B.; Ma, Y.N.; Qin, T.F.; Tang, W.L.; Qi, X.W.; Wang, X.; Liu, R.C.; Fang, H.L.; Chen, Z.Q.; Liang, C.Y.; et al. Transcriptome-wide identification and characterization of the JAZ gene family in (Mentha canadensis L). Int. J. Mol. Sci. 2021, 22, 8859. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Liu, S.; Wang, Y.; Wang, K.; Yin, C.; Yue, Y.; Hu, H. Genome-Wide Identification and Characterization of JAZ Protein Family in Two Petunia Progenitors. Plants 2019, 8, 203. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Devi, P.; Chaudhary, S.; Rani, A.; Nayyar, H. ’omics’ approaches in developing combined drought and heat tolerance in food crops. Plant Cell Rep. 2021, 41, 699–739. [Google Scholar] [CrossRef] [PubMed]
- Nawaz Sun, J.; Shabbir, S.; Khattak, W.A.; Ren, G.; Nie, X.; Bo, Y.; Javed, Q.; Du, D.; Sonne, C. A review of plants strategies to resist biotic and abiotic environmental stressors. Sci. Total Environ. 2023, 900, 165832. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A.M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef]
- An, X.H.; Hao, Y.J.; Li, E.M.; Xu, K.; Cheng, C.G. Functional identification of apple MdJAZ2 in Arabidopsis with reduced JA-sensitivity and increased stress tolerance. Plant Cell Rep. 2017, 36, 255–265. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, P.; Li, C.; Xia, G. The moss jasmonate ZIM-domain protein PnJAZ1 confers salinity tolerance via crosstalk with the abscisic acid signalling pathway. Plant Sci. 2019, 280, 1–11. [Google Scholar] [CrossRef]
- Sharma, P.; Tripathi, S.; Chaturvedi, P.; Chaurasia, D.; Chandra, R. Newly isolated Bacillus sp. PS-6 assisted phytoremediation of heavy metals using Phragmites communis: Potential application in wastewater treatment. Bioresour. Technol. 2021, 320, 124353. [Google Scholar] [CrossRef]
- Amin, H.; Arain, B.A.; Amin, F.; Surhio, M.A. Phytotoxicity of Chromium on Germination, Growth and Biochemical Attributes of Hibiscus esculentus L. Am. J. Plant Sci. 2013, 4, 2431–2439. [Google Scholar] [CrossRef]
- Ojuederie, O.B.; Babalola, O.O. Microbial and Plant-Assisted Bioremediation of Heavy Metal Polluted Environments: A Review. Int. J. Environ. Res. Public Health 2017, 14, 1504. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Zhang, W.; Chen, J.; Li, X. Methyl jasmonate alleviates cadmium toxicity in solanum nigrum by regulating metal uptake and antioxidative capacity. Biol. Plant. 2015, 59, 373–381. [Google Scholar] [CrossRef]
- Kamran, M.; Wang, D.; Alhaithloul, H.A.S.; Alghanem, S.M.; Aftab, T.; Xie, K.; Lu, Y.; Shi, C.; Sun, J.; Gu, W.; et al. Jasmonic acid-mediated enhanced regulation of oxidative, glyoxalase defense system and reduced chromium uptake contributes to alleviation of chromium (VI) toxicity in choysum (Brassica parachinensis L.). Ecotoxicol. Environ. Saf. 2021, 15, 111758. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, G.; Azzam, W.; Rebehmed, J. PyProtif: A PyMol plugin to retrieve and visualize protein motifs for structural studies. Amino Acids 2023, 55, 1429–1436. [Google Scholar] [CrossRef]
- Yan, L.; Pape, T.; Meusemann, K.; Kutty, S.N.; Meier, R.; Bayless, K.M.; Zhang, D. Monophyletic blowflies revealed by phylogenomics. BMC Biol. 2021, 19, 230. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Li, L.L.; Xiao, Y.; Wang, X.; He, Z.H.; Lv, Y.W.; Hu, X.S. The Ka /Ks and πa /πs Ratios under Different Models of Gametophytic and Sporophytic Selection. Genome Biol. Evol. 2023, 15, evad151. [Google Scholar] [CrossRef]
- Luo, D.; Zhou, Q.; Wu, Y.G.; Chai, X.T.; Liu, W.X.; Wang, Y.R.; Liu, Z.P. Full-length transcript sequencing and comparative transcriptomic analysis to evaluate the contribution of osmotic and ionic stress components towards salinity tolerance in the roots of cultivated alfalfa (Medicago sativa L.). BMC Plant Biol. 2019, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- Li, S.X.; Liu, J.L.; An, Y.R.; Cao, Y.M.; Liu, Y.S.; Zhang, J.; Geng, J.C.; Hu, T.M.; Yang, P.Z. MsPIP2; 2, a novel aquaporin gene from Medicago sativa, confers salt tolerance in transgenic Arabidopsis. Environ. Exp. Bot. 2019, 165, 39–52. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Gene ID | Chr | Protein Length (aa) | Instability Index | MW (kDa) | pI | GRAVY | α- Helix (%) | Extended Strand (%) | Random Coil (%) | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell-PLoc | |||||||||||
| MsJAZ1 | MsG0180001147.01.T01 | 1 | 327 | 45.46 | 34.29 | 9.45 | −0.15 | 9.79 | 4.28 | 85.93 | Cell membrane. Nucleus. |
| MsJAZ2 | MsG0280007019.01.T05 | 2 | 94 | 72.83 | 10.76 | 10.01 | −0.622 | 27.66 | 12.77 | 59.57 | Nucleus. |
| MsJAZ3 | MsG0280007633.01.T01 | 2 | 294 | 48.97 | 31.03 | 7.1 | −0.217 | 9.49 | 4.75 | 85.76 | Cell membrane. Nucleus. |
| MsJAZ4 | MsG0280008436.01.T01 | 2 | 250 | 44.96 | 27.15 | 8.83 | −0.443 | 12.4 | 7.2 | 80.4 | Nucleus. |
| MsJAZ5 | MsG0280008590.01.T01 | 2 | 208 | 67.64 | 22.36 | 8.41 | −0.42 | 9.62 | 5.29 | 85.1 | Nucleus. |
| MsJAZ6 | MsG0680032555.01.T01 | 6 | 209 | 78.77 | 22.41 | 8.52 | −0.409 | 11 | 6.7 | 82.3 | Nucleus. |
| MsJAZ7 | MsG0880042776.01.T01 | 8 | 337 | 44.33 | 38.03 | 8.34 | −0.436 | 12.46 | 5.04 | 82.49 | Nucleus. |
| MsJAZ8 | MsG0880047288.01.T01 | 8 | 235 | 48.71 | 25.91 | 8.34 | −0.722 | 16.17 | 5.11 | 78.72 | Nucleus. |
| MsJAZ9 | MsG0880047330.01.T01 | 8 | 178 | 58.06 | 19.33 | 8.97 | −0.433 | 9.55 | 5.06 | 85.39 | Nucleus. |
| Paralogous Pairs | Ka | Ks | Ka/Ks | Duplicate Date (Mya) | Duplicate Type |
|---|---|---|---|---|---|
| MsJAZ4/MsJAZ7 | 0.64 | 1.38 | 0.46 | 113.49 | segmental |
| MsJAZ5/MsJAZ6 | 0.02 | 0.04 | 0.47 | 3.15 | segmental |
| MsJAZ7/MsJAZ9 | 0.61 | 2.20 | 0.28 | 180.37 | segmental |
| MsJAZ5/MsJAZ8 | 0.39 | 0.58 | 0.68 | 47.87 | segmental |
| MsJAZ4/MsJAZ9 | 0.32 | 0.80 | 0.40 | 65.24 | segmental |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Bashir, A.; Yang, H.; Abbas, A.; Li, Y.; Zeng, X.; Zhu, L.; Shi, Q.; Tursunniyaz, M.; Zhang, L. Genome-Wide Analysis and Expression Profiling of the JAZ Gene Family in Response to Abiotic Stress in Alfalfa. Int. J. Mol. Sci. 2025, 26, 4684. https://doi.org/10.3390/ijms26104684
Li X, Bashir A, Yang H, Abbas A, Li Y, Zeng X, Zhu L, Shi Q, Tursunniyaz M, Zhang L. Genome-Wide Analysis and Expression Profiling of the JAZ Gene Family in Response to Abiotic Stress in Alfalfa. International Journal of Molecular Sciences. 2025; 26(10):4684. https://doi.org/10.3390/ijms26104684
Chicago/Turabian StyleLi, Xiaohong, Aneela Bashir, Huizheng Yang, Ansar Abbas, Yaoyao Li, Xin Zeng, Longkao Zhu, Qinke Shi, Mamateliy Tursunniyaz, and Lijing Zhang. 2025. "Genome-Wide Analysis and Expression Profiling of the JAZ Gene Family in Response to Abiotic Stress in Alfalfa" International Journal of Molecular Sciences 26, no. 10: 4684. https://doi.org/10.3390/ijms26104684
APA StyleLi, X., Bashir, A., Yang, H., Abbas, A., Li, Y., Zeng, X., Zhu, L., Shi, Q., Tursunniyaz, M., & Zhang, L. (2025). Genome-Wide Analysis and Expression Profiling of the JAZ Gene Family in Response to Abiotic Stress in Alfalfa. International Journal of Molecular Sciences, 26(10), 4684. https://doi.org/10.3390/ijms26104684




