Evaluation of Low-Coverage Sequencing Strategies for Whole-Genome Imputation in Pacific Abalone Haliotis discus hannai
Abstract
1. Introduction
2. Results
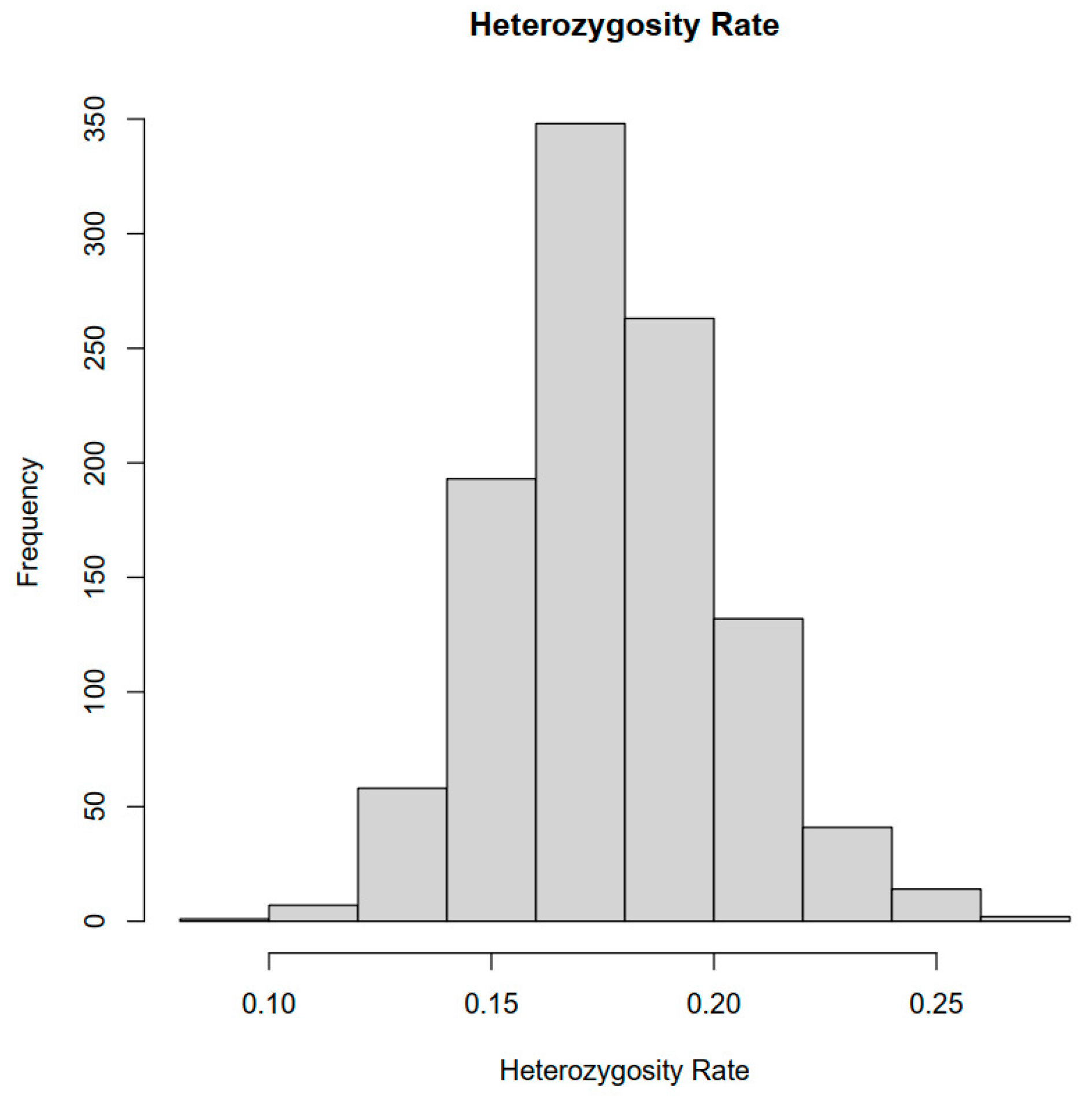
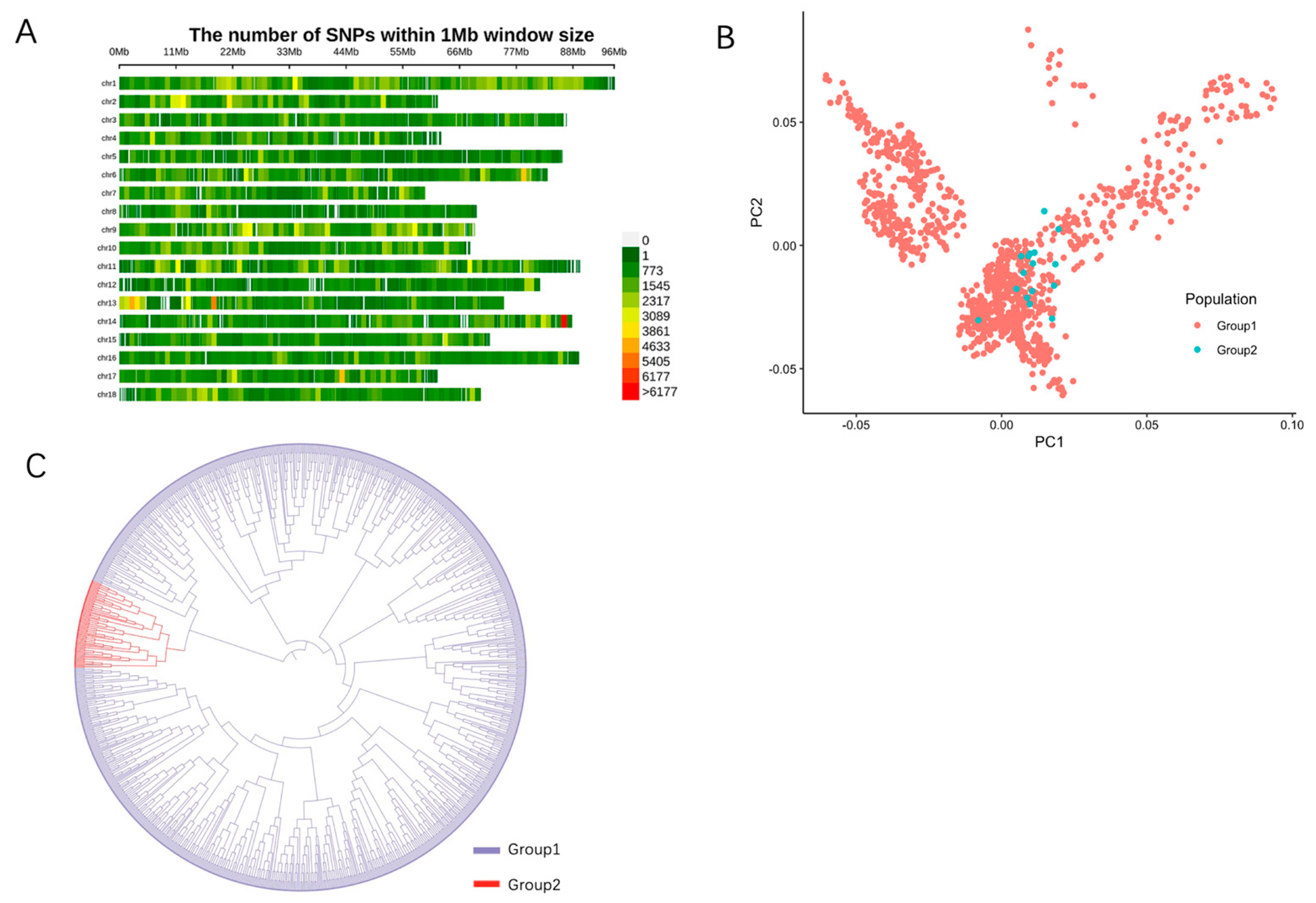
2.1. SNP Genotyping
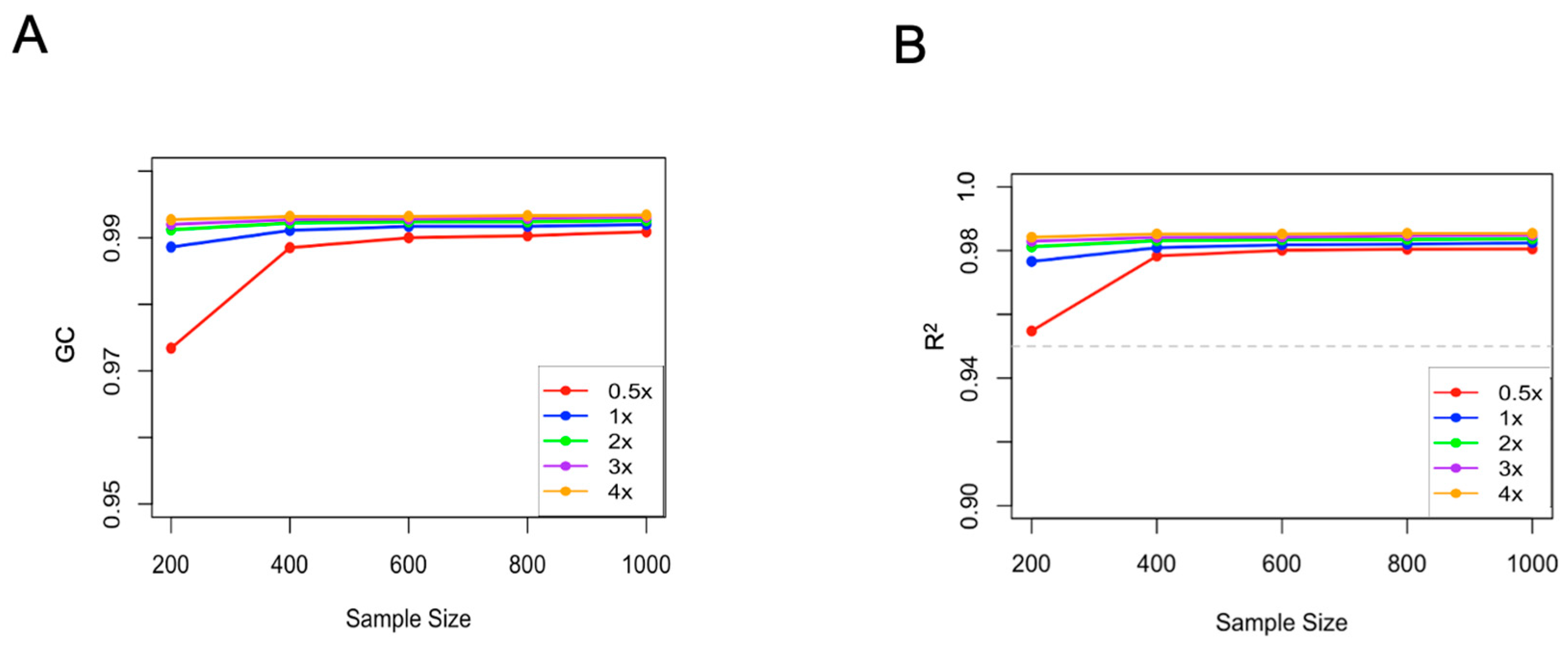
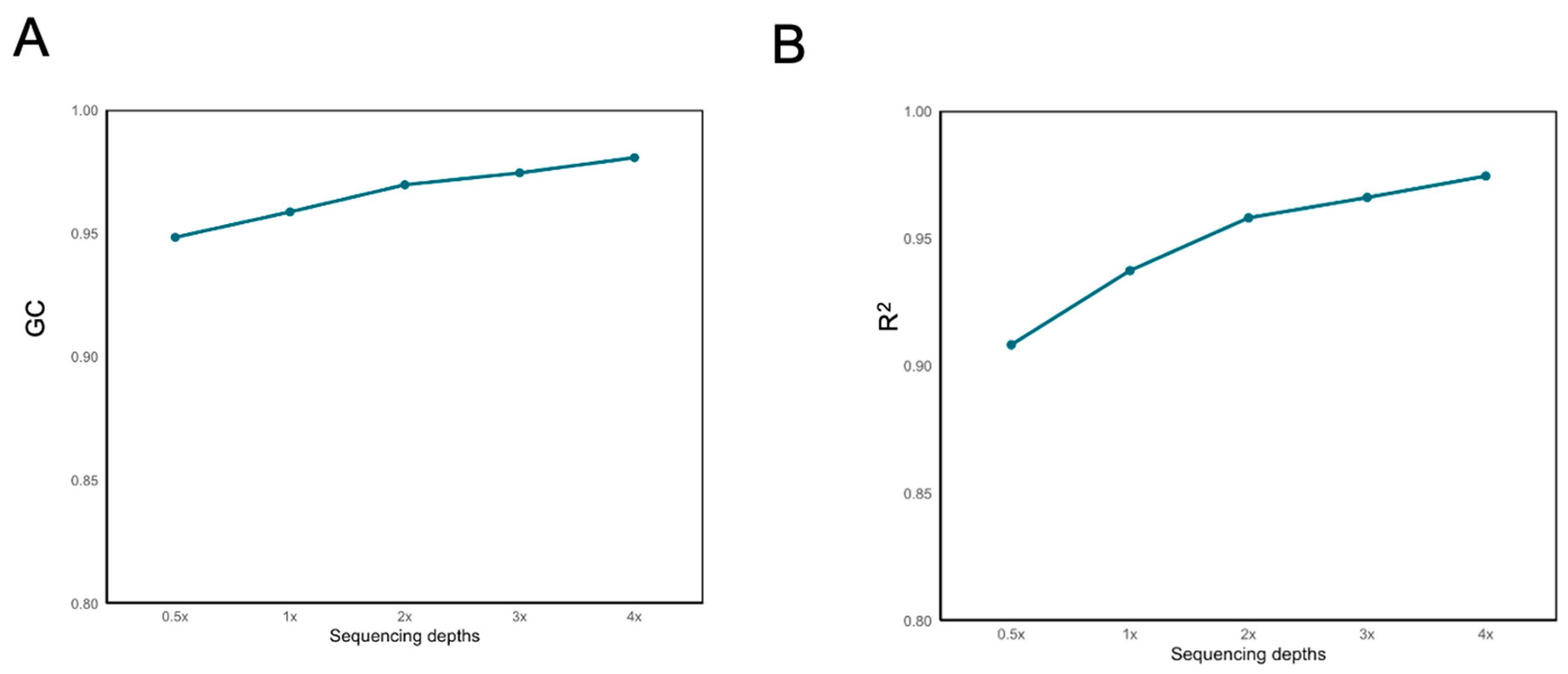
2.2. Effects of Sequencing Depth and Sample Size on Imputation Accuracy
2.3. Effects of Chromosome Length on Imputation Accuracy
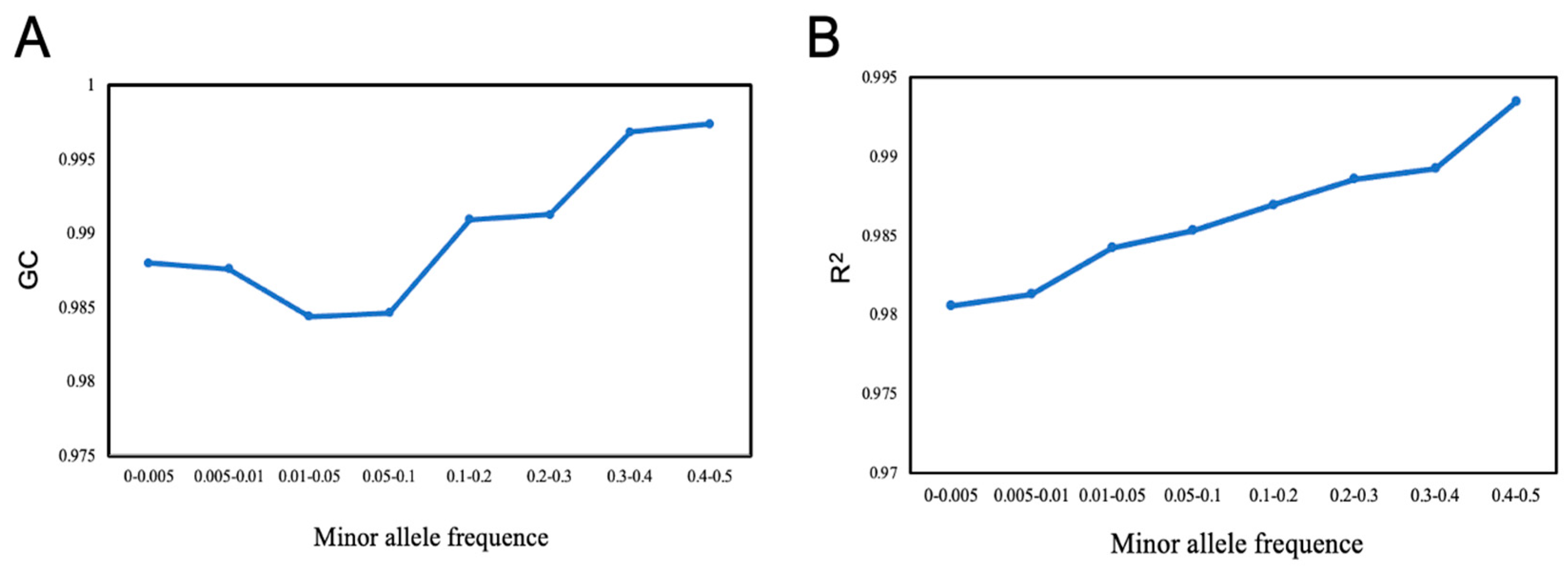
2.4. Effects of Minor Allele Frequency on Imputation Accuracy
2.5. Evaluation of STITCH and Beagle Strategy
3. Discussion
4. Materials and Methods
4.1. Sample Materials and Whole-Genome Sequencing
4.2. Sequencing Analysis and Variant Calling
4.3. Genotype Imputation
4.3.1. Evaluation of the Impact of Different Sequencing Depths and Sample Sizes on Imputation Accuracy
4.3.2. Evaluation of the Impact of Chromosome Length on Imputation Accuracy
4.3.3. Evaluation of the Effect of Allele Frequency on Imputation Accuracy
4.4. Evaluation of Imputation Accuracy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; He, Q.; Sun, H.; Liu, X. Acclimation-Dependent Expression of Heat Shock Protein 70 in Pacific Abalone (Haliotis Discus Hannai Ino) and Its Acute Response to Thermal Exposure. Chin. J. Oceanol. Limnol. 2012, 30, 146–151. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, M.; Luo, X.; You, W.; Ke, C. Transitions, Challenges and Trends in China’s Abalone Culture Industry. Rev. Aquac. 2023, 15, 1274–1293. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2020; FAO: Rome, Italy, 2020; ISBN 978-92-5-132692-3. [Google Scholar]
- Ellegren, H. Genome Sequencing and Population Genomics in Non-Model Organisms. Trends Ecol. Evol. 2014, 29, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Hon, T.; Mars, K.; Young, G.; Tsai, Y.C.; Karalius, J.W.; Landolin, J.M.; Maurer, N.; Kudrna, D.; Hardigan, M.A.; Steiner, C.C.; et al. Highly Accurate Long-Read HiFi Sequencing Data for Five Complex Genomes. Sci. Data 2020, 7, 399. [Google Scholar] [CrossRef]
- Mardis, E.R. DNA Sequencing Technologies: 2006–2016. Nat. Protoc. 2017, 12, 213–218. [Google Scholar] [CrossRef]
- Song, H.; Dong, T.; Yan, X.; Wang, W.; Tian, Z.; Sun, A.; Dong, Y.; Zhu, H.; Hu, H. Genomic Selection and Its Research Progress in Aquaculture Breeding. Rev. Aquac. 2023, 15, 274–291. [Google Scholar] [CrossRef]
- Davies, R.W.; Flint, J.; Myers, S.; Mott, R. Rapid Genotype Imputation from Sequence without Reference Panels. Nat. Genet. 2016, 48, 965–969. [Google Scholar] [CrossRef]
- Robledo, D.; Palaiokostas, C.; Bargelloni, L.; Martínez, P.; Houston, R. Applications of Genotyping by Sequencing in Aquaculture Breeding and Genetics. Rev. Aquac. 2018, 10, 670–682. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP Discovery and Genetic Mapping Using Sequenced RAD Markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Hoekstra He Double Digest RADseq: An Inexpensive Method for de Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Yang, R.; Guo, X.; Zhu, D.; Tan, C.; Bian, C.; Ren, J.; Huang, Z.; Zhao, Y.; Cai, G.; Liu, D.; et al. Accelerated Deciphering of the Genetic Architecture of Agricultural Economic Traits in Pigs Using a Low-Coverage Whole-Genome Sequencing Strategy. GigaScience 2021, 10, giab048. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M.; Null, D.J.; Sargolzaei, M.; Wiggans, G.R.; Tooker, M.E.; Cole, J.B.; Sonstegard, T.S.; Connor, E.E.; Winters, M.; van Kaam, J.B.; et al. Genomic Imputation and Evaluation Using High-Density Holstein Genotypes. J. Dairy Sci. 2013, 96, 668–678. [Google Scholar] [CrossRef]
- Li, Y.; Sidore, C.; Kang, H.M.; Boehnke, M.; Abecasis, G.R. Low-Coverage Sequencing: Implications for Design of Complex Trait Association Studies. Genome Res. 2011, 21, 940–951. [Google Scholar] [CrossRef] [PubMed]
- Zan, Y.; Payen, T.; Lillie, M.; Honaker, C.F.; Siegel, P.B.; Carlborg, Ö. Genotyping by Low-Coverage Whole-Genome Sequencing in Intercross Pedigrees from Outbred Founders: A Cost-Efficient Approach. Genet. Sel. Evol. 2019, 51, 44. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Zhang, P.; Garrick, D.; Gao, H.; Wang, L.; Zhao, F. Comparison of Genotype Imputation for SNP Array and Low-Coverage Whole-Genome Sequencing Data. Front. Genet. 2021, 12, 704118. [Google Scholar] [CrossRef]
- Nielsen, R.; Paul, J.S.; Albrechtsen, A.; Song, Y.S. Genotype and SNP Calling from Next-Generation Sequencing Data. Nat. Rev. Genet. 2011, 12, 443–451. [Google Scholar] [CrossRef]
- Fragoso, C.A.; Heffelfinger, C.; Zhao, H.; Dellaporta, S.L. Imputing Genotypes in Biallelic Populations from Low-Coverage Sequence Data. Genetics 2016, 202, 487–495. [Google Scholar] [CrossRef][Green Version]
- Ros-Freixedes, R.; Gonen, S.; Gorjanc, G.; Hickey, J.M. A Method for Allocating Low-Coverage Sequencing Resources by Targeting Haplotypes Rather than Individuals. Genet. Sel. Evol. 2017, 49, 78. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Rapid and Accurate Haplotype Phasing and Missing-Data Inference for Whole-Genome Association Studies by Use of Localized Haplotype Clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef]
- Pook, T.; Mayer, M.; Geibel, J.; Weigend, S.; Cavero, D.; Schoen, C.C.; Simianer, H. Improving Imputation Quality in BEAGLE for Crop and Livestock Data. G3 2020, 10, 177–188. [Google Scholar] [CrossRef]
- Nicod, J.; Davies, R.W.; Cai, N.; Hassett, C.; Goodstadt, L.; Cosgrove, C.; Yee, B.K.; Lionikaite, V.; McIntyre, R.E.; Remme, C.A.; et al. Genome-Wide Association of Multiple Complex Traits in Outbred Mice by Ultra-Low-Coverage Sequencing. Nat. Genet. 2016, 48, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Gilly, A.; Ritchie, G.R.; Southam, L.; Farmaki, A.E.; Tsafantakis, E.; Dedoussis, G.; Zeggini, E. Very Low-Depth Sequencing in a Founder Population Identifies a Cardioprotective APOC3 Signal Missed by Genome-Wide Imputation. Hum. Mol. Genet. 2016, 25, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Dong, T.; Wang, W.; Jiang, B.; Yan, X.; Geng, C.; Bai, S.; Xu, S.; Hu, H. Cost-Effective Genomic Prediction of Critical. Economic Traits in Sturgeons through Low-Coverage Sequencing. Genomics 2024, 116, 110874. [Google Scholar] [CrossRef] [PubMed]
- Teng, J.; Zhao, C.; Wang, D.; Chen, Z.; Tang, H.; Li, J.; Mei, C.; Yang, Z.; Ning, C.; Zhang, Q. Assessment of the Performance of Different Imputation Methods for Low-Coverage Sequencing in Holstein Cattle. J. Dairy Sci. 2022, 105, 3355–3366. [Google Scholar] [CrossRef]
- Zhao, C.; Teng, J.; Zhang, X.; Wang, D.; Zhang, X.; Li, S.; Jiang, X.; Li, H.; Ning, C.; Zhang, Q. Towards a Cost-Effective Implementation of Genomic Prediction Based on Low Coverage Whole Genome Sequencing in Dezhou Donkey. Front. Genet. 2021, 12, 728764. [Google Scholar] [CrossRef]
- Zhang, W.; Li, W.; Liu, G.; Gu, L.; Ye, K.; Zhang, Y.; Li, W.; Jiang, D.; Wang, Z.; Fang, M. Evaluation for the Effect of Low-Coverage Sequencing on Genomic Selection in Large Yellow Croaker. Aquaculture 2021, 534, 736323. [Google Scholar] [CrossRef]
- Fuller, Z.L.; Mocellin, V.J.L.; Morris, L.A.; Cantin, N.; Shepherd, J.; Sarre, L.; Peng, J.; Liao, Y.; Pickrell, J.; Andolfatto, P.; et al. Population Genetics of the Coral Acropora Millepora: Toward Genomic Prediction of Bleaching. Science 2020, 369, eaba4674. [Google Scholar] [CrossRef]
- Yang, B.; Li, Y.; Li, Q.; Liu, S. High-Throughput and Cost-Effective Genotyping by Low-Coverage Whole Genome Sequencing with Genotype Imputation in Pacific Oyster, Crassostrea Gigas. Aquaculture 2024, 591, 741134. [Google Scholar] [CrossRef]
- Sui, M.; Liu, Z.; Huang, X.; Yang, Z.; Yu, H.; Cui, C.; Hu, Y.; Wang, X.; Shen, X.; Mu, Q.; et al. Development and Evaluation of a Haplotype Reference Panel of Zhikong Scallop (Chlamys Farreri) for Genotype Imputation. Aquaculture 2024, 582, 740497. [Google Scholar] [CrossRef]
- Beemelmanns, A.; Bouchard, R.; Michaelides, S.; Normandeau, E.; Jeon, H.; Chamlian, B.; Babin, C.; Hénault, P.; Perrot, O.; Harris, L.N.; et al. Development of SNP Panels from Low-coverage Whole Genome Sequencing (lcWGS) to Support Indigenous Fisheries for Three Salmonid Species in Northern Canada. Mol. Ecol. Resour. 2025, 25, e14040. [Google Scholar] [CrossRef]
- Liu, S.; Martin, K.E.; Snelling, W.M.; Long, R.; Leeds, T.D.; Vallejo, R.L.; Wiens, G.D.; Palti, Y. Accurate Genotype Imputation from Low-Coverage Whole-Genome Sequencing Data of Rainbow Trout. G3 Genes|Genomes|Genetics 2024, 14, jkae168. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Zhao, B.; Wang, H.; Wang, M.; Teng, M.; Hu, J.; Bao, Z.; Wang, Y. Aquaculture Molecular Breeding Platform (AMBP): A Comprehensive Web Server for Genotype Imputation and Genetic Analysis in Aquaculture. Nucleic Acids Res. 2022, 50, W66–W74. [Google Scholar] [CrossRef]
- Houston, R.D.; Bean, T.P.; Macqueen, D.J.; Gundappa, M.K.; Jin, Y.H.; Jenkins, T.L.; Selly, S.L.C.; Martin, S.A.M.; Stevens, J.R.; Santos, E.M.; et al. Harnessing Genomics to Fast-Track Genetic Improvement in Aquaculture. Nat. Rev. Genet. 2020, 21, 389–409. [Google Scholar] [CrossRef] [PubMed]
- Pasaniuc, B.; Rohland, N.; McLaren, P.J.; Garimella, K.; Zaitlen, N.; Li, H.; Gupta, N.; Neale, B.M.; Daly, M.J.; Sklar, P.; et al. Extremely Low-Coverage Sequencing and Imputation Increases Power for Genome-Wide Association Studies. Nat. Genet. 2012, 44, 631–635. [Google Scholar] [CrossRef]
- Liu, J.; Peng, W.; Yu, F.; Lin, W.; Shen, Y.; Yu, W.; Gong, S.; Huang, H.; You, W.; Luo, X.; et al. Development and Validation of a 40-K Multiple-SNP Array for Pacific Abalone (Haliotis Discus Hannai). Aquaculture 2022, 558, 738393. [Google Scholar] [CrossRef]
- Lin, W.; Xiao, Q.; Yu, F.; Han, Z.; Liu, J. Development of a Low-Density SNP Genotyping Panel by a Novel Technology mGPS and Its Application in Germplasm Identification of Abalone. Aquaculture 2023, 565, 739089. [Google Scholar] [CrossRef]
- Kijas, J.; Hamilton, M.; Botwright, N.; King, H.; McPherson, L.; Krsinich, A.; McWilliam, S. Genome Sequencing of Blacklip and Greenlip Abalone for Development and Validation of a SNP Based Genotyping Tool. Front. Genet. 2018, 9, 687. [Google Scholar] [CrossRef]
- Dimond, J.L.; Bouma, J.V.; Lafarga-De la Cruz, F.; Supernault, K.J.; White, T.; Witting, D.A. Endangered Pinto/Northern Abalone (Haliotis Kamtschatkana) Are Panmictic across Their 3700 Km Range along the Pacific Coast of North America. Evol. Appl. 2024, 17, e70040. [Google Scholar] [CrossRef]
- Yang, B.; Zhai, S.Y.; Zhang, F.Q.; Wang, H.B.; Ren, L.T.; Li, Y.J.; Li, Q.; Liu, S.K. Genome-Wide Association Study toward Efficient Selection Breeding of Resistance to Vibrio Alginolyticus in Pacific Oyster, Crassostrea Gigas. Aquaculture 2022, 548, 737592. [Google Scholar] [CrossRef]
- Calus, M.; Bouwman, A.C.; Hickey, J.M.; Veerkamp, R.F.; Mulder, H.A. Evaluation of Measures of Correctness of Genotype Imputation in the Context of Genomic Prediction: A Review of Livestock Applications. Animal 2014, 8, 1743–1753. [Google Scholar] [CrossRef]
- Rutkoski, J.E.; Poland, J.; Jannink, J.L.; Sorrells, M.E. Imputation of Unordered Markers and the Impact on Genomic Selection Accuracy. G3 2013, 3, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Abecasis, G.R.; Browning, B.L. Genotype Imputation from Large Reference Panels. Annu. Rev. Genom. Hum. Genet. 2018, 19, 73–96. [Google Scholar] [CrossRef]
- Aliloo, H.; Mrode, R.; Okeyo, A.M.; Ni, G.; Goddard, M.E.; Gibson, J.P. The Feasibility of Using Low-Density Marker Panels for Genotype Imputation and Genomic Prediction of Crossbred Dairy Cattle of East Africa. J. Dairy Sci. 2018, 101, 9108–9127. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, A.; Ruiz-Lopez, F.J.; Wiggans, G.R.; Van Tassell, C.P.; Montaldo, H.H. Effect of Reference Population Size and Available Ancestor Genotypes on Imputation of Mexican Holstein Genotypes. J. Dairy Sci. 2015, 98, 3478–3484. [Google Scholar] [CrossRef] [PubMed]
- Butty, A.M.; Sargolzaei, M.; Miglior, F.; Stothard, P.; Schenkel, F.S.; Gredler-Grandl, B.; Baes, C.F. Optimizing Selection of the Reference Population for Genotype Imputation From Array to Sequence Variants. Front. Genet. 2019, 10, 510. [Google Scholar] [CrossRef]
- VanRaden, P.M.; Sun, C.; O’Connell, J.R. Fast Imputation Using Medium or Low-Coverage Sequence Data. BMC Genet. 2015, 16, 82. [Google Scholar] [CrossRef]
- Hui, R.; D’Atanasio, E.; Cassidy, L.M.; Scheib, C.L.; Kivisild, T. Evaluating Genotype Imputation Pipeline for Ultra-Low Coverage Ancient Genomes. Sci. Rep. 2020, 10, 18542. [Google Scholar] [CrossRef]
- Delomas, T.A.; Hollenbeck, C.M.; Matt, J.L.; Thompson, N.F. Evaluating Cost-Effective Genotyping Strategies for Genomic Selection in Oysters. Aquaculture 2023, 562, 738844. [Google Scholar] [CrossRef]
- Kriaridou, C.; Tsairidou, S.; Fraslin, C.; Gorjanc, G.; Looseley, M.E.; Johnston, I.A.; Houston, R.D.; Robledo, D. Evaluation of Low-Density SNP Panels and Imputation for Cost-Effective Genomic Selection in Four Aquaculture Species. Front. Genet. 2023, 14, 1194266. [Google Scholar] [CrossRef]
- Heidaritabar, M.; Calus, M.P.L.; Vereijken, A.; Groenen, M.A.M.; Bastiaansen, J.W.M. Accuracy of Imputation Using the Most Common Sires as Reference Population in Layer Chickens. BMC Genet. 2015, 16, 101. [Google Scholar] [CrossRef]
- Júnior, G.A.F.; Carvalheiro, R.; de Oliveira, H.N.; Sargolzaei, M.; Costilla, R.; Ventura, R.V.; Fonseca, L.F.S.; Neves, H.H.R.; Hayes, B.J.; de Albuquerque, L.G. Imputation Accuracy to Whole-Genome Sequence in Nellore Cattle. Genet. Sel. Evol. 2021, 53, 27. [Google Scholar] [CrossRef]
- Hayes, B.J.; Bowman, P.J.; Daetwyler, H.D.; Kijas, J.W.; van der Werf, J.H.J. Accuracy of Genotype Imputation in Sheep Breeds. Anim. Genet. 2012, 43, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Hickey, J.M.; Crossa, J.; Babu, R.; de los Campos, G. Factors Affecting the Accuracy of Genotype Imputation in Populations from Several Maize Breeding Programs. Crop Sci. 2012, 52, 654–663. [Google Scholar] [CrossRef]
- Yu, W.; Yan, S.; Zhang, S.; Ni, J.; Bin, L.; Pei, Y.; Zhang, L. Efficient Identification of Trait-associated Loss-of-function Variants in the UK Biobank Cohort by Exome-sequencing Based Genotype Imputation. Genet. Epidemiol. 2023, 47, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Boichard, D.; Chung, H.; Dassonneville, R.; David, X.; Eggen, A.; Fritz, S.; Gietzen, K.J.; Hayes, B.J.; Lawley, C.T.; Sonstegard, T.S.; et al. Design of a Bovine Low-Density SNP Array Optimized for Imputation. PLoS ONE 2012, 7, e34130. [Google Scholar] [CrossRef]
- Yuan, M.; Fang, H.; Zhang, H. Correcting for Differential Genotyping Error in Genetic Association Analysis. J. Hum. Genet. 2013, 58, 657–666. [Google Scholar] [CrossRef]
- Song, K.; Li, L.; Zhang, G. Coverage Recommendation for Genotyping Analysis of Highly Heterologous Species Using Next-Generation Sequencing Technology. Sci. Rep. 2016, 6, 35736. [Google Scholar] [CrossRef]
- Diyie, R.L.; Agyarkwa, S.K.; Armah, E.; Amonoo, N.A.; Owusu-Frimpong, I.; Osei-Atweneboana, M.Y. Genetic Variations among Different Generations and Cultured Populations of Nile Tilapia (Oreochromis Niloticus) in Ghana: Application of Microsatellite Markers. Aquaculture 2021, 544, 737070. [Google Scholar] [CrossRef]
- Sun, X.; Fei, C.; Mi, C.; Li, M.; Zhang, G.; Wu, F. Genetic Diversity and Population Structure of Pacific Abalone (Haliotis Discus Hannai) Using SNP Genotyping Data. Aquaculture 2024, 593, 741335. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A Fast and Effective Tool for Linkage Disequilibrium Decay Analysis Based on Variant Call Format Files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Plough, L.V. Genetic Load in Marine Animals: A Review. Curr. Zool. 2016, 62, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the Missing Heritability of Complex Diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.; Browning, S. A Unified Approach to Genotype Imputation and Haplotype-Phase Inference for Large Data Sets of Trios and Unrelated Individuals. Am. J. Hum. Genet. 2009, 84, 210–223. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. GigaScience 2015, 4, s13742-015-0047–0048. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-Wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Liu, S.; Huang, S.; Chen, F.; Zhao, L.; Yuan, Y.; Francis, S.S.; Fang, L.; Li, Z.; Lin, L.; Liu, R.; et al. Genomic Analyses from Non-Invasive Prenatal Testing Reveal Genetic Associations, Patterns of Viral Infections, and Chinese Population History. Cell 2018, 175, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Broad Institute. Picard Toolkit; Broad Institute of MIT and Harvard: Cambridge, MA, USA, 2019. [Google Scholar]
- Korkuć, P.; Arends, D.; Brockmann, G.A. Finding the Optimal Imputation Strategy for Small Cattle Populations. Front. Genet. 2019, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Jattawa, D.; Elzo, M.A.; Koonawootrittriron, S.; Suwanasopee, T. Imputation Accuracy from Low to Moderate Density Single Nucleotide Polymorphism Chips in a Thai Multibreed Dairy Cattle Population. Asian-Australas J. Anim. Sci. 2016, 29, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Hartz, S.M.; Zhang, Z.; Saccone, S.F.; Wang, J.; Tischfield, J.A.; Edenberg, H.J.; Kramer, J.R.; M Goate, A.; Bierut, L.J.; et al. A New Statistic to Evaluate Imputation Reliability. PLoS ONE 2010, 5, e9697. [Google Scholar] [CrossRef]
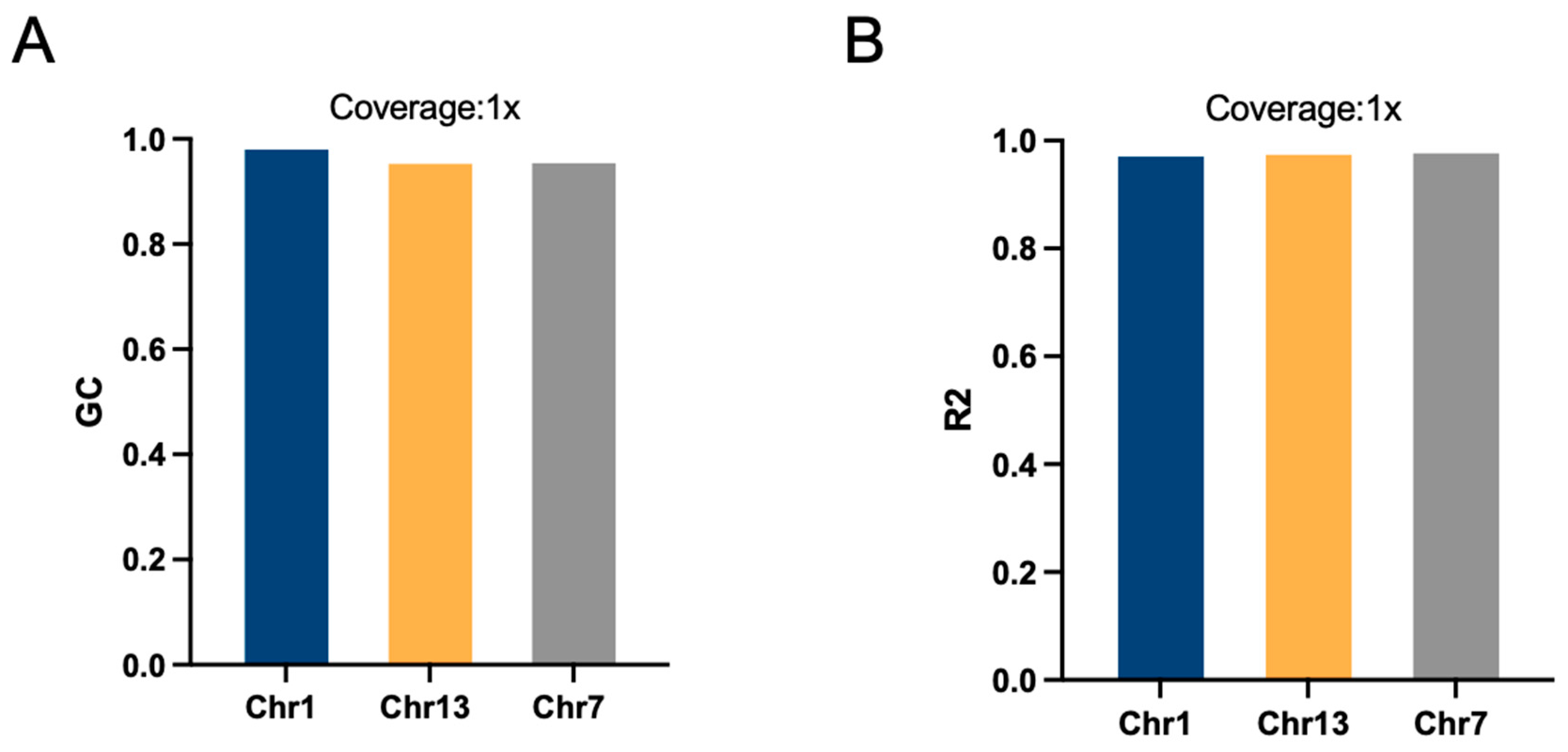
| Chr | R2 | GC | Chromosome Length | SNP Density (bp/SNP) |
|---|---|---|---|---|
| 1 | 0.982 | 0.992 | 96,096,873 | 96.983 |
| 7 | 0.985 | 0.966 | 59,286,736 | 131.189 |
| 13 | 0.988 | 0.965 | 74,588,556 | 102.116 |
| Sequencing Depth | STITCH | STITCH+BEAGLE | ||
|---|---|---|---|---|
| No. of SNPs | R2 | No. of SNPs | R2 | |
| 0.5× | 114,056 | 0.9805 | 851,527 | 0.9082 |
| 1× | 147,449 | 0.9824 | 1,369,351 | 0.9373 |
| 2× | 210,381 | 0.9837 | 1,369,982 | 0.9580 |
| 3× | 247,669 | 0.9848 | 1,369,982 | 0.9660 |
| 4× | 275,461 | 0.9854 | 1,370,182 | 0.9744 |
| Species | Sequencing Depth | Sample Size | Imputation Accuracy | SNPs Number | Publication |
|---|---|---|---|---|---|
| Crassostrea gigas | 2.8× | ≥300 | 0.860 | 11,000,000 | [29] |
| Larimichthys crocea | 0.5× | 536 | 0.795 | 5,949,426 | [27] |
| Acipenser gueldenstaedtii | 2× | ≥300 | 0.882 | >5,514,392 | [24] |
| Acropora millepora | 1.5× | 193 | 0.94 | Unknow | [28] |
| Chlamys farreri | 0.5× | 174 | 0.91 | 3,968,417 | [30] |
| Haliotis discus hannai | 1× | ≥400 | 0.98 | 147,449 | the present study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fei, C.; Zhang, S.; Chen, X.; Liu, J.; Peng, W.; Zhang, G.; You, W.; Wu, F. Evaluation of Low-Coverage Sequencing Strategies for Whole-Genome Imputation in Pacific Abalone Haliotis discus hannai. Int. J. Mol. Sci. 2025, 26, 4598. https://doi.org/10.3390/ijms26104598
Fei C, Zhang S, Chen X, Liu J, Peng W, Zhang G, You W, Wu F. Evaluation of Low-Coverage Sequencing Strategies for Whole-Genome Imputation in Pacific Abalone Haliotis discus hannai. International Journal of Molecular Sciences. 2025; 26(10):4598. https://doi.org/10.3390/ijms26104598
Chicago/Turabian StyleFei, Chengxia, Shoudu Zhang, Xiangrui Chen, Junyu Liu, Wenzhu Peng, Guofan Zhang, Weiwei You, and Fucun Wu. 2025. "Evaluation of Low-Coverage Sequencing Strategies for Whole-Genome Imputation in Pacific Abalone Haliotis discus hannai" International Journal of Molecular Sciences 26, no. 10: 4598. https://doi.org/10.3390/ijms26104598
APA StyleFei, C., Zhang, S., Chen, X., Liu, J., Peng, W., Zhang, G., You, W., & Wu, F. (2025). Evaluation of Low-Coverage Sequencing Strategies for Whole-Genome Imputation in Pacific Abalone Haliotis discus hannai. International Journal of Molecular Sciences, 26(10), 4598. https://doi.org/10.3390/ijms26104598





