Genome-Wide Identification and Expression Analysis of 1-Aminocyclopropane-1-Carboxylate Synthase (ACS) Gene Family in Myrica rubra
Abstract
1. Introduction
2. Results
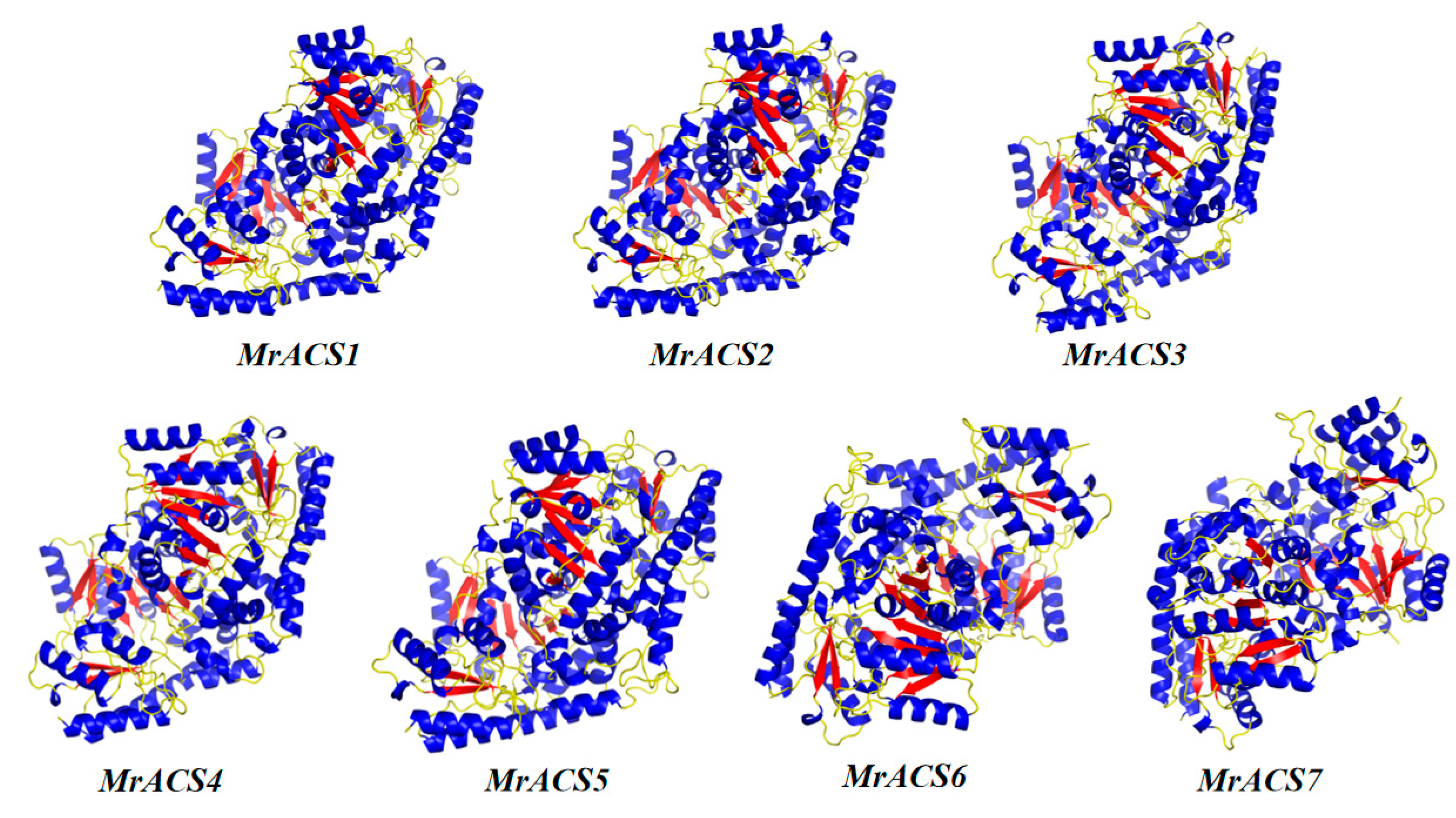
2.1. Identification of the ACS Gene Family in Myrica rubra
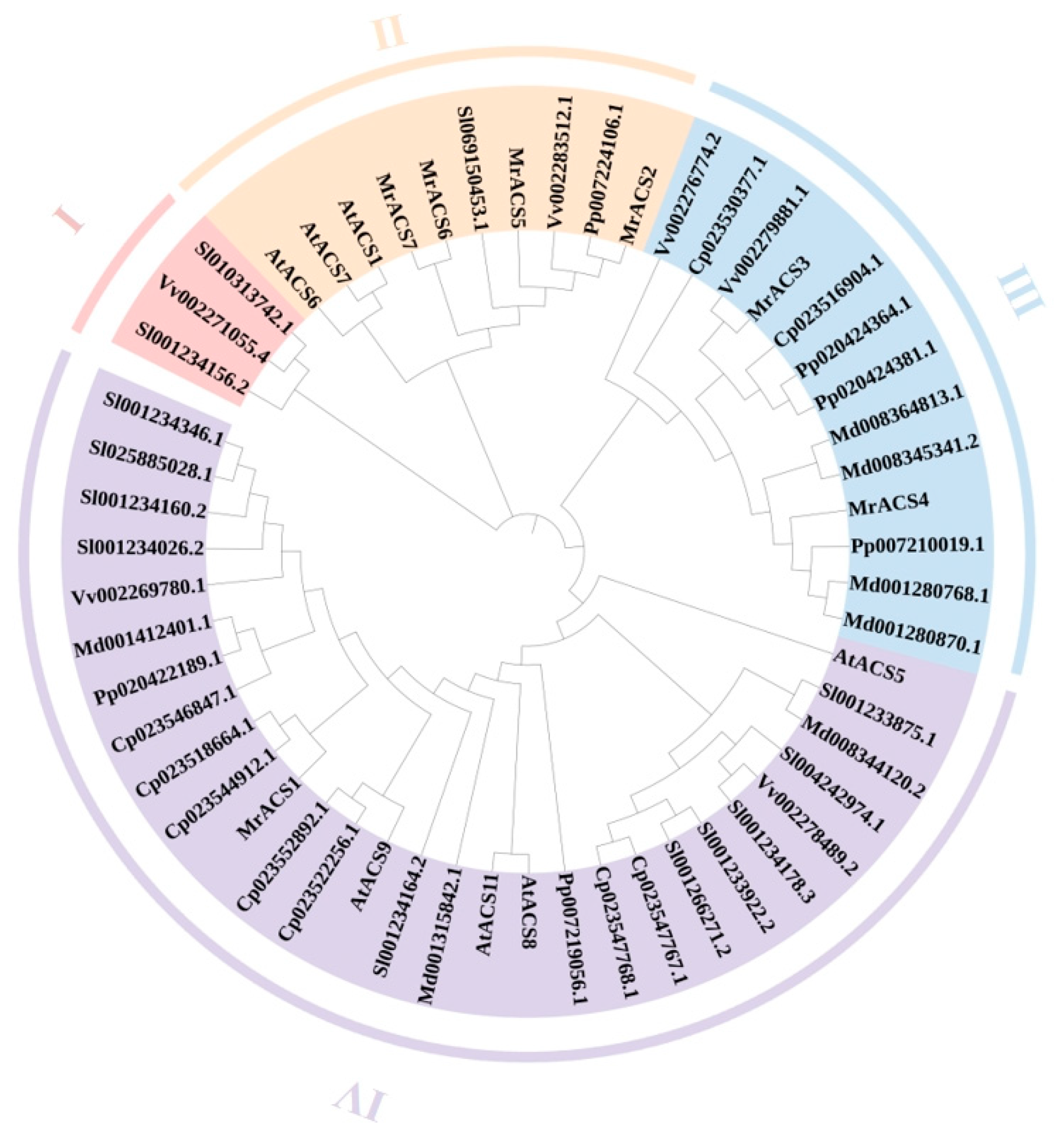
2.2. Phylogenetic Analysis of the MrACS Gene Family
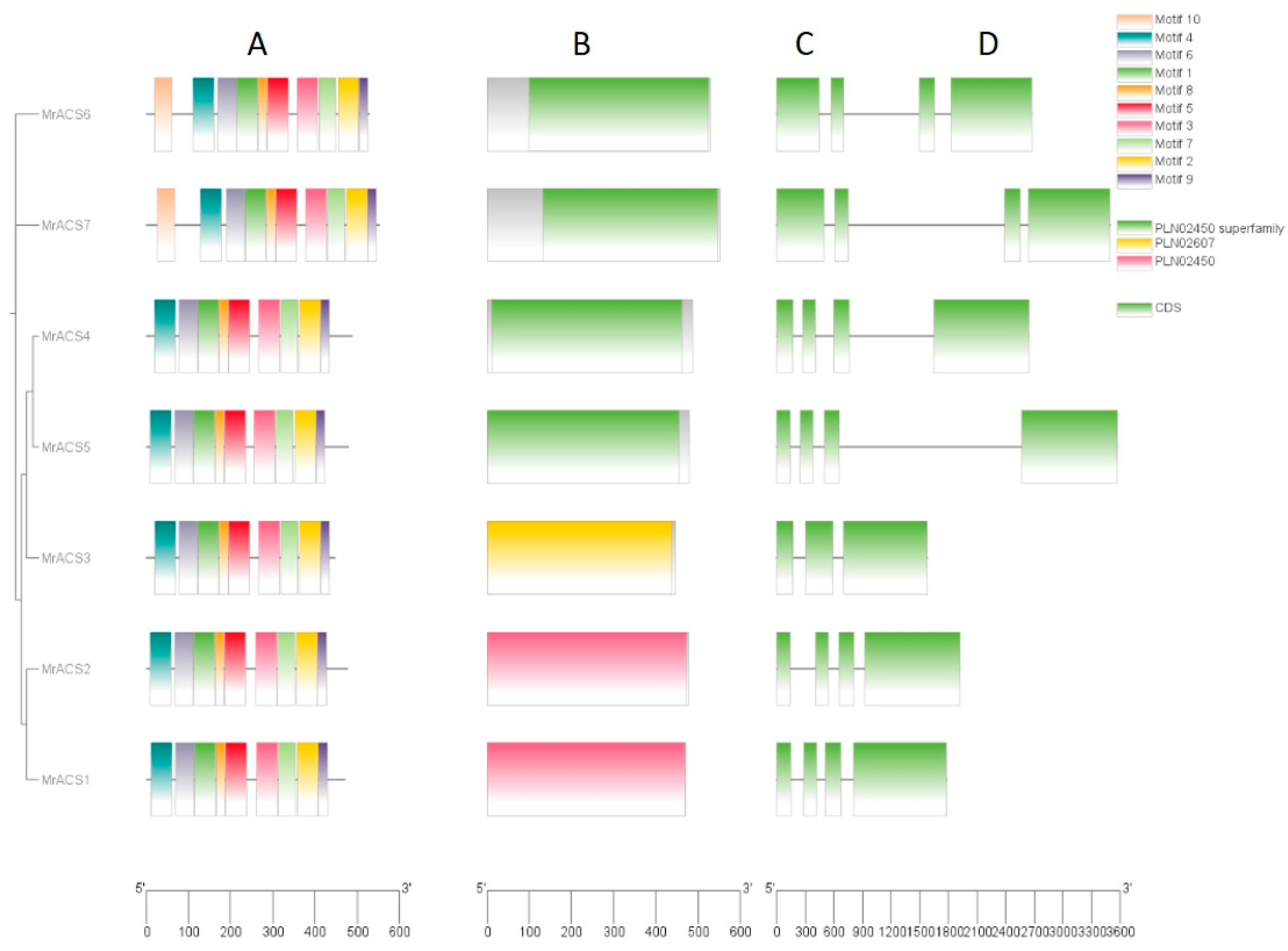
2.3. Gene Structure and Conserved Motif Analysis of MrACSs
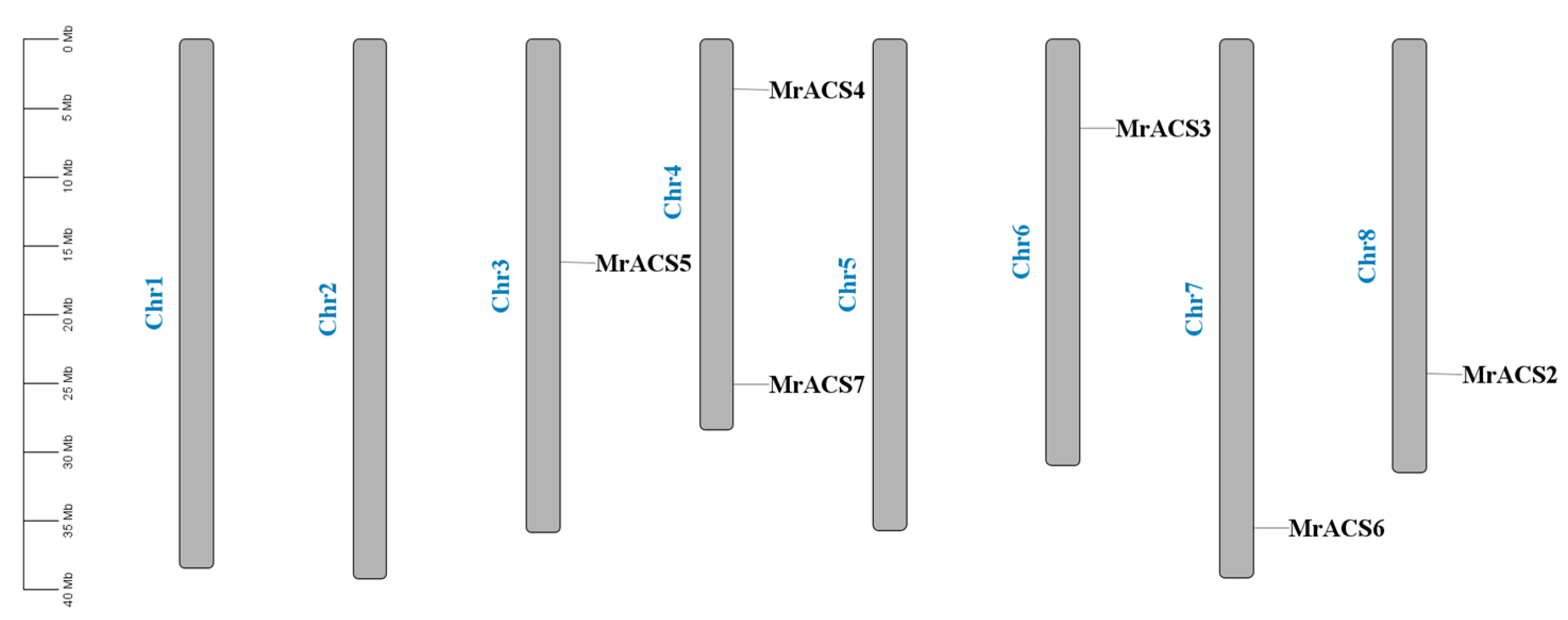
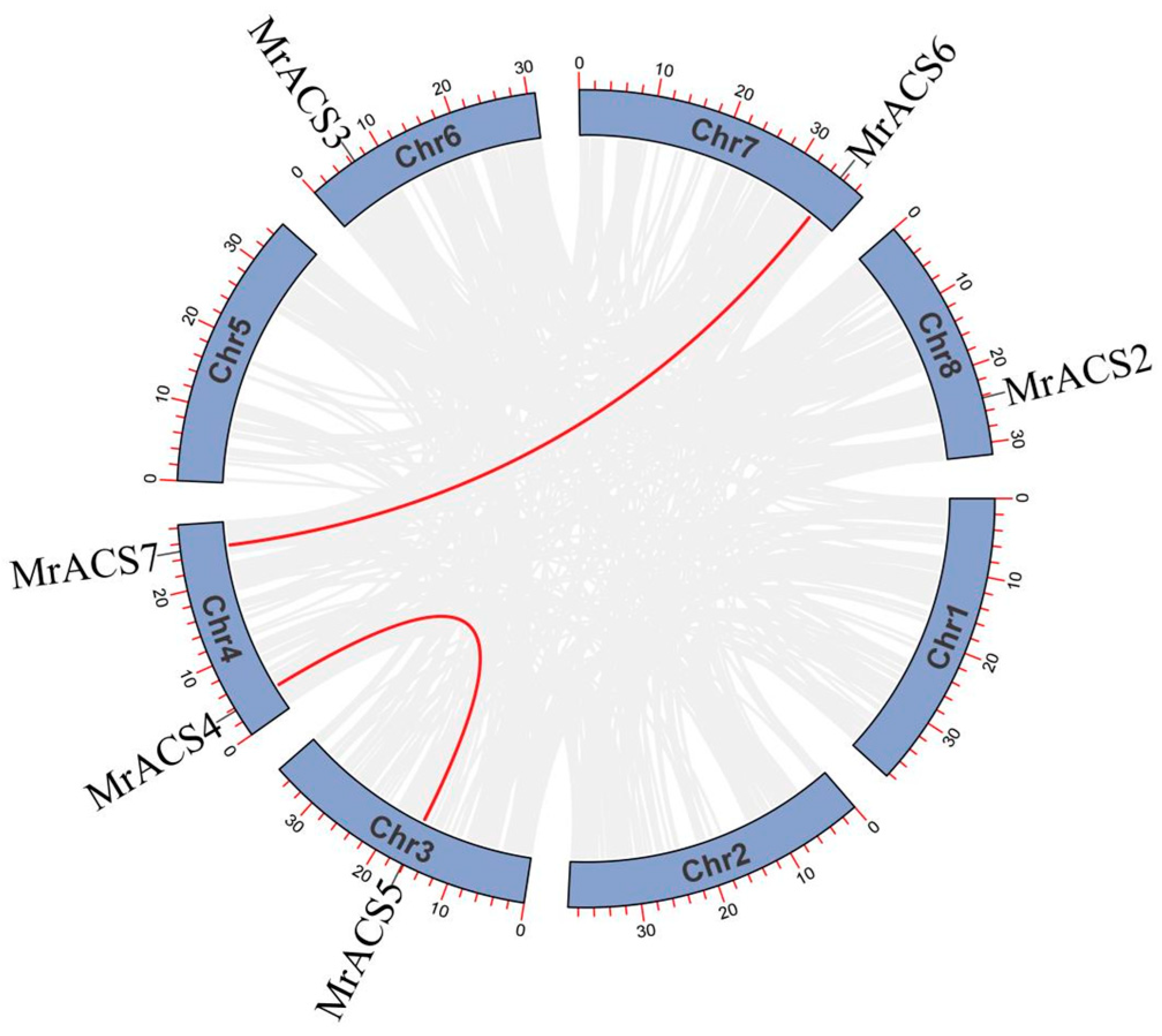
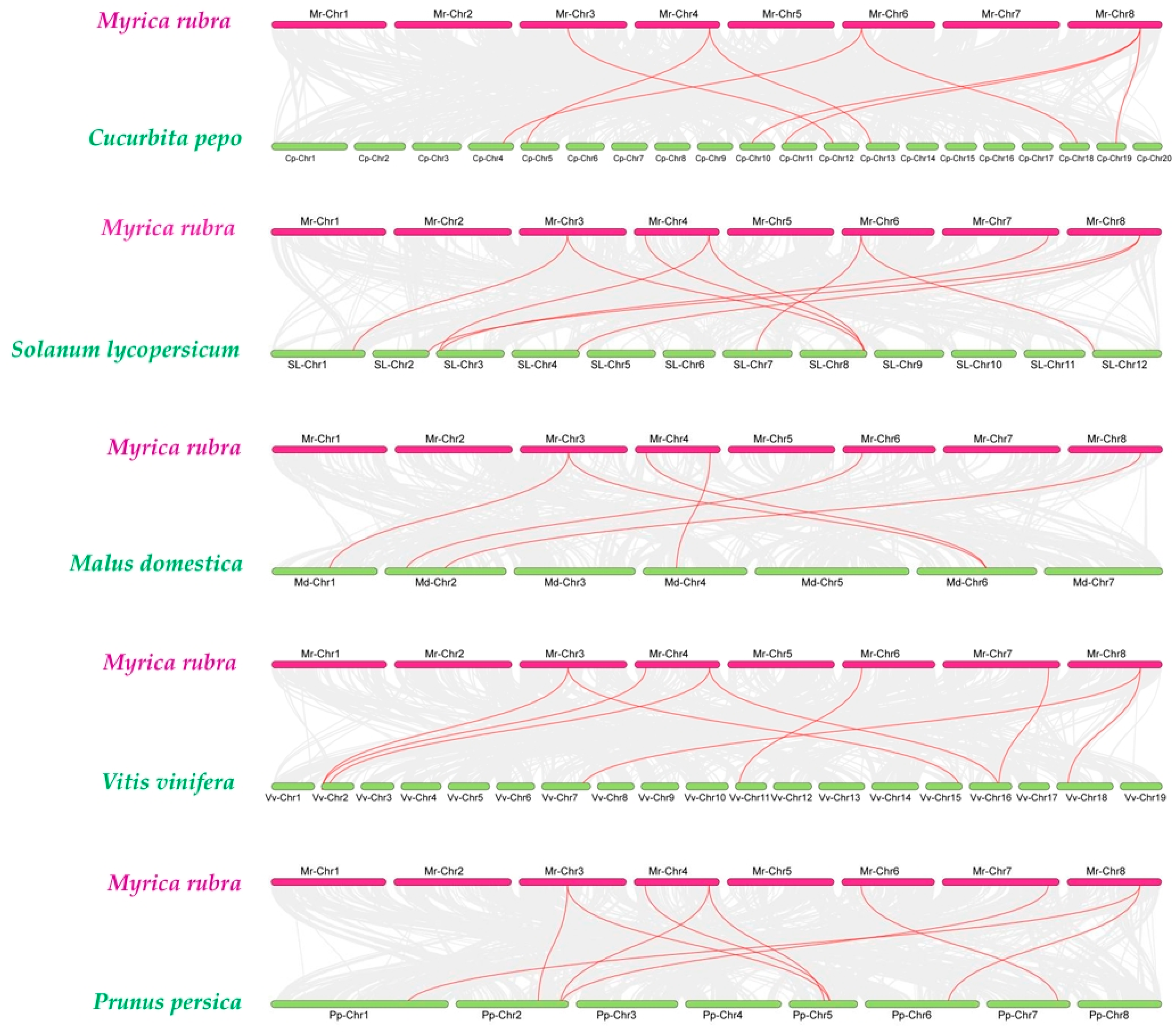
2.4. Chromosomal Localization and Collinearity Analysis of the MrACS Genes
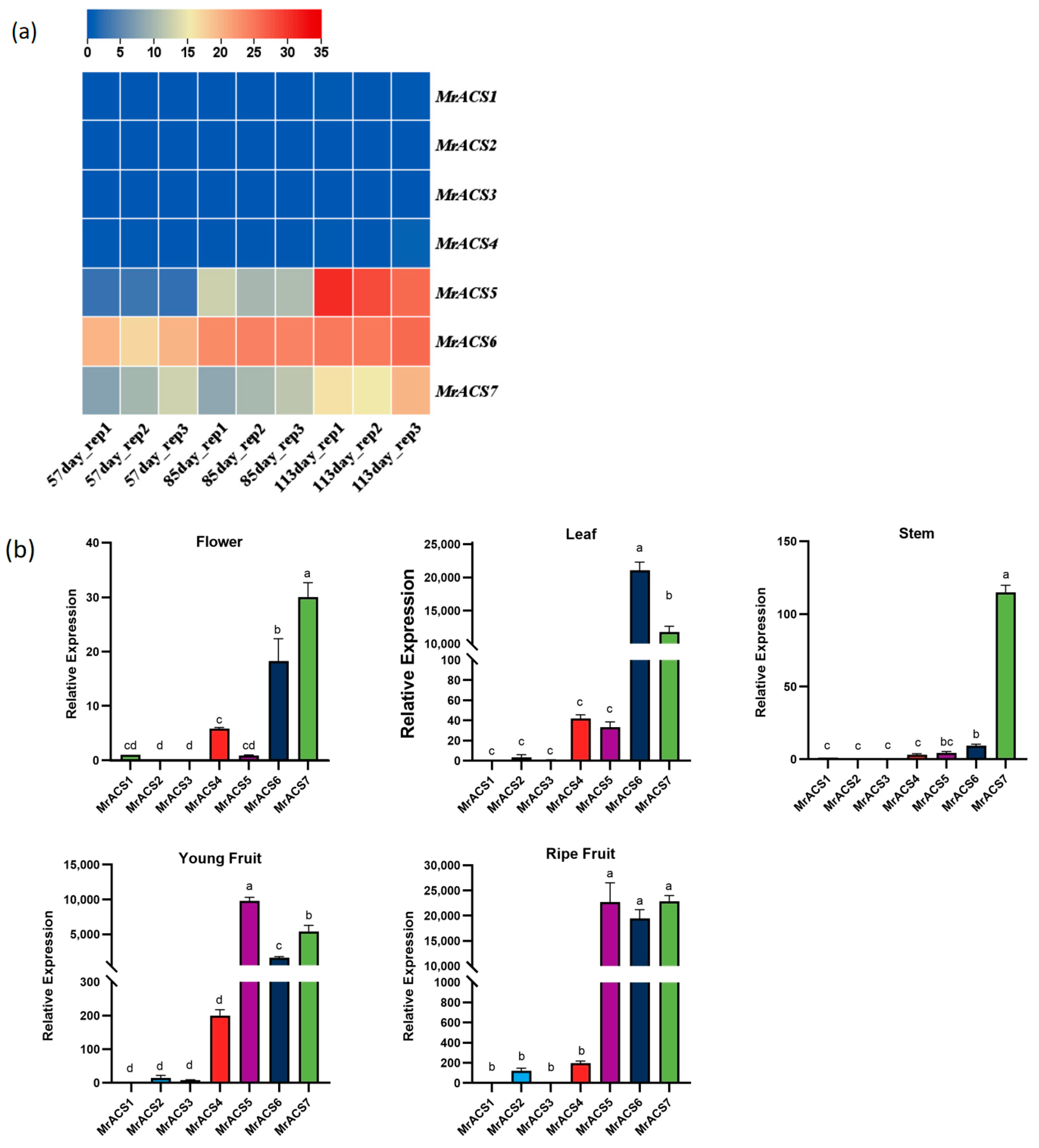
2.5. Expression Patterns of MrACS Genes in Myrica rubra
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of ACS Genes in Myrica rubra
4.3. Phylogenetic Analysis of ACS Genes in Myrica rubra
4.4. Chromosome Distribution and Collinearity Analysis of ACS Genes in Myrica rubra
4.5. Analysis of ACS Gene Structure and Conserved Motifs in Myrica rubra
4.6. Expression Profiling of MrACS Genes via Transcriptome Analysis and Quantitative Real-Time PCR
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ren, H.Y.; He, Y.H.; Qi, X.J.; Zheng, X.L.; Zhang, S.W.; Yu, Z.P.; Hu, F.R. The bayberry database: A multiomic database for Myrica rubra, an important fruit tree with medicinal value. BMC Plant Biol. 2021, 21, 452. [Google Scholar] [CrossRef]
- Zhang, S.W.; Yu, Z.P.; Sun, L.; Ren, H.Y.; Zheng, X.L.; Liang, S.M.; Qi, X.J. An overview of the nutritional value, health properties, and future challenges of Chinese bayberry. PeerJ. 2022, 10, e13070. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Gong, E.S.; Lin, Y.Y.; Li, T.; Lian, F.L.; Zheng, B.S.; Liu, R.H. Comparison of phytochemical profiles, antioxidant and antiproliferative activities in Chinese bayberry (Myrica rubra Sieb. et Zucc.) fruits. J. Food Sci. 2021, 86, 4691–4703. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.S.; Chen, J.B.; Wang, Y.; Zheng, F.H.; Qu, M.Y.; Huang, Z.W.; Yan, J.L.; Bao, F.P.; Li, X.; Sun, C.D.; et al. Cyanidin-3-O-glucoside extracted from the Chinese bayberry (Myrica rubra Sieb. et Zucc.) alleviates antibiotic-associated diarrhea by regulating gut microbiota and down-regulating inflammatory factors in NF-κB pathway. Front. Nutr. 2022, 9, 970530. [Google Scholar] [CrossRef]
- Zhang, Q.Z.; Huang, Z.J.; Wang, Y.; Wang, Y.B.; Fu, L.L.; Su, L.J. Chinese bayberry (Myrica rubra) phenolics mitigated protein glycoxidation and formation of advanced glycation end-products: A mechanistic investigation. Food Chem. 2021, 361, 130102. [Google Scholar] [CrossRef] [PubMed]
- Fenn, M.A.; Giovannoni, J.J. Phytohormones in fruit development and maturation. Plant J. 2021, 105, 446–458. [Google Scholar] [CrossRef]
- Tang, D.G.; Gallusci, P.P.; Lang, Z.B. Fruit development and epigenetic modifications. New Phytol. 2020, 228, 839–844. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Berman, A.; Shani, E. Plant hormone transport and localization: Signaling molecules on the move. Annu. Rev. Plant Biol. 2023, 22, 453–479. [Google Scholar] [CrossRef]
- Huang, J.Y.; Zhao, X.B.; Bürger, M.; Chory, J.; Wang, X.C. The role of ethylene in plant temperature stress response. Trends Plant Sci. 2023, 28, 808–824. [Google Scholar] [CrossRef]
- Zhao, H.; Yin, C.C.; Ma, B.; Chen, S.Y.; Zhang, J.S. Ethylene signaling in rice and Arabidopsis: New regulators and mechanisms. J. Integr. Plant Biol. 2021, 63, 102–125. [Google Scholar] [CrossRef]
- Yin, C.C.; Huang, Y.H.; Zhang, X.; Zhou, Y.; Chen, S.Y.; Zhang, J.S. Ethylene-mediated regulation of coleoptile elongation in rice seedlings. Plant Cell Environ. 2023, 46, 1060–1074. [Google Scholar] [CrossRef]
- Pattyn, J.; Vaughan-Hirsch, J.; Van de Poel, B. The regulation of ethylene biosynthesis: A complex multilevel control circuitry. New Phytol. 2021, 229, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Booker, M.A.; DeLong, A. Producing the ethylene signal: Regulation and diversification of ethylene biosynthetic enzymes. Plant Physiol. 2015, 169, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.Y.; Zhang, Y.X. Expression and regulation of pear 1-aminocyclopropane-1-carboxylic acid synthase gene (PpACS1a) during fruit ripening, under salicylic acid and indole-3-acetic acid treatment, and in diseased fruit. Mol. Biol. Rep. 2014, 41, 4147–4154. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.W.; Acharya, T.P.; Malladi, A.; Tsai, H.J.; NeSmith, D.S.; Doyle, J.W.; Nambeesan, S.U. Atypical Climacteric and Functional Ethylene Metabolism and Signaling During Fruit Ripening in Blueberry (Vaccinium sp.). Front. Plant Sci. 2022, 23, 932642. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Motyka, V.; Pokorna, E.; Dobrev, P.I.; Lacek, J.; Shao, J.; Lewers, K.S.; Mattoo, A.K. Comprehensive profiling of endogenous phytohormones and expression analysis of 1-aminocyclopropane-1-carboxylic acid synthase gene family during fruit development and ripening in octoploid strawberry (Fragaria × ananassa). Plant Physiol. Biochem. 2023, 196, 186–196. [Google Scholar] [CrossRef]
- Wang, A.D.; Yamakake, J.; Kudo, H.; Wakasa, Y.; Hatsuyama, Y.; Igarashi, M.; Kasai, A.; Li, T.; Harada, T. Null mutation of the MdACS3 gene, coding for a ripening-specific 1-aminocyclopropane-1-carboxylate synthase, leads to long shelf life in apple fruit. Plant Physiol. 2009, 151, 391–399. [Google Scholar] [CrossRef]
- Li, T.; Tan, D.M.; Liu, Z.; Jiang, Z.Y.; Wei, Y.; Zhang, L.C.; Li, X.Y.; Yuan, H.; Wang, A.D. Apple MdACS6 regulates ethylene biosynthesis during fruit development involving ethylene-responsive factor. Plant Cell Physiol. 2015, 56, 1909–1917. [Google Scholar] [CrossRef]
- Gupta, A.; Pal, R.K.; Rajam, M.V. Delayed ripening and improved fruit processing quality in tomato by RNAi-mediated silencing of three homologs of 1-aminopropane-1-carboxylate synthase gene. Plant Physiol. 2013, 170, 987–995. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.F.; Wei, W.; Fan, Z.Q.; Deng, W.; Li, Z.G.; Bouzayen, M.; Pirrello, J.; Lu, W.J.; Chen, J.Y. MaXB3 modulates MaNAC2, MaACS1, and MaACO1 stability to repress ethylene biosynthesis during banana fruit ripening. Plant Physiol. 2020, 184, 1153–1171. [Google Scholar] [CrossRef]
- Park, C.; Lee, H.Y.; Yoon, G.M. The regulation of ACC synthase protein turnover: A rapid route for modulating plant development and stress responses. Curr. Opin. Plant Biol. 2021, 63, 102046. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Cheng, K.; Lu, Y.; Wen, H.Y.; Ma, L.Q.; Zhang, C.J.; Suprun, A.R.; Zhu, H.L. Regulation of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) expression and its functions in plant life. Plant Horm. 2025, 1, e002. [Google Scholar] [CrossRef]
- Tang, X.L.; Liu, R.; Mei, Y.Y.; Wang, D.; He, K.X.; Wang, N.N. Identification of key ubiquitination sites involved in the proteasomal degradation of AtACS7 in Arabidopsis. Int. J. Mol. Sci. 2024, 25, 2931. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.; Seo, Y.S.; Kim, S.M.; Hong, D.K.; Park, K.Y.; Kim, W.T. Brassinosteroid induction of AtACS4 encoding an auxin-responsive 1-aminocyclopropane-1-carboxylate synthase 4 in Arabidopsis seedlings. Physiologia Plantarum. 2006, 126, 453–461. [Google Scholar] [CrossRef]
- Wang, N.N.; Shih, M.C.; Li, N. The GUS reporter-aided analysis of the promoter activities of Arabidopsis ACC synthase genes AtACS4, AtACS5, and AtACS7 induced by hormones and stresses. J. Exp. Bot. 2005, 56, 909–920. [Google Scholar] [CrossRef]
- Cao, H.H.; Chen, J.; Yue, M.; Xu, C.; Jian, W.; Liu, Y.D.; Song, B.Q.; Gao, Y.Q.; Cheng, Y.L.; Li, Z.G. Tomato transcriptional repressor MYB70 directly regulates ethylene-dependent fruit ripening. Plant J. 2020, 104, 1568–1581. [Google Scholar] [CrossRef]
- Wang, Z.; Yadav, V.; Yan, X.; Cheng, D.; Wei, C.; Zhang, X. Systematic genome-wide analysis of the ethylene-responsive ACS gene family: Contributions to sex form differentiation and development in melon and watermelon. Gene. 2021, 805, 145910. [Google Scholar] [CrossRef]
- Cui, Y.; Ji, X.; Yu, W.; Liu, Y.; Bai, Q.; Su, S. Genome-Wide Characterization and Functional Validation of the ACS Gene Family in the Chestnut Reveals Its Regulatory Role in Ovule Development. Int. J. Mol. Sci. 2024, 25, 4454. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.; Qi, R.; Zhao, H.; Yang, H.; Liao, N.; Ali, A.; Malangisha, G.K.; Ma, Y.; Zhang, K.; Zhou, Y.; et al. An allelic variant in the ACS7 gene promotes primary root growth in watermelon. Theor. Appl. Genet. 2022, 135, 3357–3373. [Google Scholar] [CrossRef]
- Wang, Z.; Wei, X.; Cui, X.; Wang, J.; Wang, Y.; Sun, M.; Zhao, P.; Yang, B.; Wang, Q.; Jiang, Y.Q. The transcription factor WRKY22 modulates ethylene biosynthesis and root development through transactivating the transcription of ACS5 and ACO5 in Arabidopsis. Physiol. Plant. 2024, 176, e14371. [Google Scholar] [CrossRef]
- Yu, Y.; Qi, Y.; Xu, J.; Dai, X.; Chen, J.; Dong, C.H.; Xiang, F. Arabidopsis WRKY71 regulates ethylene-mediated leaf senescence by directly activating EIN2, ORE1 and ACS2 genes. Plant J. 2021, 107, 1819–1836. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Q.; Lei, C.; Zhu, Z.H.; Li, M.Z.; Chen, Z.P.; He, W.; Liu, B.; Chen, L.P.; Li, X.J.; Xie, Y.Z. Genome-wide analysis and identification of 1-aminocyclopropane-1-carboxylate synthase (ACS) gene family in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2023, 24, 11158. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.J.; Li, W.L.; Chen, F.Y.; Cheng, Y.Q.; Huang, X.; Zou, B.X.; Wang, Y.L.; Xu, W.L.; Qu, S.P. Genome-wide identification and characterization of members of the ACS gene family in Cucurbita maxima and their transcriptional responses to the specific treatments. Int. J. Mol. Sci. 2022, 23, 8476. [Google Scholar] [CrossRef]
- Van Der Straeten, D.; Zhou, Z.; Prinsen, E.; Van Onckelen, H.A.; Van Montagu, M.C. A comparative molecular-physiological study of submergence response in lowland and deepwater rice. Plant Physiol. 2001, 125, 955–968. [Google Scholar] [CrossRef]
- Tabassum, N.; Shafiq, M.; Fatima, S.; Tahir, S.; Tabassum, B.; Ali, Q.; Javed, M.A. Genome-wide in-silico analysis of ethylene biosynthesis gene family in Musa acuminata L. and their response under nutrient stress. Sci. Rep. 2024, 14, 558. [Google Scholar] [CrossRef]
- Li, L.; Li, X.; Yang, C.; Cheng, Y.; Cai, Z.; Nian, H.; Ma, Q. GsERF1 enhances Arabidopsis thaliana aluminum tolerance through an ethylene-mediated pathway. BMC Plant Biol. 2022, 22, 258. [Google Scholar] [CrossRef]
- Datta, R.; Kumar, D.; Sultana, A.; Hazra, S.; Bhattacharyya, D.; Chattopadhyay, S. Glutathione regulates 1-aminocyclopropane-1-carboxylate synthase transcription via WRKY33 and 1-aminocyclopropane-1-carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol. 2015, 169, 2963–2981. [Google Scholar] [PubMed]
- Wang, Z.; Li, F. Potential Roles of 1-Aminocyclopropane-1-carboxylic Acid Synthase Genes in the Response of Gossypium Species to Abiotic Stress by Genome-Wide Identification and Expression Analysis. Plants 2022, 11, 1524. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, X.; Zhang, N.; Gao, X.; Feng, X.; Zeng, Q.; Chen, X.L.; Wu, J.Y.; Qi, Y.W. Identification of ACC synthetase genes in saccharum and their expression profiles during plant growth and in response to low-nitrogen stress. Trop. Plant Biol. 2022, 15, 197–210. [Google Scholar] [CrossRef]
- Iqbal, N.; Trivellini, A.; Masood, A.; Ferrante, A.; Khan, N.A. Current understanding on ethylene signaling in plants: The influence of nutrient availability. Plant Physiol. Biochem. 2013, 73, 128–138. [Google Scholar] [CrossRef]
- Zeng, W.F.; Pan, L.; Liu, H.; Niu, L.; Lu, Z.H.; Cui, G.C.; Wang, Z.Q. Characterization of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) genes during nectarine fruit development and ripening. Tree Genet. Genomes 2015, 11, 18. [Google Scholar] [CrossRef]
- Chen, H.T.; Bai, S.L.; Kusano, M.; Ezura, H.; Wang, N. Increased ACS enzyme dosage causes Initiation of climacteric ethylene production in tomato. Int. J. Mol. Sci. 2022, 23, 10788. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Zhang, Y.R.; Yu, Z.; Wang, H.Y.; Ping, B.; Liu, Y.X.; Liang, J.K.; Ma, F.W.; Zou, Y.J.; Zhao, T. Insights into ACO genes across Rosaceae: Evolution, expression, and regulatory networks in fruit development. Genes. Genom. 2024, 46, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.C.; Qiao, Q.; Zhong, Y. Detecting adaptive evolution and functional divergence in aminocyclopropane-1-carboxylate synthase (ACS) gene family. Comput. Biol. Chem. 2012, 38, 10–16. [Google Scholar] [CrossRef]
- Ahmadizadeh, M.; Chen, J.T.; Hasanzadeh, S.; Ahmar, S.; Heidari, P. Insights into the genes involved in the ethylene biosynthesis pathway in Arabidopsis thaliana and Oryza sativa. J. Genet. Eng. Biotechnol. 2020, 18, 62. [Google Scholar] [CrossRef]
- Liu, Y.D.; Shi, Y.; Su, D.D.; Lu, W.; Li, Z. SlGRAS4 accelerates fruit ripening by regulating ethylene biosynthesis genes and SlMADS1 in tomato. Hortic. Res. 2021, 8, 3. [Google Scholar] [CrossRef]
- Gao, Y.; Wei, W.; Zhao, X.D.; Tan, X.L.; Fan, Z.Q.; Zhang, Y.P.; Jing, Y.; Meng, L.H.; Zhu, B.Z.; Zhu, H.L.; et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 2018, 5, 75. [Google Scholar] [CrossRef]
- Dougherty, L.; Zhu, Y.D.; Xu, K.N. Assessing the allelotypic effect of two aminocyclopropane carboxylic acid synthase-encoding genes MdACS1 and MdACS3a on fruit ethylene production and softening in Malus. Hortic. Res. 2016, 3, 16024. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Gupta, S.; Sarma, S.; Rai, M.; Sreelakshmi, Y.; Sharma, R. Mutations in tomato 1-aminocyclopropane carboxylic acid synthase 2 uncover its role in development beside fruit ripening. Plant J. 2021, 106, 95–112. [Google Scholar] [CrossRef]
- Kim, S.H.; Yang, S.H.; Kim, T.J.; Han, J.S.; Suh, J.W. Hypertonic stress increased extracellular ATP levels and the expression of stress-responsive genes in Arabidopsis thaliana seedlings. Biosci. Biotechnol. Biochem. 2009, 73, 1252–1256. [Google Scholar] [CrossRef]
- Giovannoni, J.; Nguyen, C.; Ampofo, B.; Zhong, S.; Fei, Z. The epigenome and transcriptional dynamics of fruit ripening. Annu. Rev. Plant Biol. 2017, 68, 61–84. [Google Scholar] [CrossRef]
- Ji, Y.L.; Wang, A.D. Recent advances in epigenetic triggering of climacteric fruit ripening. Plant Physiol. 2023, 192, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.C.; Pirrello, J.; Chervin, C.; Roustan, J.P.; Bouzayen, M. Ethylene control of fruit ripening: Revisiting the complex network of transcriptional regulation. Plant Physiol. 2015, 169, 2380–2390. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Shima, Y.; Nakamura, N.; Kotake-Nara, E.; Kawasaki, S.; Toki, S. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat. Plants. 2017, 3, 866–874. [Google Scholar] [CrossRef]
- Gao, Y.; Wei, W.; Fan, Z.Q.; Zhao, X.D.; Zhang, Y.P.; Jing, Y.; Zhu, B.Z.; Zhu, H.L.; Shan, W.; Chen, J.Y.; et al. Re-evaluation of the nor mutation and the role of the NAC-NOR transcription factor in tomato fruit ripening. J. Exp. Bot. 2020, 71, 3560–3574. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Jiang, Z.Y.; Zhang, L.C.; Tan, D.M.; Wei, Y.; Yuan, H.; Li, T.L.; Wang, A.D. Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 2016, 88, 735–748. [Google Scholar] [CrossRef]
- Shekhawat, K.; Fröhlich, K.; García-Ramírez, G.X.; Trapp, M.A.; Hirt, H. Ethylene: A master regulator of Plant-Microbe interactions under abiotic stresses. Cells 2022, 12, 31. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A master regulator of salinity stress tolerance in plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef]
- Zhang, B.G.; Liu, H.F.; Ding, X.H.; Qiu, J.J.; Zhang, M.; Chu, Z. Arabidopsis thaliana ACS8 plays a crucial role in the early biosynthesis of ethylene elicited by Cu2+ ions. J. Cell Sci. 2018, 131, jcs202424. [Google Scholar] [CrossRef]
- Poulaki, E.G.; Tsolakidou, M.D.; Gkizi, D.; Pantelides, I.S.; Tjamos, S.E. The ethylene biosynthesis genes ACS2 and ACS6 modulate disease severity of Verticillium dahliae. Plants 2020, 9, 907. [Google Scholar] [CrossRef]
- Zarembinski, T.I.; Theologis, A. Expression characteristics of OS-ACS1 and OS-ACS2, two members of the 1-aminocyclopropane-1-carboxylate synthase gene family in rice (Oryza sativa L. cv. Habiganj Aman II) during partial submergence. Plant Mol. Biol. 1997, 33, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Huang, Y.Y.; Ge, W.N.; Jia, Z.H.; Song, S.S.; Zhang, L.; Huang, Y.L. Involvement of jasmonic acid, ethylene and salicylic acid signaling pathways behind the systemic resistance induced by Trichoderma longibrachiatum H9 in cucumber. BMC Genom. 2019, 20, 144. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.F.; Zhang, H.Q.; Allan, A.C.; Wang, W.Q.; Yin, X.R. Transcriptional and post-transcriptional regulation of ethylene biosynthesis by exogenous acetylsalicylic acid in kiwifruit. Hortic. Res. 2022, 9, uhac116. [Google Scholar] [CrossRef]
- Xie, L.M.; Wang, Y.N.; Tao, Y.T.; Chen, L.X.; Lin, H.Y.; Qi, Z.C.; Li, J.M. Genome-wide identification and analysis of anthocyanin synthesis-related R2R3-MYB genes in Fragaria pentaphylla. BMC Genomics. 2024, 25, 952. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.Y.; Yu, H.Y.; Zhang, S.W.; Liang, S.M.; Zheng, X.L.; Zhang, S.J.; Yao, P.; Zheng, H.K.; Qi, X.J. Genome sequencing provides insights into the evolution and antioxidant activity of Chinese bayberry. BMC Genom. 2019, 20, 458. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Hernandez, M.; Berardini, T.Z.; Chen, G.H.; Crist, D.; Doyle, A.; Huala, E.; Knee, E.; Lambrecht, M.; Miller, N.; Mueller, L.A.; et al. TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genom. 2002, 2, 239–253. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.; Tosatto, S.; Paladin, L.; Raj, S.; Richardson, L.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, 412–419. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, X.; Shi, L.Y.; Shao, J.R.; Chen, W.; Zheng, Y.H.; Yang, Z.F. Molecular cloning and expression analysis of MrFRK2 in Chinese bayberry during fruit ripening. Acta Hortic. Sinica. 2016, 43, 1585. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Number of Amino Acid | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| MrACS1 | 470 | 52.77 | 8.22 | 45.56 | 83.21 | −0.269 | Cytoplasm |
| MrACS2 | 477 | 53.71 | 5.84 | 47.28 | 82.6 | −0.24 | Nucleus |
| MrACS3 | 446 | 50.18 | 5.79 | 50.83 | 83.77 | −0.336 | Cytoplasm |
| MrACS4 | 486 | 54.60 | 6.6 | 43.83 | 82.84 | −0.18 | Cytoplasm |
| MrACS5 | 479 | 53.64 | 6.21 | 50.03 | 83.63 | −0.209 | Cytoplasm |
| MrACS6 | 528 | 58.41 | 8.4 | 50.54 | 89.6 | −0.084 | Nucleus |
| MrACS7 | 551 | 60.34 | 8.33 | 48.73 | 89 | −0.07 | Cytoplasm |
| Ka | Ks | Ka/Ks | ||
|---|---|---|---|---|
| MrACS4 | MrACS5 | 0.17033069606252893 | 1.9638562679829152 | 0.08673277104819707 |
| MrACS6 | MrACS7 | 0.23165164492644064 | 1.4133469890885786 | 0.16390288210528203 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, H.; Liu, X.; Liu, Y.; Wu, F.; Jin, W. Genome-Wide Identification and Expression Analysis of 1-Aminocyclopropane-1-Carboxylate Synthase (ACS) Gene Family in Myrica rubra. Int. J. Mol. Sci. 2025, 26, 4580. https://doi.org/10.3390/ijms26104580
Huang H, Liu X, Liu Y, Wu F, Jin W. Genome-Wide Identification and Expression Analysis of 1-Aminocyclopropane-1-Carboxylate Synthase (ACS) Gene Family in Myrica rubra. International Journal of Molecular Sciences. 2025; 26(10):4580. https://doi.org/10.3390/ijms26104580
Chicago/Turabian StyleHuang, Huanhui, Xintong Liu, Yiqing Liu, Fangli Wu, and Weibo Jin. 2025. "Genome-Wide Identification and Expression Analysis of 1-Aminocyclopropane-1-Carboxylate Synthase (ACS) Gene Family in Myrica rubra" International Journal of Molecular Sciences 26, no. 10: 4580. https://doi.org/10.3390/ijms26104580
APA StyleHuang, H., Liu, X., Liu, Y., Wu, F., & Jin, W. (2025). Genome-Wide Identification and Expression Analysis of 1-Aminocyclopropane-1-Carboxylate Synthase (ACS) Gene Family in Myrica rubra. International Journal of Molecular Sciences, 26(10), 4580. https://doi.org/10.3390/ijms26104580






