Expression of the CpXTH6 and CpXTH23 Genes in Carica papaya Fruits
Abstract
1. Introduction
2. Results
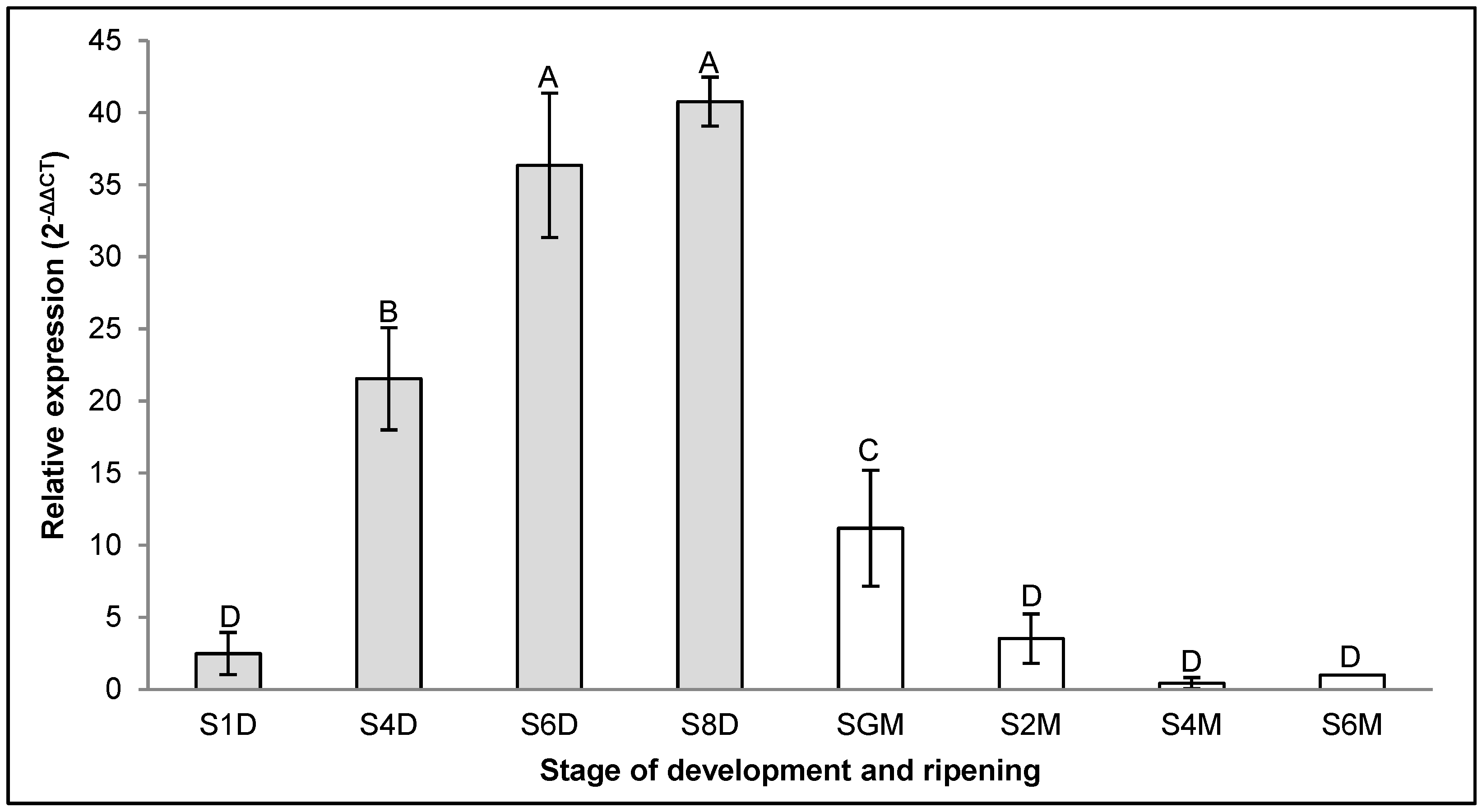
2.1. Expression of CpXTH6 and CpXTH23 Genes in C. papaya Fruits
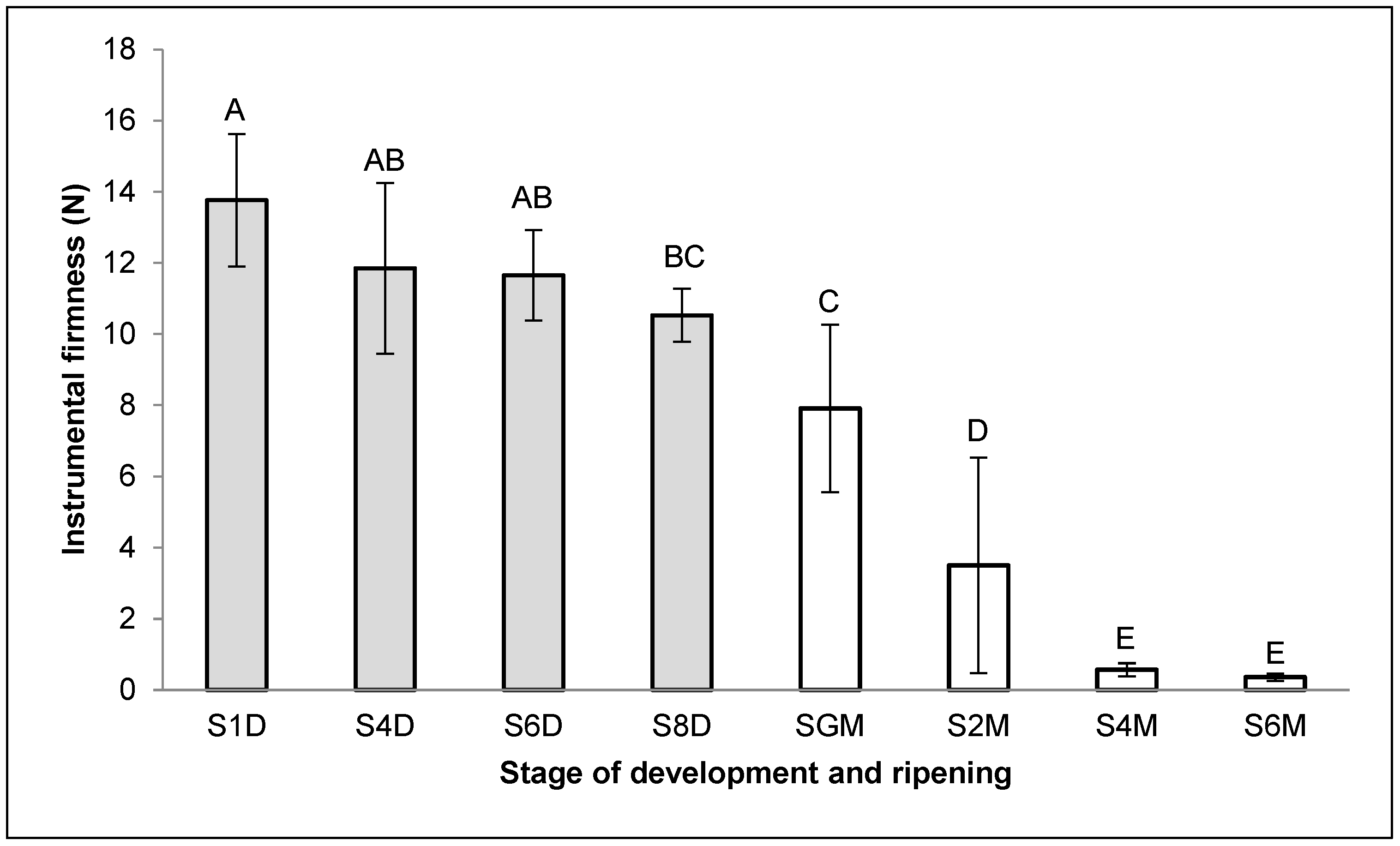
2.2. Firmness of C. papaya Fruits
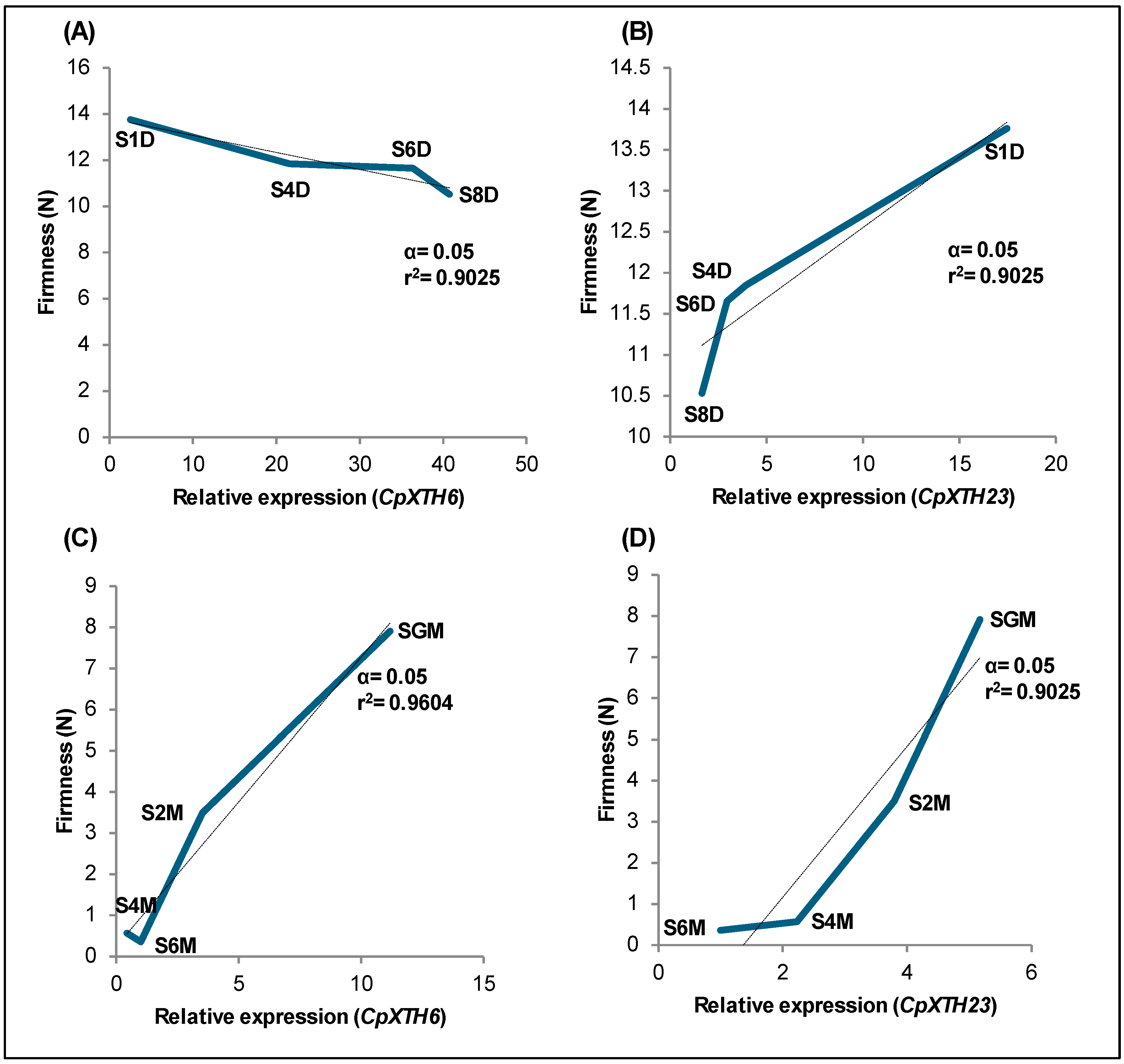
2.3. Correlation Between Relative Expression of CpXTH Genes and Fruit Firmness
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. Total RNA Extraction
4.3. cDNA Synthesis
4.4. Oligonucleotides and Probes Used
4.5. Gene Expression Analysis by qPCR
4.6. Instrumental Firmness of C. papaya Fruits
4.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Redagrícola. Crece Producción de Papaya Mexicana 48.3% en Diez Años. 2024. Available online: https://redagricola.com/crece-produccion-de-papaya-mexicana-48-3-en-diez-anos/ (accessed on 25 April 2025).
- Payasi, A.; Mishra, N.N.; Chaves, A.L.S.; Singh, R. Biochemistry of fruit softening: An overview. Physiol. Mol. Biol. Plants 2009, 15, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Brummell, D.A. Cell wall disassembly in ripening fruit. Funct. Plant Biol. 2006, 33, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Prado, S.B.; Melfi, P.R.; Castro-Alves, V.C.; Broetto, S.G.; Araújo, E.S.; do Nascimento, J.R.; Fabi, J.P. Physiological degradation of pectin in papaya cell walls: Release of long chains galacturonans derived from insoluble fractions during postharvest fruit ripening. Front. Plant Sci. 2016, 7, 1120. [Google Scholar] [CrossRef]
- Ray, S.; Vigouroux, J.; Quémener, B.; Bonnin, E.; Lahaye, M. Novel and diverse fine structures in LiCl–DMSO extracted apple hemicelluloses. Carbohydr. Polym. 2014, 108, 46–57. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 2015, 67, 463–476. [Google Scholar] [CrossRef]
- Miedes, E.; Zarra, I.; Hoson, T.; Herbers, K.; Sonnewald, U.; Lorences, E.P. Xyloglucan endotransglucosylase and cell wall extensibility. J. Plant Physiol. 2011, 168, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Lorences, E.P. Xyloglucan endotransglucosylase/hydrolases (XTHs) during tomato fruit growth and ripening. J. Plant Physiol. 2009, 166, 489–498. [Google Scholar] [CrossRef]
- Muñoz-Bertomeu, J.; Miedes, E.; Lorences, E.P. Expression of xyloglucan endotransglucosylase/hydrolase (XTH) genes and XET activity in ethylene treated apple and tomato fruits. J. Plant Physiol. 2013, 170, 1194–1201. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Johnston, S.L.; Yauk, Y.-K.; Sharma, N.N.; Schröder, R. Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwi fruit and apple. Postharvest Biol. Technol. 2009, 51, 149–157. [Google Scholar] [CrossRef]
- Goulao, L.F.; Cosgrove, D.J.; Oliveira, C.M. Cloning, characterization, and expression analyses of cDNA clones encoding cell wall-modifying enzymes isolated from ripe apples. Postharvest Biol. Technol. 2008, 48, 37–51. [Google Scholar] [CrossRef]
- Dheilly, E.; Gall, S.L.; Guillou, M.C.; Renou, J.-P.; Bonnin, E.; Orsel, M.; Lahaye, M. Cell wall dynamics during apple development and storage involves hemicellulose modifications and related expressed genes. BMC Plant Biol. 2016, 16, 201. [Google Scholar] [CrossRef]
- Han Ye Ban, Q.; Hou, Y.; Meng, K.; Suo, J.; Rao, J. Isolation and characterization of two persimmon xyloglucan endotransglycosylase/hydrolase (XTH) genes that have divergent functions in cell wall modification and fruit postharvest softening. Front. Plant Sci. 2016, 7, 624. [Google Scholar] [CrossRef]
- Han Ye Zhu, Q.; Zhang, Z.; Meng, K.; Hou, Y.; Ban, Q.; Rao, J. Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) genes and diverse roles of isoenzymes during persimmon fruit development and postharvest softening. PLoS ONE 2015, 10, e0123668. [Google Scholar] [CrossRef]
- Saladié, M.; Rose, J.K.; Cosgrove, D.J.; Catala, C. Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. Plant J. 2006, 47, 282–295. [Google Scholar] [CrossRef]
- Opazo, M.C.; Figueroa, C.R.; Henríquez, J.; Herrera, R.; Bruno, C.; Valenzuela, P.D.; Moya-León, M.A. Characterization of two divergent cDNAs encoding xyloglucan endotransglycosylase/hydrolase (XTH) expressed in Fragaria chiloensis fruit. Plant Sci. 2010, 179, 479–488. [Google Scholar] [CrossRef]
- Witasari, L.D.; Huang, F.C.; Hoffmann, T.; Rozhon, W.; Fry, S.C.; Schwab, W. Higher expression of the strawberry xyloglucan endotransglucosylase/hydrolase genes FvXTH9 and FvXTH6 accelerates fruit ripening. Plant J. 2019, 100, 1237–1253. [Google Scholar] [CrossRef]
- Su, Q.; Li, X.; Wang, L.; Wang, B.; Feng, Y.; Yang, H.; Zhao, Z. Variation in Cell Wall Metabolism and Flesh Firmness of Four Apple Cultivars during fruit development. Foods 2022, 11, 3518. [Google Scholar] [CrossRef]
- Nishiyama, K.; Guis, M.; Rose, J.K.; Kubo, Y.; Bennett, K.A.; Wangjin, L.; Bouzayen, M. Ethylene regulation of fruit softening and cell wall disassembly in Charentais melon. J. Exp. Bot. 2007, 58, 1281–1290. [Google Scholar] [CrossRef]
- Zhai, Z.; Feng, C.; Wang, Y.; Sun, Y.; Peng, X.; Xiao, Y.; Zhang, X.; Zhou, X.; Jiao, J.; Wang, W.; et al. Genome-wide identification of the xyloglucan endotransglucosylase/hydrolase (XTH) and polygalacturonase (PG) genes and characterization of their role in fruit softening of sweet cherry. Int. J. Mol. Sci. 2021, 22, 12331. [Google Scholar] [CrossRef]
- Aguilar-Velázquez, B.; Rosas-Quijano, R.; Vázquez-Ovando, A.; Salvador-Figueroa, M.; Adriano-Anaya, L.; Gálvez-López, D. CpXTH2 and CpXTH5 genes are expressed during fruit development and ripening in Carica papaya L. Fruits 2021, 76, 3–10. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, N.; Jiang, S.; Xu, H.; Wang, Y.; Wang, C.; Chen, X. Analysis of the xyloglucan endotransglucosylase/hydrolase gene family during apple fruit ripening and softening. J. Agric. Food Chem. 2017, 65, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, H.; Yin, C.; Wang, X.; Jiang, Q.; Zhang, R.; Yang, L. Genome-wide identification and characterization of xyloglucan endotransglycosylase/hydrolase in Ananas comosus during Development. Genes 2019, 10, 537. [Google Scholar] [CrossRef] [PubMed]
- Vissenberg, K.; Fry, S.C.; Pauly, M.; Höfte, H.; Verbelen, J.P. XTH acts at the microfibril–matrix interface during cell elongation. J. Exp. Bot. 2005, 56, 673–683. [Google Scholar] [CrossRef]
- Santamaría Basulto, F.; Díaz Plaza, R.; Sauri Duch, E.; Espadas y Gil, F.; Santamaría Fernández, J.M.; Larqué Saavedra, A. Características de calidad de frutos de papaya Maradol en la madurez de consumo. Agric. Técnica México 2009, 35, 347–353. [Google Scholar]
- Zheng, Y.; Duan, L.; Ran, Y.; Zhang, P.; Jiang, Y.; Zhao, Z.; Li, Z.; Chen, L.; Tang, Y.; Li, X.; et al. Transcriptomics analysis reveals molecular mechanism of softening and cell wall polysaccharides-disassembling in peaches treated by flow microcirculation of sodium nitroprusside medium. Postharvest Biol. Technol. 2023, 196, 112190. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Gene | Tm (°C) | F (5′-3′) | R (5′-3′) | TaqMan® Probes |
|---|---|---|---|---|
| CpXTH6 | 56 | TGGACGAAGTTCCGATCAGAGT | CCATGGGTTGGAATTTTGGA | 6FAMAAGAACAACGAAGCCAGAAACATCCCGT-QSY |
| CpXTH23 | 59 | TTGATATGCAGCTCAAGCTTGTC | AAGTTGAGAGGCATACATAATAAGCAGTA | 6FAMCGGAAACTCCGCTGGCACCG-QSY |
| Actin | 60 | TGGTGAAGGCTGGATTTGCT | GTCTAGGACGGCCCACAATACT | VICATGATGCTCCCAGGGCAGTTTTCCC-QSY |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zúñiga-Hernández, M.E.; Rosas-Quijano, R.; Salvador-Figueroa, M.; Vázquez-Ovando, A.; Gálvez-López, D. Expression of the CpXTH6 and CpXTH23 Genes in Carica papaya Fruits. Int. J. Mol. Sci. 2025, 26, 4490. https://doi.org/10.3390/ijms26104490
Zúñiga-Hernández ME, Rosas-Quijano R, Salvador-Figueroa M, Vázquez-Ovando A, Gálvez-López D. Expression of the CpXTH6 and CpXTH23 Genes in Carica papaya Fruits. International Journal of Molecular Sciences. 2025; 26(10):4490. https://doi.org/10.3390/ijms26104490
Chicago/Turabian StyleZúñiga-Hernández, Melvin E., Raymundo Rosas-Quijano, Miguel Salvador-Figueroa, Alfredo Vázquez-Ovando, and Didiana Gálvez-López. 2025. "Expression of the CpXTH6 and CpXTH23 Genes in Carica papaya Fruits" International Journal of Molecular Sciences 26, no. 10: 4490. https://doi.org/10.3390/ijms26104490
APA StyleZúñiga-Hernández, M. E., Rosas-Quijano, R., Salvador-Figueroa, M., Vázquez-Ovando, A., & Gálvez-López, D. (2025). Expression of the CpXTH6 and CpXTH23 Genes in Carica papaya Fruits. International Journal of Molecular Sciences, 26(10), 4490. https://doi.org/10.3390/ijms26104490







