Association of Klotho Gene Polymorphism and Serum Level of α Klotho Protein with Different Tumor Grades, Overall Survival and Cytokine Profile in Glioma Patients
Abstract
1. Introduction
2. Results
2.1. Characteristics of Glioma Patients and Selected Variables
2.2. Analysis of Klotho rs1207568 and rs564481 Polymorphisms in Patients with Gliomas and Healthy Control Group
2.3. Association of Klotho rs1207568 and rs564481 Genotypes with Overall Survival in Glioma Patients
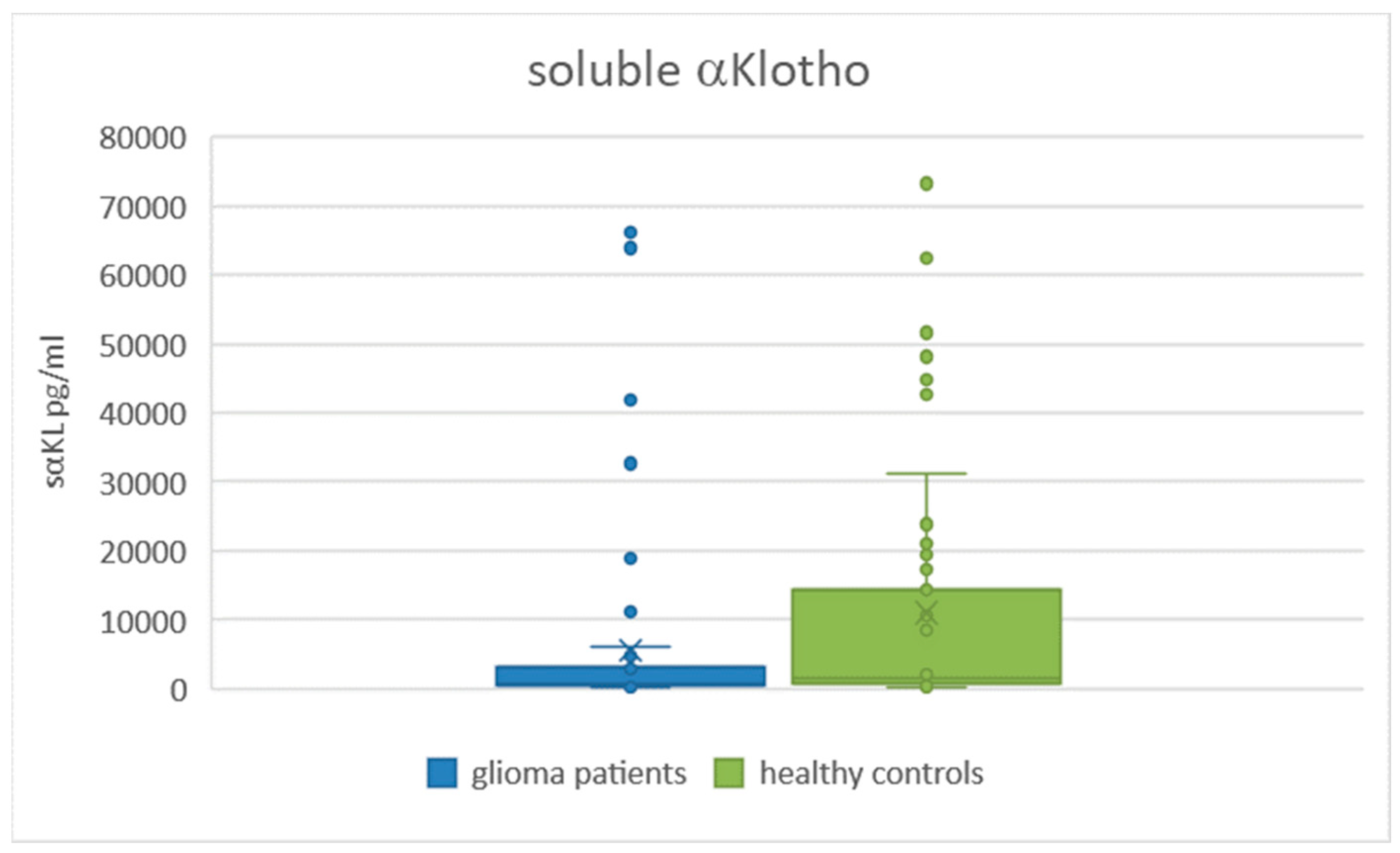
2.4. Comparison of Serum Levels of sαKL in Glioma Patients and Healthy Controls
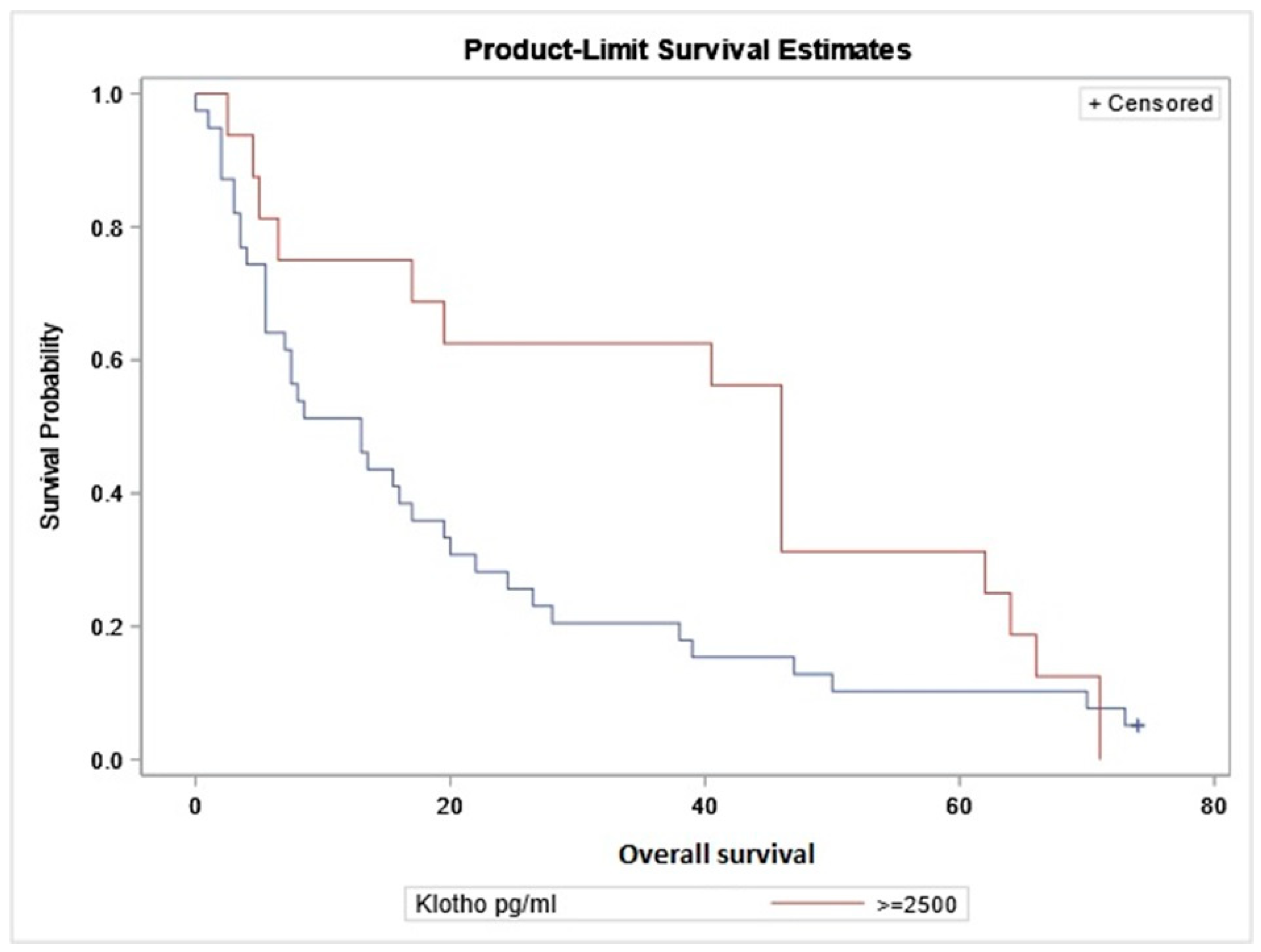
2.5. Correlation of Serum Level of sαKL with Overall Survival
2.6. Correlation of Serum Levels of sαKL with the Levels of Candidate Biomarkers
3. Discussion
4. Materials and Methods
4.1. Study Groups
4.2. Genotyping
4.3. Analysis of Serum/Plasma Levels of Investigated Biomarkers
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro-Oncology 2022, 24, v1–v95. [Google Scholar] [CrossRef] [PubMed]
- Cantrell, J.N.; Waddle, M.R.; Rotman, M.; Peterson, J.L.; Ruiz-Garcia, H.; Heckman, M.G.; Quiñones-Hinojosa, A.; Rosenfeld, S.S.; Brown, P.D.; Trifiletti, D.M. Progress Toward Long-Term Survivors of Glioblastoma. Mayo Clin. Proc. 2019, 94, 1278–1286. [Google Scholar] [CrossRef]
- Tykocki, T.; Eltayeb, M. Ten-year survival in glioblastoma. A systematic review. J. Clin. Neurosci. 2018, 54, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Gusyatiner, O.; Hegi, M.E. Glioma epigenetics: From subclassification to novel treatment options. Semin. Cancer Biol. 2018, 51, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Thomas, D.L. 2021 updates to the World Health Organization classification of adult-type and pediatric-type diffuse gliomas: A clinical practice review. Chin. Clin. Oncol. 2023, 12, 7. [Google Scholar] [CrossRef]
- Hayashi, H.; Iwashita, H.; Tateishi, K. Circumscribed Astrocytic Gliomas. No Shinkei Geka 2023, 51, 884–891. [Google Scholar] [PubMed]
- Kuro-o, M.; Matsumura, Y.; Aizawa, H.; Kawaguchi, H.; Suga, T.; Utsugi, T.; Ohyama, Y.; Kurabayashi, M.; Kaname, T.; Kume, E.; et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 1997, 390, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Kuro-o, M. Klotho as a regulator of oxidative stress and senescence. Biol. Chem. 2008, 389, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Aizawa, H.; Shiraki-Iida, T.; Nagai, R.; Kuro-o, M.; Nabeshima, Y. Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem. Biophys. Res. Commun. 1998, 242, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Fujimori, T.; Furuya, A.; Satoh, J.; Nabeshima, Y.; Nabeshima, Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J. Clin. Investig. 2005, 115, 2202–2208. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Okino, N.; Kakuta, Y.; Shikanai, T.; Tani, M.; Narimatsu, H.; Ito, M. Klotho-related protein is a novel cytosolic neutral beta-glycosylceramidase. J. Biol. Chem. 2007, 282, 30889–30900. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sun, Z. Molecular basis of Klotho: From gene to function in aging. Endocr Rev 2015, 36, 174–193. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Z. Current understanding of klotho. Ageing Res. Rev. 2009, 8, 43–51. [Google Scholar] [CrossRef]
- Kuro-o, M. Klotho. Pflugers Arch. 2010, 459, 333–343. [Google Scholar] [CrossRef]
- Urakawa, I.; Yamazaki, Y.; Shimada, T.; Iijima, K.; Hasegawa, H.; Okawa, K.; Fujita, T.; Fukumoto, S.; Yamashita, T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 2006, 444, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Ishikawa, K.; Matsukawa, N.; Kida, I.; Ohta, J.; Ikushima, M.; Chihara, Y.; Rui, X.; Rakugi, H.; Ogihara, T. Klotho protein activates the PKC pathway in the kidney and testis and suppresses 25-hydroxyvitamin D3 1alpha-hydroxylase gene expression. Endocrine 2004, 25, 229–234. [Google Scholar] [CrossRef]
- Kuro-o, M. Klotho and aging. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2009, 1790, 1049–1058. [Google Scholar] [CrossRef]
- Yamamoto, M.; Clark, J.D.; Pastor, J.V.; Gurnani, P.; Nandi, A.; Kurosu, H.; Miyoshi, M.; Ogawa, Y.; Castrillon, D.H.; Rosenblatt, K.P.; et al. Regulation of oxidative stress by the anti-aging hormone klotho. J. Biol. Chem. 2005, 280, 38029–38034. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Imura, A.; Urakawa, I.; Shimada, T.; Murakami, J.; Aono, Y.; Hasegawa, H.; Yamashita, T.; Nakatani, K.; Saito, Y.; et al. Establishment of sandwich ELISA for soluble alpha-Klotho measurement: Age-dependent change of soluble alpha-Klotho levels in healthy subjects. Biochem. Biophys. Res. Commun. 2010, 398, 513–518. [Google Scholar] [CrossRef]
- Kurosu, H.; Yamamoto, M.; Clark, J.D.; Pastor, J.V.; Nandi, A.; Gurnani, P.; McGuinness, O.P.; Chikuda, H.; Yamaguchi, M.; Kawaguchi, H.; et al. Suppression of aging in mice by the hormone Klotho. Science 2005, 309, 1829–1833. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Yamada, K.; Kim, H.C.; Kim, Y.S.; Noda, Y.; Imura, A.; Nabeshima, Y.; Nabeshima, T. Cognition impairment in the genetic model of aging klotho gene mutant mice: A role of oxidative stress. Faseb J. 2003, 17, 50–52. [Google Scholar] [CrossRef] [PubMed]
- Mytych, J. Actions of Klotho on hippocampal neuronal cells. Vitam. Horm. 2022, 118, 223–246. [Google Scholar]
- Cheng, M.F.; Chen, L.J.; Niu, H.S.; Yang, T.T.; Lin, K.C.; Cheng, J.T. Signals mediating Klotho-induced neuroprotection in hippocampal neuronal cells. Acta Neurobiol. Exp. (Wars) 2015, 75, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wang, X.; Zhao, W.; Wu, J. Klotho inhibits growth and promotes apoptosis in human lung cancer cell line A549. J. Exp. Clin. Cancer. Res. 2010, 29, 99. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Zhou, J.; Yuan, L.; Ren, F.; Liu, D.C.; Li, Q.; Shu, G. Epigenetic silencing of Klotho expression correlates with poor prognosis of human hepatocellular carcinoma. Hum. Pathol. 2013, 44, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Rubinek, T.; Wolf, I. The Role of Alpha-Klotho as a Universal Tumor Suppressor. Vitam. Horm. 2016, 101, 197–214. [Google Scholar]
- Abboud, M.; Merenbakh-Lamin, K.; Volkov, H.; Ben-Neriah, S.; Ligumsky, H.; Bronfeld, S.; Keren-Khadmy, N.; Giladi, M.; Shomron, N.; Wolf, I.; et al. Revealing the tumor suppressive sequence within KL1 domain of the hormone Klotho. Oncogene 2024, 43, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Doi, S.; Zou, Y.; Togao, O.; Pastor, J.V.; John, G.B.; Wang, L.; Shiizaki, K.; Gotschall, R.; Schiavi, S.; Yorioka, N.; et al. Klotho inhibits transforming growth factor-beta1 (TGF-beta1) signaling and suppresses renal fibrosis and cancer metastasis in mice. J. Biol. Chem. 2011, 286, 8655–8665. [Google Scholar] [CrossRef]
- Mota, J.; Lima, A.M.M.; Gomes, J.I.S.; Souza de Andrade, M.; Brito, H.O.; Silva, M.; Faustino-Rocha, A.I.; Oliveira, P.A.; Lopes, F.F.; Gil da Costa, R.M. Klotho in Cancer: Potential Diagnostic and Prognostic Applications. Diagnostics 2023, 13, 3357. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Peshes-Yeloz, N.; Ungar, L.; Wohl, A.; Jacoby, E.; Fisher, T.; Leitner, M.; Nass, D.; Rubinek, T.; Wolf, I.; Cohen, Z.R. Role of Klotho Protein in Tumor Genesis, Cancer Progression, and Prognosis in Patients with High-Grade Glioma. World Neurosurg. 2019, 130, e324–e332. [Google Scholar] [CrossRef]
- Su, J.; Ma, Q.; Long, W.; Tang, H.; Wu, C.; Luo, M.; Wang, X.; Xiao, K.; Li, Y.; Xiao, Q.; et al. LCTL Is a Prognostic Biomarker and Correlates With Stromal and Immune Infiltration in Gliomas. Front. Oncol. 2019, 9, 1083. [Google Scholar] [CrossRef]
- National Library of Medicine (US). Genetics Home Reference; The Library: Bethesda, MD, USA, 2013; September 16. Available online: https://www.ncbi.nlm.nih.gov/snp/rs1207568 (accessed on 27 May 2024).
- National Library of Medicine (US). Genetics Home Reference; The Library: Bethesda, MD, USA, 2013; September 16. Available online: https://www.ncbi.nlm.nih.gov/snp/rs564481 (accessed on 31 May 2024).
- Liu, C.; Cui, W.; Wang, L.; Yan, L.; Ruan, X.; Liu, Y.; Jia, X.; Zhang, X. Klotho gene polymorphisms are related to colorectal cancer susceptibility. Int. J. Clin. Exp. Pathol. 2015, 8, 7446–7449. [Google Scholar]
- Kamal, A.; Salama, M.; Kamal, A.; Mohsen, A.; Siam, I. Klotho (rs1207568 and rs564481) gene variants and colorectal cancer risk. Turk. J. Gastroenterol. 2020, 31, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Kuro-o, M. Klotho and the aging process. Korean J. Intern. Med. 2011, 26, 113–122. [Google Scholar] [CrossRef]
- Shimoyama, Y.; Taki, K.; Mitsuda, Y.; Tsuruta, Y.; Hamajima, N.; Niwa, T. KLOTHO gene polymorphisms G-395A and C1818T are associated with low-density lipoprotein cholesterol and uric acid in Japanese hemodialysis patients. Am. J. Nephrol. 2009, 30, 383–388. [Google Scholar] [CrossRef]
- Chang, B.; Kim, J.; Jeong, D.; Jeong, Y.; Jeon, S.; Jung, S.I.; Yang, Y.; Kim, K.I.; Lim, J.S.; Kim, C.; et al. Klotho inhibits the capacity of cell migration and invasion in cervical cancer. Oncol. Rep. 2012, 28, 1022–1028. [Google Scholar] [CrossRef]
- Chen, B.; Ma, X.; Liu, S.; Zhao, W.; Wu, J. Inhibition of lung cancer cells growth, motility and induction of apoptosis by Klotho, a novel secreted Wnt antagonist, in a dose-dependent manner. Cancer Biol. Ther. 2012, 13, 1221–1228. [Google Scholar] [CrossRef]
- Xie, B.; Chen, J.; Liu, B.; Zhan, J. Klotho acts as a tumor suppressor in cancers. Pathol. Oncol. Res. 2013, 19, 611–617. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, X. Klotho: A novel biomarker for cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Wolf, I.; Levanon-Cohen, S.; Bose, S.; Ligumsky, H.; Sredni, B.; Kanety, H.; Kuro-o, M.; Karlan, B.; Kaufman, B.; Koeffler, H.P.; et al. Klotho: A tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene 2008, 27, 7094–7105. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, A.; Gouge, J.; Kontovounisios, C.; Nikolaou, S.; Ashworth, A.; Lim, K.; Chong, I. Klotho and the Treatment of Human Malignancies. Cancers 2020, 12, 1665. [Google Scholar] [CrossRef]
- Ligumsky, H.; Merenbakh-Lamin, K.; Keren-Khadmy, N.; Wolf, I.; Rubinek, T. The role of αKlotho in human cancer: Molecular and clinical aspects. Oncogene 2022, 41, 4487–4497. [Google Scholar] [CrossRef] [PubMed]
- Gigante, M.; Lucarelli, G.; Divella, C.; Netti, G.S.; Pontrelli, P.; Cafiero, C.; Grandaliano, G.; Castellano, G.; Rutigliano, M.; Stallone, G.; et al. Soluble Serum αKlotho Is a Potential Predictive Marker of Disease Progression in Clear Cell Renal Cell Carcinoma. Medicine 2015, 94, e1917. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Wang, Y.; Fan, Z.; Ji, G.; Wang, M.; Lin, J.; Huang, S.; Meltzer, S.J. Klotho: A tumor suppressor and modulator of the Wnt/β-catenin pathway in human hepatocellular carcinoma. Lab. Investig. 2016, 96, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Fan, Z.; Wang, Y.; Ji, G.; Wang, M.; Lin, J.; Huang, S. Expression of klotho and β-catenin in esophageal squamous cell carcinoma, and their clinicopathological and prognostic significance. Dis. Esophagus 2016, 29, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Pako, J.; Bikov, A.; Barta, I.; Matsueda, H.; Puskas, R.; Galffy, G.; Kerpel-Fronius, A.; Antus, B.; Horvath, I. Assessment of the circulating klotho protein in lung cancer patients. Pathol. Oncol. Res. 2020, 26, 233–238. [Google Scholar] [CrossRef]
- Ashkan, K.; Baig Mirza, A.; Soumpasis, C.; Syrris, C.; Kalaitzoglou, D.; Sharma, C.; James, Z.J.; Khoja, A.K.; Ahmed, R.; Vastani, A.; et al. MGMT Promoter Methylation: Prognostication beyond Treatment Response. J. Pers. Med. 2023, 13, 999. [Google Scholar] [CrossRef]
- Bucova, M.; Kluckova, K.; Kozak, J.; Rychly, B.; Suchankova, M.; Svajdler, M.; Matejcik, V.; Steno, J.; Zsemlye, E.; Durmanova, V. HLA-G 14bp Ins/Del Polymorphism, Plasma Level of Soluble HLA-G, and Association with IL-6/IL-10 Ratio and Survival of Glioma Patients. Diagnostics 2022, 12, 1099. [Google Scholar] [CrossRef] [PubMed]
- Saaid, A.; Monticelli, M.; Ricci, A.A.; Orlando, G.; Botta, C.; Zeppa, P.; Bianconi, A.; Osella-Abate, S.; Bruno, F.; Pellerino, A.; et al. Prognostic Analysis of the IDH1 G105G (rs11554137) SNP in IDH-Wildtype Glioblastoma. Genes 2022, 13, 1439. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Liu, F.; Peng, Y.; Wang, P.; Ma, B.; Li, L.; Si, C.; Wang, X.; Zhang, M.; Song, F. Association of serum Klotho levels with cancer and cancer mortality: Evidence from National Health and Nutrition Examination Survey. Cancer Med. 2023, 12, 1922–1934. [Google Scholar] [CrossRef]
- Villa, C.; Miquel, C.; Mosses, D.; Bernier, M.; Di Stefano, A.L. The 2016 World Health Organization classification of tumours of the central nervous system. Presse Med. 2018, 47, e187–e200. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
| Patients | n | Mean Age ± SD |
| All gliomas | 55 | 54.38 ± 15.22 |
| Sex (male/female) | 32/23 | 49.63 ± 15.25/61.09 ± 12.55 |
| Grades (male/female) | ||
| G. II | 11/4 | 40.33 ± 13.54 |
| G. III | 7/4 | 50.44 ± 10.14 |
| G. IV | 14/15 | 64.14 ± 9.59 |
| Primary diagnosis | 42 | |
| Recidivism | 9 | |
| Not analyzed | 4 | |
| Survival time | (months) | |
| Overall | 25.3 ± 24.36 | |
| Grade II | 50.47 ± 24.40 | |
| Grade III | 32.45 ± 20.14 | |
| Grade IV | 9.57 ± 9.28 |
| SNP/ Model | Allele/ Genotype | Gliomas (n = 55) | Controls (n = 140) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|---|---|
| p | OR (95% CI) | p | OR (95% CI) | ||||
| rs1207568 | G | 91 (82.73%) | 244 (87.14%) | ||||
| -395G/A | A | 19 (17.27%) | 36 (12.86%) | 0.33 | 1.42 (0.77–2.59) | - | - |
| GG | 38 (69.09%) | 108 (77.14%) | 1.00 | 1.00 | |||
| Co-dominant | GA | 15 (27.27%) | 28 (20.00%) | 0.51 | 1.52 (0.74–3.15) | 0.80 | 1.33 (0.57–3.10) |
| AA | 2 (3.64%) | 4 (2.86%) | 1.42 (0.25–8.07 | 0.94 (0.12–7.60) | |||
| GG | 38 (69.09%) | 108 (77.14%) | 1.00 | 1.00 | |||
| Dominant | GA + AA | 17 (30.91%) | 32 (22.86%) | 0.25 | 1.51 (0.75–3.02) | 0.55 | 1.28 (0.57–2.86) |
| GG + GA | 53 (96.36%) | 136 (97.14%) | 1.00 | 1.00 | |||
| Recessive | AA | 2 (3.64%) | 4 (2.86%) | 0.78 | 1.28 (0.23–7.21) | 0.90 | 0.87 (0.11–7.07) |
| GG + AA | 40 (72.73%) | 112 (80.00%) | 1.00 | 1.00 | |||
| Over-dominant | GA | 15 (27.27%) | 28 (20.00%) | 0.28 | 1.50 (0.73–3.09) | 0.50 | 1.34 (0.58–3.09) |
| HWE (χ2/p) | 0.11/0.73 | 1.62/0.20 | |||||
| rs564481 | C | 61 (55.45%) | 179 (63.93%) | ||||
| 1818C/T | T | 49 (44.55%) | 101 (36.07%) | 0.15 | 1.42 (0.91–2.23) | - | - |
| CC | 15 (27.27%) | 56 (40.00%) | 1.00 | 1.00 | |||
| Co-dominant | CT | 31 (56.36%) | 67 (47.86%) | 0.23 | 1.73 (0.85–3.52) | 0.16 | 2.05 (0.91–4.61) |
| TT | 9 (16.36%) | 17 (12.14%) | 1.98 (0.74–5.31) | 2.19 (0.72–6.66) | |||
| CC | 15 (27.27%) | 56 (40.00%) | 1.00 | 1.00 | |||
| Dominant | CT + TT | 40 (72.73%) | 84 (60.00%) | 0.09 | 1.78 (0.90–3.52) | 0.058 | 2.08 (0.96–4.52) |
| CC + CT | 46 (83.64%) | 123 (87.86%) | 1.00 | 1.00 | |||
| Recessive | TT | 9 (16.36%) | 17 (12.14%) | 0.44 | 1.42 (0.59–3.40) | 0.49 | 1.42 (0.53–3.79) |
| CC + TT | 24 (43.63%) | 73 (52.14%) | 1.00 | 1.00 | |||
| Over-dominant | CT | 31 (56.36%) | 67 (47.86%) | 0.28 | 1.41 (0.75–2.64) | 0.19 | 1.61 (0.79–3.29) |
| HWE (χ2/p) | 1.09/0.29 | 0.19/0.66 | |||||
| SNP/ Model | Allele/ Genotype | Grade IV (n = 29) | Grade II + III (n = 26) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|---|---|
| p | OR (95% CI) | p | OR (95% CI) | ||||
| rs1207568 | G | 42 (72.41%) | 49 (94.23%) | ||||
| -395G/A | A | 16 (27.59%) | 3 (5.77%) | 0.006 | 6.22 (1.69–22.84) | - | - |
| GG | 15 (51.72%) | 23 (88.46%) | 1.00 | 1.00 | |||
| Co-dominant | GA | 12 (41.38%) | 3 (11.54%) | 0.0064 | 6.13 (1.48–25.44) | 0.029 | 10.90 (1.39–85.53) |
| AA | 2 (6.90%) | 0 (0.00%) | NA (0.00—NA) | NA (0.00—NA) | |||
| GG | 15 (51.72%) | 23 (88.46%) | 1.00 | 1.00 | |||
| Dominant | GA + AA | 14 (48.28%) | 3 (11.54%) | 0.0023 | 7.16 (1.75–29.20) | 0.0085 | 11.59 (1.51–89.19) |
| GG + GA | 27 (93.10%) | 26 (100.00%) | 1.00 | 1.00 | |||
| Recessive | AA | 2 (6.90%) | 0 (0.00%) | 0.10 | NA (0.00—NA) | 0.43 | NA (0.00—NA) |
| GG + AA | 17 (58.62%) | 23 (88.46%) | 1.00 | 1.00 | |||
| Over-dominant | GA | 12 (41.38%) | 3 (11.54%) | 0.011 | 5.41 (1.32–22.21) | 0.015 | 9.99 (1.25–79.81) |
| Parameter rs1207568 | G/G (n = 38) | G/A (n = 15) | A/A (n = 2) | p/p * CM | p/p * DM | p/p * RM | p/p * OD |
| Overall survival, months ± SD | 27.95 ± 25.80 | 18.53 ± 20.06 | 25.75 ± 28.64 | 0.46/0.31 | 0.23/0.12 | 0.98/0.65 | 0.21/0.16 |
| Parameter rs564481 | C/C (n = 15) | C/T (n = 31) | T/T (n = 9) | p/p * CM | p/p * DM | p/p * RM | p/p * OD |
| Overall survival, months ± SD | 25.17 ± 26.06 | 25.21 ± 24.50 | 25.83 ± 23.78 | 1.00/0.93 | 0.98/0.70 | 0.94/0.89 | 0.98/0.81 |
| Patients (n = 55) | Controls (n = 47) | p | |
|---|---|---|---|
| sαKL (pg/mL) (median (IQR)) | 672.16 (2583.88) | 1517.85 (13,591.67) | 0.0036 |
| Grade II (n = 15) (Median (IQR)) | Grade III (n = 11) (Median (IQR)) | Grade IV (n = 29) (Median (IQR)) | p (II vs. III) | p (II vs. IV) | p (III vs. IV) | |
|---|---|---|---|---|---|---|
| sαKL (pg/mL) | 997.2 (4449.1) | 363.32 (2580.55) | 598.1 (588.3) | 0.24 | 0.47 | 0.43 |
| Grade II (n = 15) | Grade III (n = 11) | Grade IV (n = 29) | Controls (n = 47) | p (II vs. ctr) | p (III vs. ctr) | p (IV vs. ctr) | |
|---|---|---|---|---|---|---|---|
| sαKL (pg/mL) (median (IQR)) | 997.2 (4449.1) | 363.32 (2580.55) | 598.1 (588.3) | 1517.85 (13,591.67) | 0.21 | 0.034 | 0.0083 |
| (a) | ||||
| Spearman r | 95% CI | p | ||
| sαKL (pg/mL) | 672.16 (2583.88) | 0.195 | −0.082–0.444 | 0.15 |
| Survival time (months) | 25.3 ± 24.36 | |||
| (b) | ||||
| Spearman r | 95% CI | p | ||
| sαKL (pg/mL) | 1238.44 (4741.26) | 0.211 | −0.352–0.662 | 0.45 |
| Survival time (months) | 64 (32.5) | |||
| (c) | ||||
| Spearman r | 95% CI | p | ||
| sαKL (pg/mL) | 589.75 (941.36) | −0.067 | −0.38–0.258 | 0.68 |
| Survival time (months) | 8.5 (16.5) | |||
| Molecules | Median (IQR) |
|---|---|
| VEGF (pg/mL) | 92.76 (409.1) |
| Fractalkine (pg/mL) | 582 (529.75) |
| sTREM-1 (pg/mL) | 33.2 (37.16) |
| IFN-γ (pg/mL) | 18.29 (112.12) |
| sHLA-G (U/mL) | 27.69 (39.95) |
| GDNF (pg/mL) | 46.27 (340.18) |
| IL-6 (pg/mL) | 3 (9.37) |
| IL-4 (pg/mL) | 216.6 (298.4) |
| IL-13 (pg/mL) | 31.46 (175.64) |
| Correlation of | Spearman r | 95% CI | p |
|---|---|---|---|
| sαKL with VEGF | 0.475 | 0.215–0.673 | 0.0008 |
| sαKL with fractalkine | 0.431 | 0.188–0.625 | 0.0009 |
| sαKL with sTREM-1 | −0.0247 | −0.2955–0.249 | 0.858 |
| sαKL with IFN-γ | 0.408 | 0.141–0.619 | 0.003 |
| sαKL with sHLA-G | 0.0436 | −0.249–0.329 | 0.766 |
| sαKL with GDNF | 0.338 | 0.032–0.585 | 0.0268 |
| sαKL with IL-6 | 0.302 | 0.015–0.543 | 0.0347 |
| sαKL with IL-4 | 0.424 | 0.140–0.644 | 0.0037 |
| sαKL with IL-13 | 0.509 | 0.245–0.703 | 0.0004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zsemlye, E.; Durmanova, V.; Kluckova, K.; Kozak, J.; Rychly, B.; Svajdler, M.; Matejcik, V.; Homolova, M.; Steno, J.; Hunakova, L.; et al. Association of Klotho Gene Polymorphism and Serum Level of α Klotho Protein with Different Tumor Grades, Overall Survival and Cytokine Profile in Glioma Patients. Int. J. Mol. Sci. 2025, 26, 330. https://doi.org/10.3390/ijms26010330
Zsemlye E, Durmanova V, Kluckova K, Kozak J, Rychly B, Svajdler M, Matejcik V, Homolova M, Steno J, Hunakova L, et al. Association of Klotho Gene Polymorphism and Serum Level of α Klotho Protein with Different Tumor Grades, Overall Survival and Cytokine Profile in Glioma Patients. International Journal of Molecular Sciences. 2025; 26(1):330. https://doi.org/10.3390/ijms26010330
Chicago/Turabian StyleZsemlye, Eszter, Vladimira Durmanova, Kristina Kluckova, Jan Kozak, Boris Rychly, Marian Svajdler, Viktor Matejcik, Monika Homolova, Juraj Steno, Luba Hunakova, and et al. 2025. "Association of Klotho Gene Polymorphism and Serum Level of α Klotho Protein with Different Tumor Grades, Overall Survival and Cytokine Profile in Glioma Patients" International Journal of Molecular Sciences 26, no. 1: 330. https://doi.org/10.3390/ijms26010330
APA StyleZsemlye, E., Durmanova, V., Kluckova, K., Kozak, J., Rychly, B., Svajdler, M., Matejcik, V., Homolova, M., Steno, J., Hunakova, L., & Bucova, M. (2025). Association of Klotho Gene Polymorphism and Serum Level of α Klotho Protein with Different Tumor Grades, Overall Survival and Cytokine Profile in Glioma Patients. International Journal of Molecular Sciences, 26(1), 330. https://doi.org/10.3390/ijms26010330






