Spinocerebellar Ataxia Type 3 Pathophysiology—Implications for Translational Research and Clinical Studies
Abstract
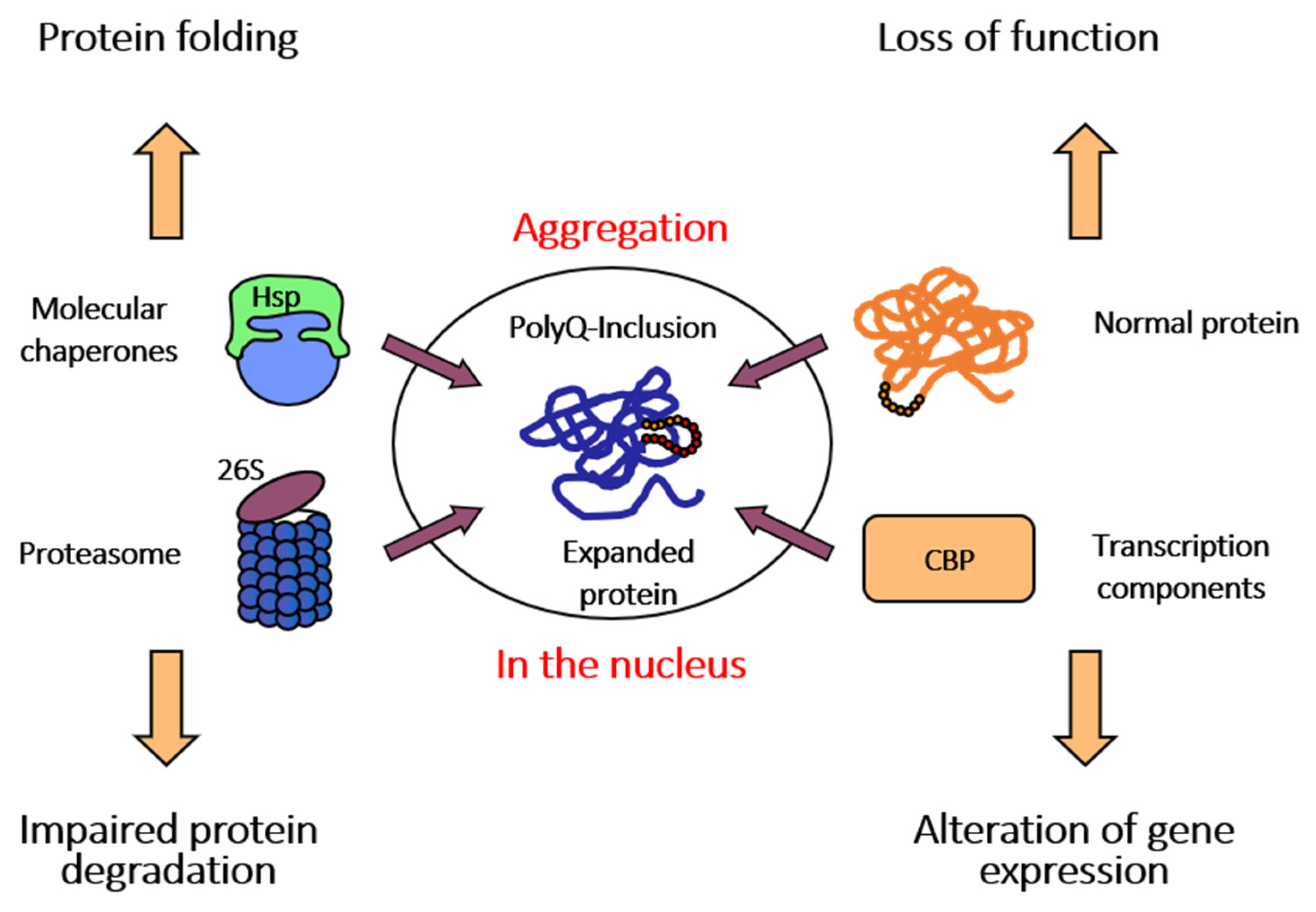
1. Spinocerebellar Ataxia Type 3
2. Ataxin-3 Gene
3. Transcriptional and Epigenetic Regulation
4. Posttranscriptional Regulation
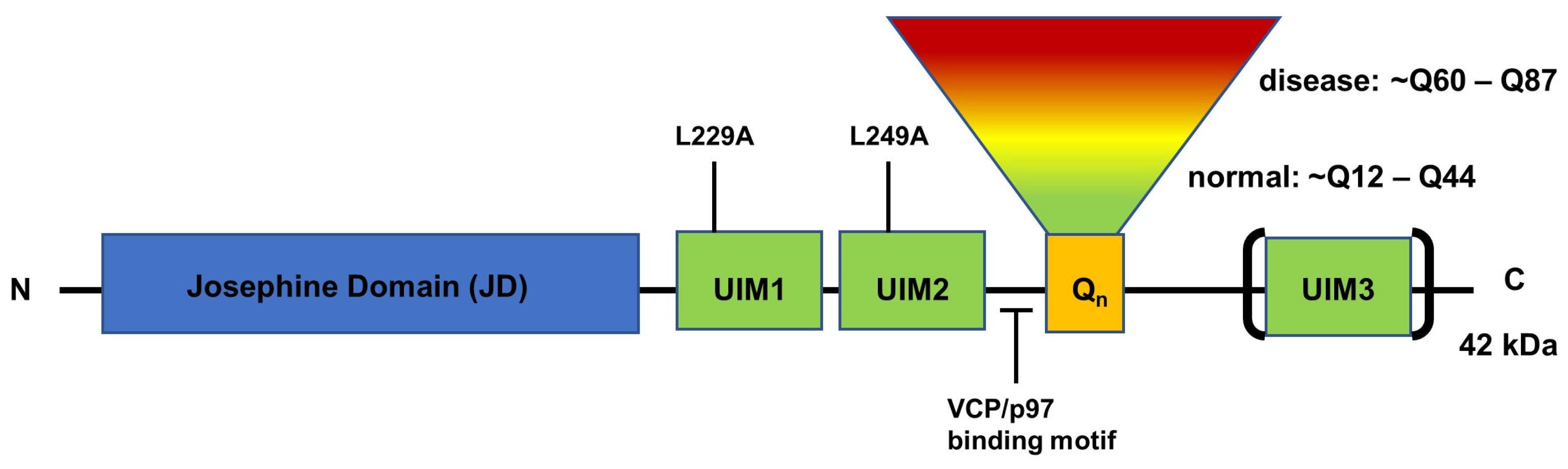
5. Structure and Function of ATXN3 Protein
6. Current Approaches for Treatment of SCA3
6.1. ASO- and miRNA-Based Approaches for Treatment of SCA3
6.2. CRISPR/CAS-Based Approaches
7. Current Clinical Trials for SCA3
7.1. BHV-4157 (Troriluzole)
7.2. SLS-005 (IV Trehalose)
7.3. ASO (VO659) and BIIB132
7.4. Stem Cell Therapy (MSC and UC-MSC Therapy)
8. New Targets for Treatments
8.1. Genetic Modifier of ATXN3 Expression
8.2. Compound Modifier of ATXN3 Expression
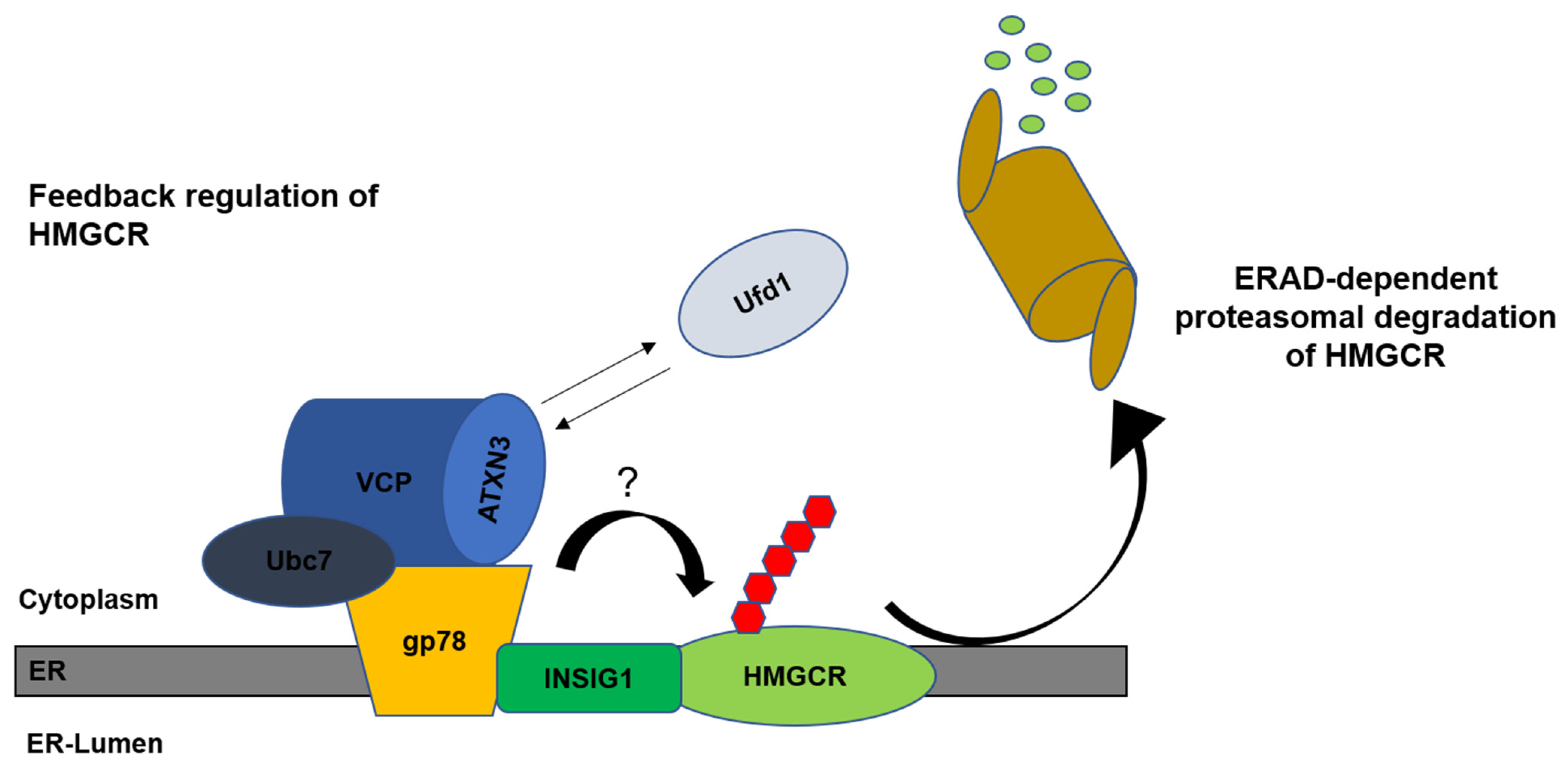
9. Cholesterol in SCA3
Funding
Conflicts of Interest
References
- Klockgether, T.; Mariotti, C.; Paulson, H.L. Spinocerebellar ataxia. Nat. Rev. Dis. Primers 2019, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; de Almeida, A.V.; Macedo-Ribeiro, S. Polyglutamine expansion diseases: More than simple repeats. J. Struct. Biol. 2018, 201, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Paulson, H.L.; Perez, M.K.; Trottier, Y.; Trojanowski, J.Q.; Subramony, S.H.; Das, S.S.; Vig, P.; Mandel, J.L.; Fischbeck, K.H.; Pittman, R.N. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 1997, 19, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Evert, B.O.; Wullner, U.; Schulz, J.B.; Weller, M.; Groscurth, P.; Trottier, Y.; Brice, A.; Klockgether, T. High level expression of expanded full-length ataxin-3 in vitro causes cell death and formation of intranuclear inclusions in neuronal cells. Hum. Mol. Genet. 1999, 8, 1169–1176. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kawaguchi, Y.; Okamoto, T.; Taniwaki, M.; Aizawa, M.; Inoue, M.; Katayama, S.; Kawakami, H.; Nakamura, S.; Nishimura, M.; Akiguchi, I.; et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994, 8, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, I.; Evert, B.O.; Khazneh, H.; Klockgether, T.; Wuellner, U. The human MJD gene: Genomic structure and functional characterization of the promoter region. Gene 2003, 314, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, C.; Santos, C.; Montiel, R.; Costa Mdo, C.; Cruz-Morales, P.; Santos, L.R.; Simoes, N.; Kay, T.; Vasconcelos, J.; Maciel, P.; et al. Increased transcript diversity: Novel splicing variants of Machado-Joseph disease gene (ATXN3). Neurogenetics 2010, 11, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Landwehrmeyer, G.B.; Schmitt, I.; Trottier, Y.; Auburger, G.; Laccone, F.; Klockgether, T.; Volpel, M.; Epplen, J.T.; Schols, L.; et al. An isoform of ataxin-3 accumulates in the nucleus of neuronal cells in affected brain regions of SCA3 patients. Brain Pathol. 1998, 8, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Peng, H.; Li, J.; Ding, D.; Chen, Z.; Long, Z.; Peng, Y.; Zhou, X.; Ye, W.; Li, K.; et al. Alteration of methylation status in the ATXN3 gene promoter region is linked to the SCA3/MJD. Neurobiol. Aging 2017, 53, 192.e5–192.e10. [Google Scholar] [CrossRef]
- Carmona, V.; Cunha-Santos, J.; Onofre, I.; Simoes, A.T.; Vijayakumar, U.; Davidson, B.L.; Pereira de Almeida, L. Unravelling Endogenous MicroRNA System Dysfunction as a New Pathophysiological Mechanism in Machado-Joseph Disease. Mol. Ther. 2017, 25, 1038–1055. [Google Scholar] [CrossRef]
- Krauss, S.; Nalavade, R.; Weber, S.; Carter, K.; Evert, B.O. Upregulation of miR-25 and miR-181 Family Members Correlates with Reduced Expression of ATXN3 in Lymphocytes from SCA3 Patients. Microrna 2019, 8, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Krauss, S.; Evert, B.O. The Role of MicroRNAs in Spinocerebellar Ataxia Type 3. J. Mol. Biol. 2019, 431, 1729–1742. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, H.S.; Moore, L.R.; Paulson, H.L. Pathogenesis of SCA3 and implications for other polyglutamine diseases. Neurobiol. Dis. 2020, 134, 104635. [Google Scholar] [CrossRef] [PubMed]
- Boeddrich, A.; Gaumer, S.; Haacke, A.; Tzvetkov, N.; Albrecht, M.; Evert, B.O.; Muller, E.C.; Lurz, R.; Breuer, P.; Schugardt, N.; et al. An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J. 2006, 25, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.V.; Williams, D.R.; Cocklin, S.; Loll, P.J. Interaction between the AAA(+) ATPase p97 and its cofactor ataxin3 in health and disease: Nucleotide-induced conformational changes regulate cofactor binding. J. Biol. Chem. 2017, 292, 18392–18407. [Google Scholar] [CrossRef] [PubMed]
- Sicorello, A.; Rozycki, B.; Konarev, P.V.; Svergun, D.I.; Pastore, A. Capturing the Conformational Ensemble of the Mixed Folded Polyglutamine Protein Ataxin-3. Structure 2021, 29, 70–81.e5. [Google Scholar] [CrossRef] [PubMed]
- Sicorello, A.; Kelly, G.; Oregioni, A.; Novacek, J.; Sklenar, V.; Pastore, A. The Structural Properties in Solution of the Intrinsically Mixed Folded Protein Ataxin-3. Biophys. J. 2018, 115, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Costa Mdo, C.; Paulson, H.L. Toward understanding Machado-Joseph disease. Prog. Neurobiol. 2012, 97, 239–257. [Google Scholar] [CrossRef]
- Trottier, Y.; Cancel, G.; An-Gourfinkel, I.; Lutz, Y.; Weber, C.; Brice, A.; Hirsch, E.; Mandel, J.L. Heterogeneous intracellular localization and expression of ataxin-3. Neurobiol. Dis. 1998, 5, 335–347. [Google Scholar] [CrossRef]
- Luo, H.; Todi, S.V.; Paulson, H.L.; Costa, M.D.C. Regional and age-dependent changes in ubiquitination in cellular and mouse models of spinocerebellar ataxia type 3. Front. Mol. Neurosci. 2023, 16, 1154203. [Google Scholar] [CrossRef]
- Schmitt, I.; Linden, M.; Khazneh, H.; Evert, B.O.; Breuer, P.; Klockgether, T.; Wuellner, U. Inactivation of the mouse Atxn3 (ataxin-3) gene increases protein ubiquitination. Biochem. Biophys. Res. Commun. 2007, 362, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.J.; Coppola, G.; Santos, C.; Costa Mdo, C.; Ailion, M.; Sequeiros, J.; Geschwind, D.H.; Maciel, P. Functional genomics and biochemical characterization of the C. elegans orthologue of the Machado-Joseph disease protein ataxin-3. FASEB J. 2007, 21, 1126–1136. [Google Scholar] [CrossRef]
- Scheel, H.; Tomiuk, S.; Hofmann, K. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum. Mol. Genet. 2003, 12, 2845–2852. [Google Scholar] [CrossRef] [PubMed]
- Burnett, B.; Li, F.; Pittman, R.N. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 2003, 12, 3195–3205. [Google Scholar] [CrossRef]
- Burnett, B.G.; Pittman, R.N. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc. Natl. Acad. Sci. USA 2005, 102, 4330–4335. [Google Scholar] [CrossRef]
- Maya, V.R.; Kimberly, C.G.; Prajakta, D.M.; Patrick, J.L. Ubiquitin Substrates Dramatically Increase Ataxin3 Deubiquitinating Activity: Allosteric crosstalk connects three distinct sites. bioRxiv, 2020; bioRxiv:2020.05.05.078998. [Google Scholar] [CrossRef]
- Durcan, T.M.; Kontogiannea, M.; Thorarinsdottir, T.; Fallon, L.; Williams, A.J.; Djarmati, A.; Fantaneanu, T.; Paulson, H.L.; Fon, E.A. The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum. Mol. Genet. 2011, 20, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Jana, N.R.; Dikshit, P.; Goswami, A.; Kotliarova, S.; Murata, S.; Tanaka, K.; Nukina, N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 2005, 280, 11635–11640. [Google Scholar] [CrossRef]
- Connell, P.; Ballinger, C.A.; Jiang, J.; Wu, Y.; Thompson, L.J.; Hohfeld, J.; Patterson, C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001, 3, 93–96. [Google Scholar] [CrossRef]
- Ballinger, C.A.; Connell, P.; Wu, Y.; Hu, Z.; Thompson, L.J.; Yin, L.Y.; Patterson, C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell Biol. 1999, 19, 4535–4545. [Google Scholar] [CrossRef]
- Scaglione, K.M.; Zavodszky, E.; Todi, S.V.; Patury, S.; Xu, P.; Rodriguez-Lebron, E.; Fischer, S.; Konen, J.; Djarmati, A.; Peng, J.; et al. Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell 2011, 43, 599–612. [Google Scholar] [CrossRef]
- Doss-Pepe, E.W.; Stenroos, E.S.; Johnson, W.G.; Madura, K. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol. Cell Biol. 2003, 23, 6469–6483. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Pittman, R.N. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 2006, 15, 2409–2420. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.L.; Libohova, K.; Blount, J.R.; Sujkowski, A.L.; Prifti, M.V.; Tsou, W.L.; Todi, S.V. Targeting the VCP-binding motif of ataxin-3 improves phenotypes in Drosophila models of Spinocerebellar Ataxia Type 3. Neurobiol. Dis. 2021, 160, 105516. [Google Scholar] [CrossRef] [PubMed]
- Ristic, G.; Sutton, J.R.; Libohova, K.; Todi, S.V. Toxicity and aggregation of the polyglutamine disease protein, ataxin-3 is regulated by its binding to VCP/p97 in Drosophila melanogaster. Neurobiol. Dis. 2018, 116, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Johnston, H.E.; Samant, R.S. Alternative systems for misfolded protein clearance: Life beyond the proteasome. FEBS J. 2021, 288, 4464–4487. [Google Scholar] [CrossRef] [PubMed]
- Vijiaratnam, N.; Simuni, T.; Bandmann, O.; Morris, H.R.; Foltynie, T. Progress towards therapies for disease modification in Parkinson’s disease. Lancet Neurol. 2021, 20, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R.; Yau, W.Y.; O’Connor, E.; Houlden, H. Spinocerebellar ataxia: An update. J. Neurol. 2019, 266, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Kingwell, K. Double setback for ASO trials in Huntington disease. Nat. Rev. Drug Discov. 2021, 20, 412–413. [Google Scholar] [CrossRef]
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.E.; Lang, A.E. Parkinson disease. Nat. Rev. Dis. Primers 2017, 3, 17013. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Roy, S. Gene-based therapies for neurodegenerative diseases. Nat. Neurosci. 2021, 24, 297–311. [Google Scholar] [CrossRef]
- Kantor, B.; Tagliafierro, L.; Gu, J.; Zamora, M.E.; Ilich, E.; Grenier, C.; Huang, Z.Y.; Murphy, S.; Chiba-Falek, O. Downregulation of SNCA Expression by Targeted Editing of DNA Methylation: A Potential Strategy for Precision Therapy in PD. Mol. Ther. 2018, 26, 2638–2649. [Google Scholar] [CrossRef] [PubMed]
- Hauser, S.; Helm, J.; Kraft, M.; Korneck, M.; Hubener-Schmid, J.; Schols, L. Allele-specific targeting of mutant ataxin-3 by antisense oligonucleotides in SCA3-iPSC-derived neurons. Mol. Ther. Nucleic Acids 2022, 27, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Martier, R.; Sogorb-Gonzalez, M.; Stricker-Shaver, J.; Hubener-Schmid, J.; Keskin, S.; Klima, J.; Toonen, L.J.; Juhas, S.; Juhasova, J.; Ellederova, Z.; et al. Development of an AAV-Based MicroRNA Gene Therapy to Treat Machado-Joseph Disease. Mol. Ther. Methods Clin. Dev. 2019, 15, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Hu, S.; Wu, Z.; He, M. Therapeutic effects of engineered exosome-based miR-25 and miR-181a treatment in spinocerebellar ataxia type 3 mice by silencing ATXN3. Mol. Med. 2023, 29, 96. [Google Scholar] [CrossRef]
- McLoughlin, H.S.; Gundry, K.; Rainwater, O.; Schuster, K.H.; Wellik, I.G.; Zalon, A.J.; Benneyworth, M.A.; Eberly, L.E.; Oz, G. Antisense Oligonucleotide Silencing Reverses Abnormal Neurochemistry in Spinocerebellar Ataxia 3 Mice. Ann. Neurol. 2023, 94, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wang, S.; Peng, L.; Zhao, H.; Li, S.; Han, X.; Habimana, J.D.; Chen, Z.; Wang, C.; Peng, Y.; et al. CRISPR/Cas9 mediated gene correction ameliorates abnormal phenotypes in spinocerebellar ataxia type 3 patient-derived induced pluripotent stem cells. Transl. Psychiatry 2021, 11, 479. [Google Scholar] [CrossRef]
- Li, T.; Yang, Y.; Qi, H.; Cui, W.; Zhang, L.; Fu, X.; He, X.; Liu, M.; Li, P.F.; Yu, T. CRISPR/Cas9 therapeutics: Progress and prospects. Signal Transduct. Target Ther. 2023, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Abdallah, M.O.E.; Breuer, P.; Stahl, F.; Bakhit, Y.; Potthoff, A.L.; Pregler, B.E.F.; Schneider, M.; Waha, A.; Wullner, U.; et al. Downregulation of MGMT expression by targeted editing of DNA methylation enhances temozolomide sensitivity in glioblastoma. Neoplasia 2023, 44, 100929. [Google Scholar] [CrossRef]
- Zaltzman, R.; Elyoseph, Z.; Lev, N.; Gordon, C.R. Trehalose in Machado-Joseph Disease: Safety, Tolerability, and Efficacy. Cerebellum 2020, 19, 672–679. [Google Scholar] [CrossRef]
- Vo, S.H.; Butzlaff, M.; Pru, S.K.; Ni Charthaigh, R.A.; Karsten, P.; Lankes, A.; Hamm, S.; Simons, M.; Adryan, B.; Schulz, J.B.; et al. Large-scale screen for modifiers of ataxin-3-derived polyglutamine-induced toxicity in Drosophila. PLoS ONE 2012, 7, e47452. [Google Scholar] [CrossRef]
- Fardghassemi, Y.; Maios, C.; Parker, J.A. Small Molecule Rescue of ATXN3 Toxicity in C. elegans via TFEB/HLH-30. Neurotherapeutics 2021, 18, 1151–1165. [Google Scholar] [CrossRef]
- Costa, M.D.C.; Ashraf, N.S.; Fischer, S.; Yang, Y.; Schapka, E.; Joshi, G.; McQuade, T.J.; Dharia, R.M.; Dulchavsky, M.; Ouyang, M.; et al. Unbiased screen identifies aripiprazole as a modulator of abundance of the polyglutamine disease protein, ataxin-3. Brain 2016, 139, 2891–2908. [Google Scholar] [CrossRef]
- Ashraf, N.S.; Sutton, J.R.; Yang, Y.; Ranxhi, B.; Libohova, K.; Shaw, E.D.; Barget, A.J.; Todi, S.V.; Paulson, H.L.; Costa, M.D.C. Druggable genome screen identifies new regulators of the abundance and toxicity of ATXN3, the Spinocerebellar Ataxia type 3 disease protein. Neurobiol. Dis. 2020, 137, 104697. [Google Scholar] [CrossRef]
- Naveed, M.; Ali, N.; Aziz, T.; Hanif, N.; Fatima, M.; Ali, I.; Alharbi, M.; Alasmari, A.F.; Albekairi, T.H. The natural breakthrough: Phytochemicals as potent therapeutic agents against spinocerebellar ataxia type 3. Sci. Rep. 2024, 14, 1529. [Google Scholar] [CrossRef]
- Fardghassemi, Y.; Parker, J.A. Overexpression of FKH-2/FOXG1 is neuroprotective in a C. elegans model of Machado-Joseph disease. Exp. Neurol. 2021, 337, 113544. [Google Scholar] [CrossRef]
- Stahl, F.; Schmitt, I.; Denner, P.; de Boni, L.; Wullner, U.; Breuer, P. High throughput compound screening in neuronal cells identifies statins as activators of ataxin 3 expression. Sci. Rep. 2023, 13, 14911. [Google Scholar] [CrossRef]
- Endo, A.; Kuroda, M.; Tanzawa, K. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett. 1976, 72, 323–326. [Google Scholar] [CrossRef]
- Brown, M.S.; Radhakrishnan, A.; Goldstein, J.L. Retrospective on Cholesterol Homeostasis: The Central Role of Scap. Annu. Rev. Biochem. 2018, 87, 783–807. [Google Scholar] [CrossRef]
- Toonen, L.J.A.; Overzier, M.; Evers, M.M.; Leon, L.G.; van der Zeeuw, S.A.J.; Mei, H.; Kielbasa, S.M.; Goeman, J.J.; Hettne, K.M.; Magnusson, O.T.; et al. Transcriptional profiling and biomarker identification reveal tissue specific effects of expanded ataxin-3 in a spinocerebellar ataxia type 3 mouse model. Mol. Neurodegener. 2018, 13, 31. [Google Scholar] [CrossRef]
- Nobrega, C.; Mendonca, L.; Marcelo, A.; Lamaziere, A.; Tome, S.; Despres, G.; Matos, C.A.; Mechmet, F.; Langui, D.; den Dunnen, W.; et al. Restoring brain cholesterol turnover improves autophagy and has therapeutic potential in mouse models of spinocerebellar ataxia. Acta Neuropathol. 2019, 138, 837–858. [Google Scholar] [CrossRef] [PubMed]
- Pikuleva, I.A.; Cartier, N. Cholesterol Hydroxylating Cytochrome P450 46A1: From Mechanisms of Action to Clinical Applications. Front. Aging Neurosci. 2021, 13, 696778. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.M.; DeBose-Boyd, R.A. Posttranslational Regulation of HMG CoA Reductase, the Rate-Limiting Enzyme in Synthesis of Cholesterol. Annu. Rev. Biochem. 2021, 90, 659–679. [Google Scholar] [CrossRef]
- DeBose-Boyd, R.A. Feedback regulation of cholesterol synthesis: Sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008, 18, 609–621. [Google Scholar] [CrossRef] [PubMed]
| Scheme | Readout | Compound/Gene | Action | Organism | Author, Year |
|---|---|---|---|---|---|
| Druggable genome screen | Overexpression induced toxicity | FBXL3 (and others) | SKP1–Cullin–F-box (SCF) ubiquitin ligase complex Overexpression FBXL3 reduced ATXN3 level | HEK 293 reporter cells Drosophila | Ashraf et al., 2020 [55] |
| RNAi screen | Overexpression induced toxicity | Several genes | Involved in protein turnover/quality control; nuclear import/export and mRNA transport, editing, translation | Drosophila | Vossfeldt et al., 2012 [52] |
| RNAi TF screen | Overexpression induced toxicity | FKH-2/FOXG1 | Ortholog of human forkhead box G1 (FOXG1), associated rare disorder with impaired development and structural brain abnormalities Reduced overexpression-induced phenotypes | C. elegans | Fardghassemi and Parker 2021 [57] |
| Drug screen | Overexpression induced toxicity | Aripiprazole | Increased longevity in flies, reduction in aggregated ATXN3 species in flies and mice brains | HEK 293 reporter cells Drosophila Mice | Costa et al., 2016 [54] |
| Drug screen | Overexpression induced toxicity | Chenodiol, fenbufen, and sulfaphenazole | Modulators of TFEB/HLH-30 TF Reduced overexpression-induced phenotypes | C. elegans | Fardghassemi et al., 2021 [53] |
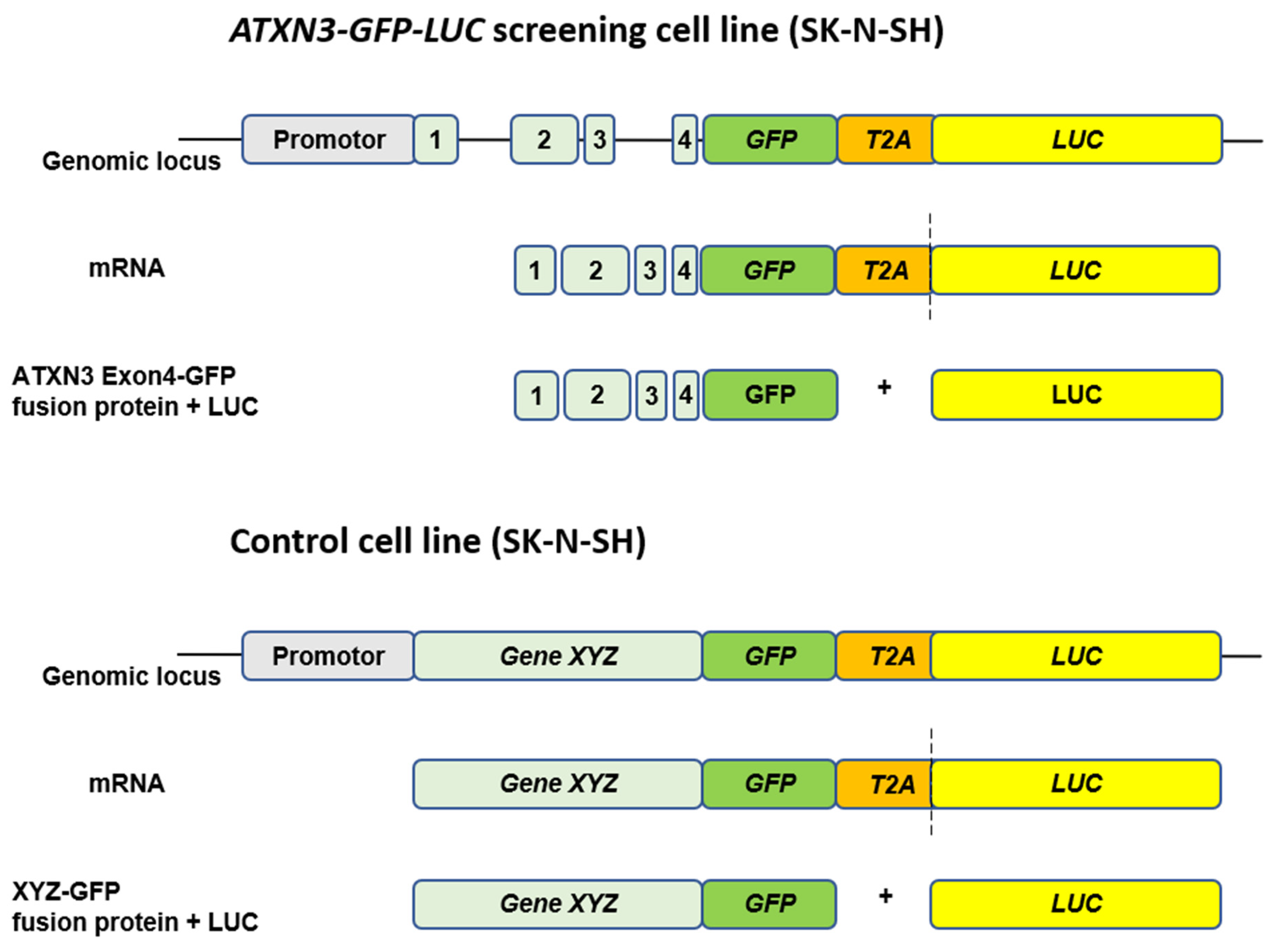
| Drug screen | Endogenous expression of ATXN3 LUC signal (primary screen) | Simvastatin | Activator of ATXN3 expression | Endogenous SK-N-SH ATXN3-GFP-LUC fusion reporter cell line, SK-N-SH wild type and SCA3 patient-derived NSC | Stahl et al., 2023 [58] |
| In-silico modelling | ------ | Flavonoid chamanetin | Highly active against ATXN3 | In-vitro/vivo studies are pending | Naveed et al., 2024 [56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stahl, F.; Evert, B.O.; Han, X.; Breuer, P.; Wüllner, U. Spinocerebellar Ataxia Type 3 Pathophysiology—Implications for Translational Research and Clinical Studies. Int. J. Mol. Sci. 2024, 25, 3984. https://doi.org/10.3390/ijms25073984
Stahl F, Evert BO, Han X, Breuer P, Wüllner U. Spinocerebellar Ataxia Type 3 Pathophysiology—Implications for Translational Research and Clinical Studies. International Journal of Molecular Sciences. 2024; 25(7):3984. https://doi.org/10.3390/ijms25073984
Chicago/Turabian StyleStahl, Fabian, Bernd O. Evert, Xinyu Han, Peter Breuer, and Ullrich Wüllner. 2024. "Spinocerebellar Ataxia Type 3 Pathophysiology—Implications for Translational Research and Clinical Studies" International Journal of Molecular Sciences 25, no. 7: 3984. https://doi.org/10.3390/ijms25073984
APA StyleStahl, F., Evert, B. O., Han, X., Breuer, P., & Wüllner, U. (2024). Spinocerebellar Ataxia Type 3 Pathophysiology—Implications for Translational Research and Clinical Studies. International Journal of Molecular Sciences, 25(7), 3984. https://doi.org/10.3390/ijms25073984






