Gene Expression Profiling Reveals Fundamental Sex-Specific Differences in SIRT3-Mediated Redox and Metabolic Signaling in Mouse Embryonic Fibroblasts
Abstract
1. Introduction
2. Results
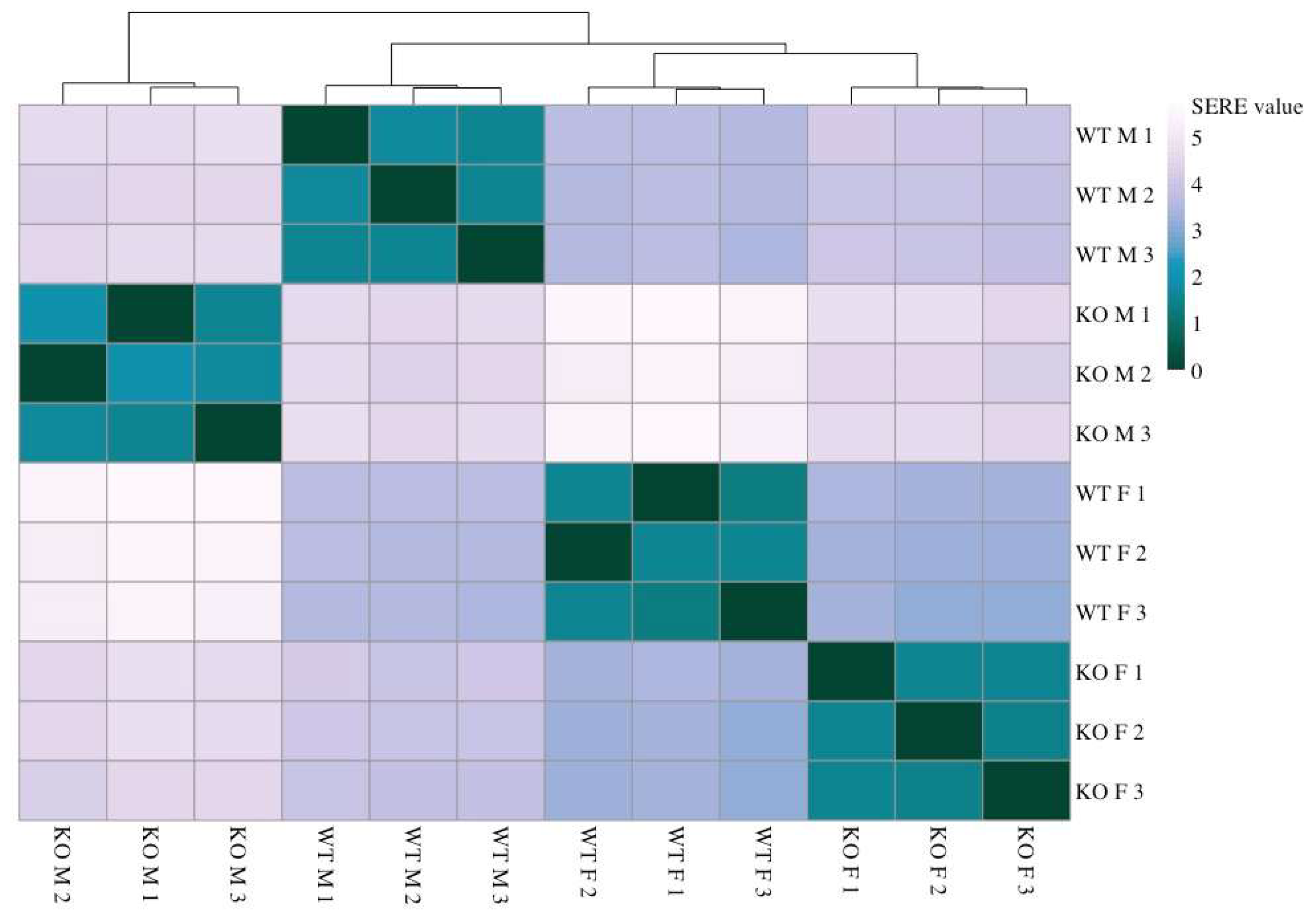
2.1. Sirt-3-Dependent Changes in Gene Expression
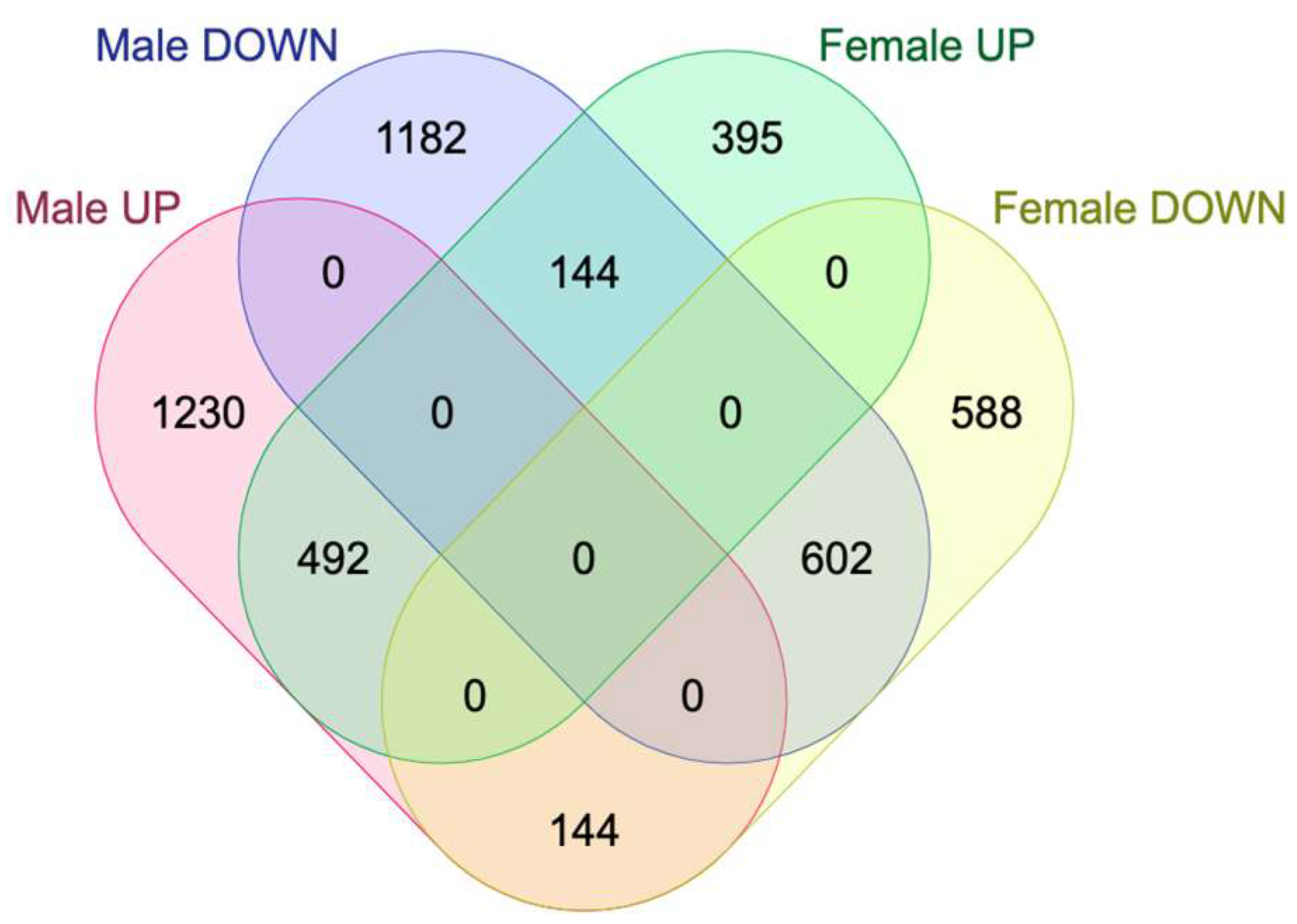
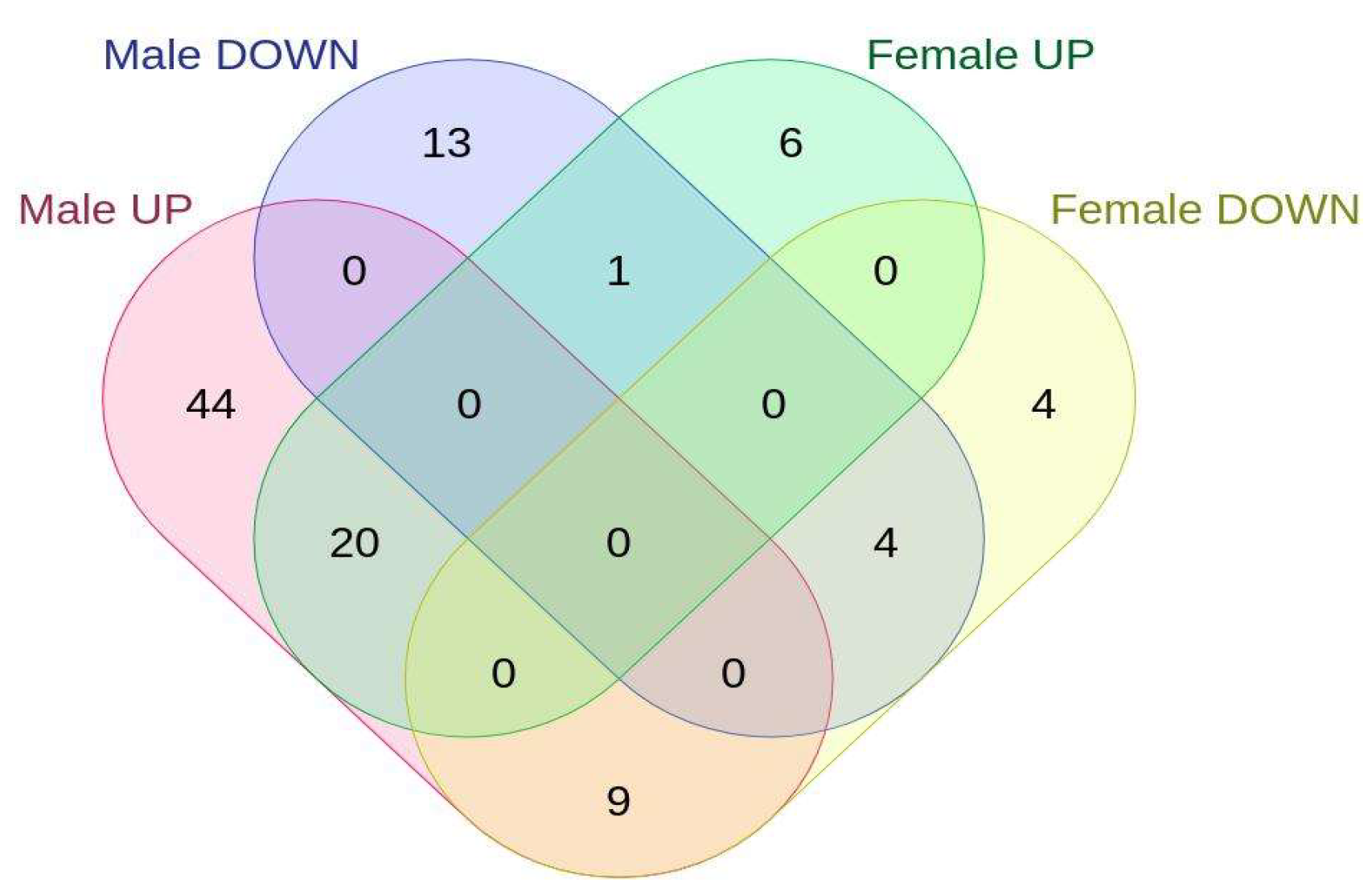
2.2. Sex-Specific Changes in Gene Expression
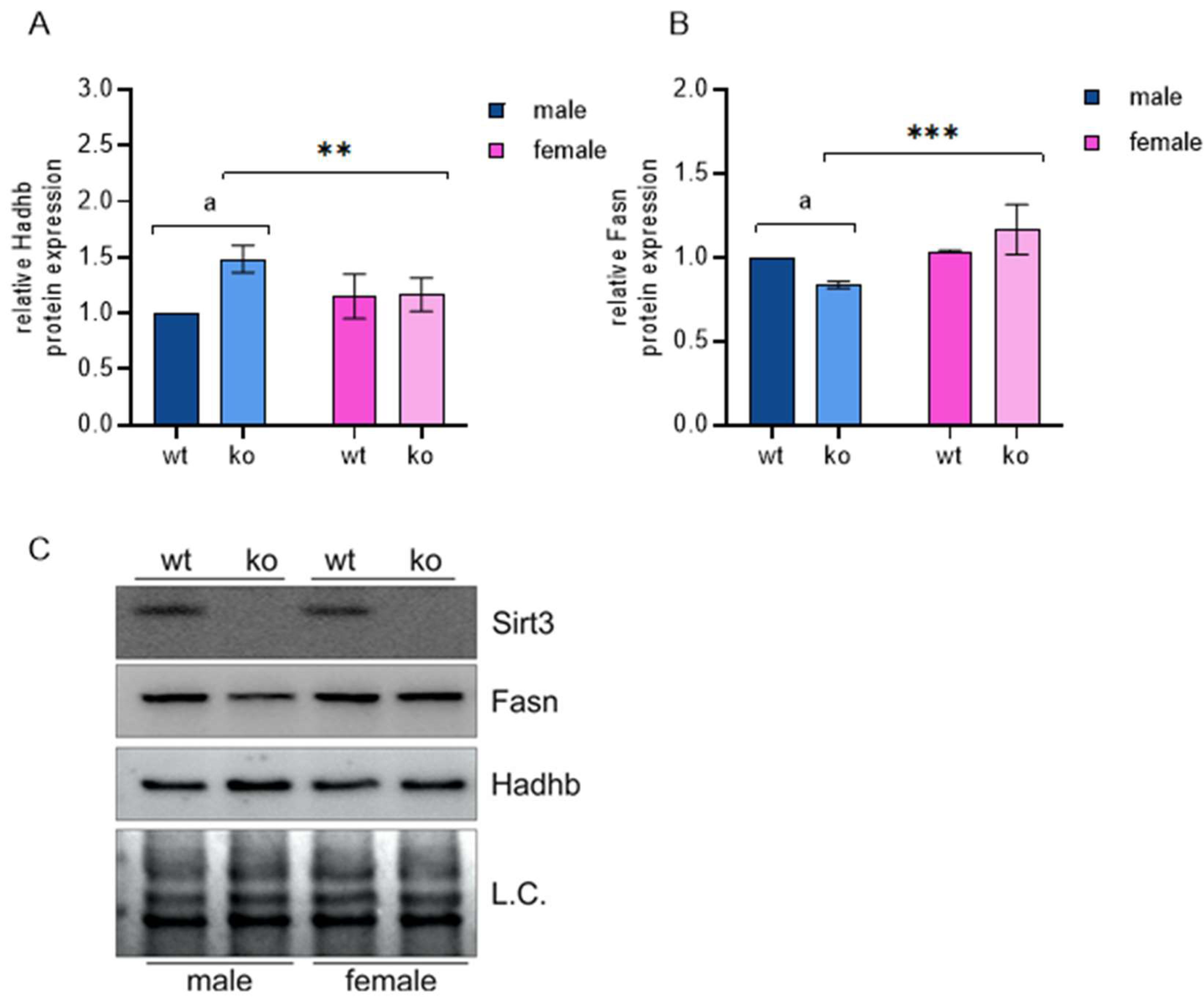
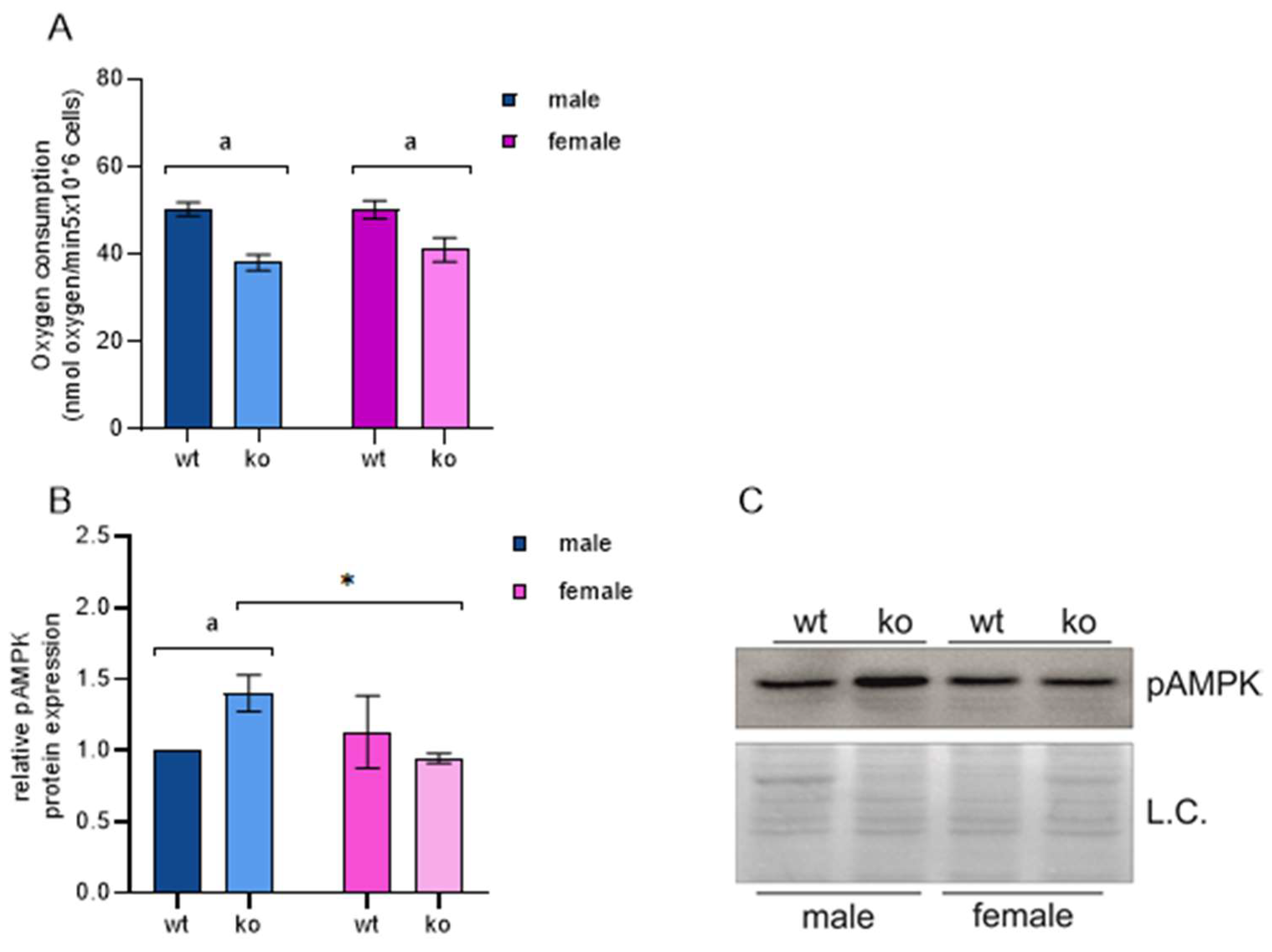
2.3. OXPHOS and MEF Energy Status
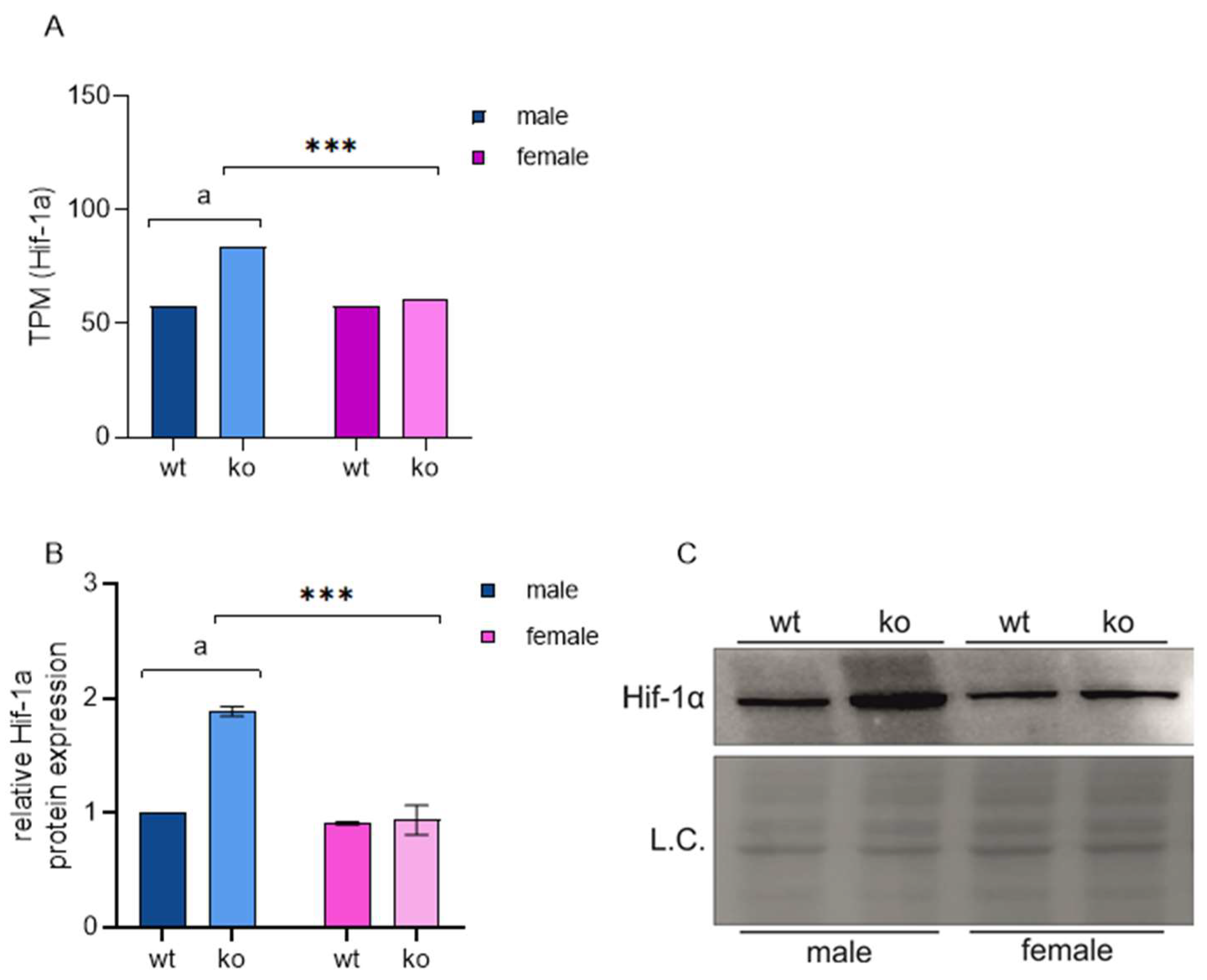
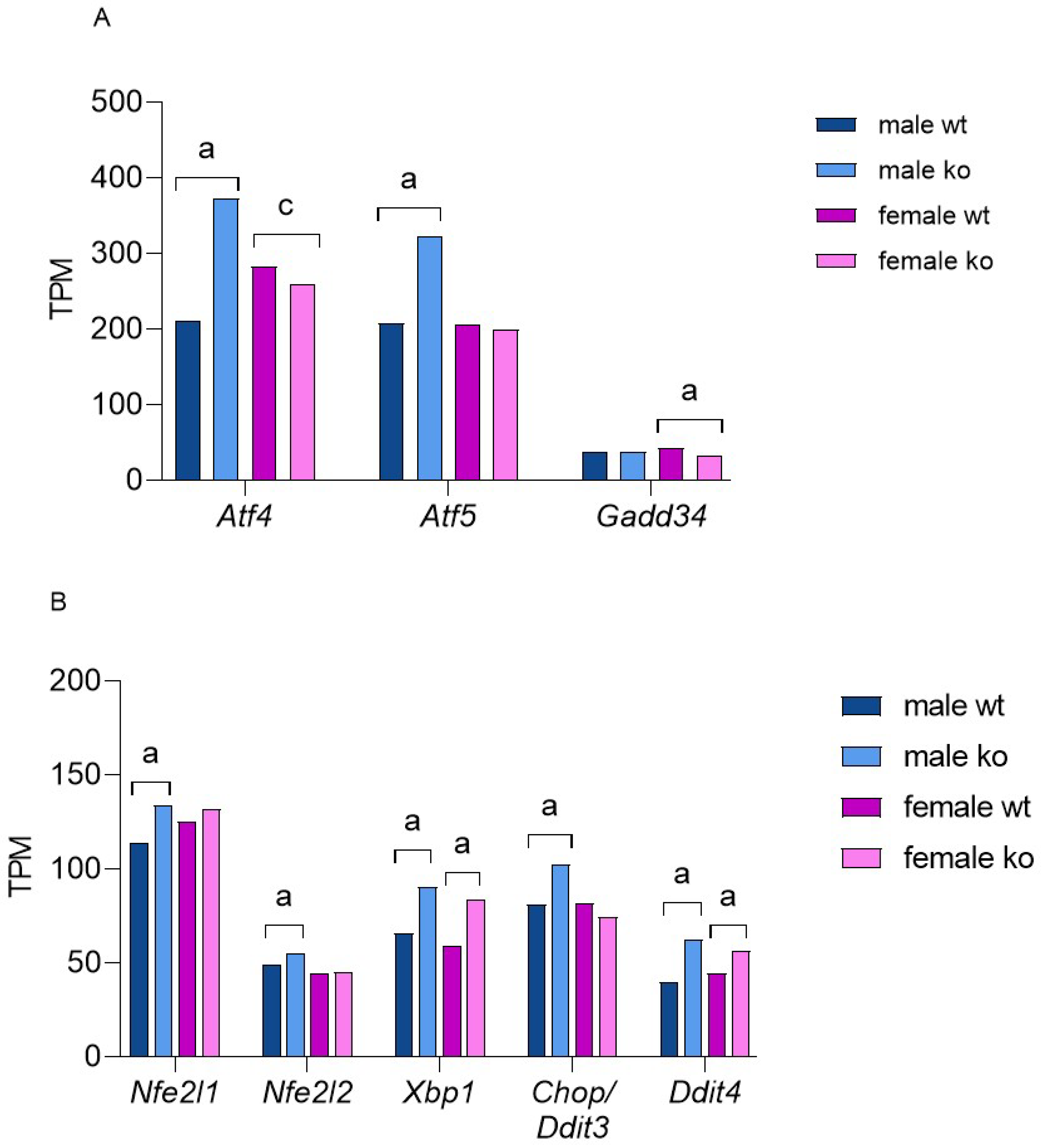
2.4. Sirt-3 Loss Leads to Male-Restricted, Atf-4-Mediated ISR
2.5. Sirt-3 KO Induces UPRER Independent of Sex
2.6. Loss of Sirt-3 Activates the UPRmt Stress Response in Male MEF
2.7. Male-Restricted ISR Induction as a Consequence of Oxidative Stress
3. Discussion
Perspectives & Significance
4. Materials and Methods
4.1. Cell Lines
4.2. RNA Sequencing
4.3. Western Blot Analysis
4.4. Oxygen Consumption
4.5. RNA-seq Data Analysis
4.6. Statistical Analysis
5. Conclusions
- Male and female MEFs mount significantly different adaptive metabolic responses to loss of Sirt3 function and the resulting mitochondrial dysfunction;
- Female MEFs compensate for Sirt-3 loss, at least in part, through increased antioxidant enzyme expression; in male cells, Sirt3 knock-out induces pseudohypoxia resulting in chronic oxidative stress, shift to glycolysis and fatty acid oxidation;
- In male MEFs, loss of Sirt3 increases Hif-1a and induces an ATF4-mediated integrated stress response;
- In the MEF (hormone-independent) experimental model, female cells exhibit a higher level of protection from oxidative stress induced by impaired mitochondrial function, maintaining the normal metabolic and oxidative states;
- Through maintenance of mitochondrial function, Sirt3 is involved in aging and neurodegenerative and cardiovascular diseases; significant differences between sexes should be accounted for in future research.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ngo, S.T.; Steyn, F.J.; McCombe, P.A. Gender differences in autoimmune disease. Front. Neuroendocrinol. 2014, 35, 347–369. [Google Scholar] [CrossRef] [PubMed]
- Merz, A.A.; Cheng, S. Sex differences in cardiovascular ageing. Heart 2016, 102, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.B.; Lagas, J.S.; Broestl, L.; Sponagel, J.; Rockwell, N.; Rhee, G.; Rosen, S.F.; Chen, S.; Klein, R.S.; Imoukhuede, P.; et al. Sex differences in cancer mechanisms. Biol. Sex Differ. 2020, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Zarulli, V.; Barthold Jones, J.A.; Oksuzyan, A.; Lindahl-Jacobsen, R.; Christensen, K.; Vaupel, J.W. Women live longer than men even during severe famines and epidemics. Proc. Natl. Acad. Sci. USA 2018, 115, E832–E840. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F. Sex differences in metabolic homeostasis, diabetes, and obesity. Biol. Sex Differ. 2015, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Schiebinger, L.; Stefanick, M.L.; Cahill, L.; Danska, J.; de Vries, G.J.; Kibbe, M.R.; McCarthy, M.M.; Mogil, J.S.; Woodruff, T.K.; et al. Opinion: Sex inclusion in basic research drives discovery. Proc. Natl. Acad. Sci. USA 2015, 112, 5257–5258. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, B.J.; Onishi, K.G.; Zucker, I. Female mice liberated for inclusion in neuroscience and biomedical research. Neurosci. Biobehav. Rev. 2014, 40, 1–5. [Google Scholar] [CrossRef]
- Itoh, Y.; Arnold, A.P. Are females more variable than males in gene expression? Meta-analysis of microarray datasets. Biol. Sex Differ. 2015, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Schwer, B.; North, B.J.; Frye, R.A.; Ott, M.; Verdin, E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide–dependent deacetylase. J. Cell Biol. 2002, 158, 647–657. [Google Scholar] [CrossRef]
- Ansari, A.; Rahman, M.S.; Saha, S.K.; Saikot, F.K.; Deep, A.; Kim, K.H. Function of the SIRT3 mitochondrial deacetylase in cellular physiology, cancer, and neurodegenerative disease. Aging Cell 2016, 16, 4–16. [Google Scholar] [CrossRef]
- Giralt, A.; Villarroya, F. SIRT3, a pivotal actor in mitochondrial functions: Metabolism, cell death and aging. Biochem. J. 2012, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pinterić, M.; Podgorski, I.I.; Hadžija, M.P.; Bujak, I.T.; Dekanić, A.; Bagarić, R.; Farkaš, V.; Sobočanec, S.; Balog, T. Role of sirt3 in differential sex-related responses to a high-fat diet in mice. Antioxidants 2020, 9, 174. [Google Scholar] [CrossRef] [PubMed]
- Pinterić, M.; Podgorski, I.I.; Popović Hadžija, M.; Tartaro Bujak, I.; Tadijan, A.; Balog, T.; Sobočanec, S. Chronic high fat diet intake impairs hepatic metabolic parameters in ovariectomized sirt3 ko mice. Int. J. Mol. Sci. 2021, 22, 4277. [Google Scholar] [CrossRef] [PubMed]
- Lombard, D.B.; Alt, F.W.; Cheng, H.L.; Bunkenborg, J.; Streeper, R.S.; Mostoslavsky, R.; Schwer, B. Mammalian Sir2 Homolog SIRT3 Regulates Global Mitochondrial. Mol. Cell Biol. 2007, 27, 8807–8814. [Google Scholar] [CrossRef] [PubMed]
- Barger, J.L.; Anderson, R.M.; Newton, M.A.; da Silva, C.; Vann, J.A.; Pugh, T.D.; Someya, S.; Prolla, T.A.; Weindruch, R. A conserved transcriptional signature of delayed aging and reduced disease vulnerability is partially mediated by SIRT3. PLoS ONE 2015, 10, e0120738. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chiang, Y.L.; Lyssiotis, C.A.; Teater, M.R.; Hong, J.Y.; Shen, H.; Melnick, A.M. Non-oncogene Addiction to SIRT3 Plays a Critical Role in Lymphomagenesis. Cancer Cell 2019, 35, 916–931.e9. [Google Scholar] [CrossRef]
- Zhu, S.; Donovan, E.L.; Makosa, D.; Mehta-D’souza, P.; Jopkiewicz, A.; Batushansky, A.; Cortassa, D.; Simmons, A.D.; Lopes, E.B.P.; Kinter, M.; et al. Sirt3 Promotes Chondrogenesis, Chondrocyte Mitochondrial Respiration and the Development of High-Fat Diet-Induced Osteoarthritis in Mice. J. Bone Min. Res. Off. J. Am. Soc. Bone Min. Res. 2022, 37, 2531–2547. [Google Scholar] [CrossRef]
- Bell, E.L.; Emerling, B.M.; Ricoult, S.J.; Guarente, L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene 2011, 30, 2986–2996. [Google Scholar] [CrossRef]
- Hirschey, M.D.; Shimazu, T.; Goetzman, E.; Jing, E.; Schwer, B.; Lombard, D.B.; Grueter, C.A.; Harris, C.; Biddinger, S.; Ilkayeva, O.R.; et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 2010, 464, 121–126. [Google Scholar] [CrossRef]
- Rider, M.H.; Bertrand, L.; Vertommen, D.; Michels, P.A.; Rousseau, G.G.; Hue, L. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: Head-to-head with a bifunctional enzyme that controls glycolysis. Biochem. J. 2004, 381, 561–579. [Google Scholar] [CrossRef]
- Marcus, J.M.; Andrabi, S.A. SIRT3 Regulation Under Cellular Stress: Making Sense of the Ups and Downs. Front. Neurosci. 2018, 12, 799. [Google Scholar] [CrossRef] [PubMed]
- van Waveren, C.; Moraes, C.T. Transcriptional co-expression and co-regulation of genes coding for components of the oxidative phosphorylation system. BMC Genom. 2008, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.X.; Thompson, K.; Taylor, R.W.; Oláhová, M. Mitochondrial OXPHOS Biogenesis: Co-Regulation of Protein Synthesis, Import, and Assembly Pathways. Int. J. Mol. Sci. 2020, 21, 3820. [Google Scholar] [CrossRef] [PubMed]
- Stram, A.R.; Payne, R.M. Post-translational modifications in mitochondria: Protein signaling in the powerhouse. Cell Mol. life Sci. C 2016, 73, 4063–4073. [Google Scholar] [CrossRef] [PubMed]
- Kadenbach, B. Complex IV-The regulatory center of mitochondrial oxidative phosphorylation. Mitochondrion 2021, 58, 296–302. [Google Scholar] [CrossRef]
- Lin, J.; Puigserver, P.; Donovan, J.; Tarr, P.; Spiegelman, B.M. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002, 277, 1645–1648. [Google Scholar] [CrossRef] [PubMed]
- Piantadosi, C.A.; Carraway, M.S.; Babiker, A.; Suliman, H.B. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ. Res. 2008, 103, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. AMP-activated protein kinase: An energy sensor that regulates all aspects of cell function. Genes Dev. 2011, 25, 1895–1908. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-C.; Hardie, D.G. AMPK: Sensing Glucose as well as Cellular Energy Status. Cell Metab. 2018, 27, 299–313. [Google Scholar] [CrossRef]
- Kim, H.S.; Patel, K.; Muldoon-Jacobs, K.; Bisht, K.S.; Aykin-Burns, N.; Pennington, J.D.; van der Meer, R.; Nguyen, P.; Savage, J.; Owens, K.M.; et al. SIRT3 Is a Mitochondria-Localized Tumor Suppressor Required for Maintenance of Mitochondrial Integrity and Metabolism during Stress. Cancer Cell 2010, 17, 41–52. [Google Scholar] [CrossRef]
- Hayashi, Y.; Yokota, A.; Harada, H.; Huang, G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1α in cancer. Cancer Sci. 2019, 110, 1510–1517. [Google Scholar] [CrossRef] [PubMed]
- Finley, L.W.; Carracedo, A.; Lee, J.; Souza, A.; Egia, A.; Zhang, J.; Teruya-Feldstein, J.; Moreira, P.I.; Cardoso, S.M.; Clish, C.B.; et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1?? Destabilization. Cancer Cell 2011, 19, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hydroxylation of HIF-1: Oxygen sensing at the molecular level. Physiology 2004, 19, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Barp, J.; Araújo, A.S.R.; Fernandes, T.R.G.; Rigatto, K.V.; Llesuy, S.; Belló-Klein, A.; Singal, P. Myocardial antioxidant and oxidative stress changes due to sex hormones. Braz. J. Med. Biol. Res. 2002, 35, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, K.; Elmarakby, A.A.; El-Remessy, A.B.; Sullivan, J.C. Oxidative stress contributes to sex differences in angiotensin II-mediated hypertension in spontaneously hypertensive rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R274–R282. [Google Scholar] [CrossRef] [PubMed]
- Kander, M.C.; Cui, Y.; Liu, Z. Gender difference in oxidative stress: A new look at the mechanisms for cardiovascular diseases. J. Cell Mol. Med. 2017, 21, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The integrated stress response. EMBO Rep. 2016, 17, 1374–1395. [Google Scholar] [CrossRef] [PubMed]
- Suragani, R.N.; Zachariah, R.S.; Velazquez, J.G.; Liu, S.; Sun, C.W.; Townes, T.M.; Chen, J.J. Heme-regulated eIF2α kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood 2012, 119, 5276–5284. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.M.; Nargund, A.M.; Sun, T.; Haynes, C.M. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 2012, 8, e1002760. [Google Scholar] [CrossRef]
- Pyo, C.-W.; Lee, S.-H.; Choi, S.-Y. Oxidative stress induces PKR-dependent apoptosis via IFN-gamma activation signaling in Jurkat T cells. Biochem. Biophys. Res. Commun. 2008, 377, 1001–1006. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 2003, 11, 619–633. [Google Scholar] [CrossRef] [PubMed]
- B’chir, W.; Maurin, A.-C.; Carraro, V.; Averous, J.; Jousse, C.; Muranishi, Y.; Parry, L.; Stepien, G.; Fafournoux, P.; Bruhat, A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013, 41, 7683–7699. [Google Scholar] [CrossRef] [PubMed]
- Kilberg, M.S.; Shan, J.; Su, N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 2009, 20, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Fiorese, C.J.; Schulz, A.M.; Lin, Y.F.; Rosin, N.; Pellegrino, M.W.; Haynes, C.M. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol. 2016, 26, 2037–2043. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, Y.; Katsuoka, F.; Funayama, R.; Nagashima, T.; Nishida, Y.; Nakayama, K.; Engel, J.D.; Yamamoto, M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012, 40, 10228–10239. [Google Scholar] [CrossRef]
- Kreß, J.K.C.; Jessen, C.; Hufnagel, A.; Schmitz, W.; Xavier da Silva, T.N.; Ferreira Dos Santos, A.; Mosteo, L.; Goding, C.R.; Friedmann Angeli, J.P.; Meierjohann, S. The integrated stress response effector ATF4 is an obligatory metabolic activator of NRF2. Cell Rep. 2023, 42, 112724. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.; Huang, C.; Snider, M.D.; Komar, A.A.; Tanaka, J.; Kaufman, R.J.; Krokowski, D.; Hatzoglou, M. A novel feedback loop regulates the response to endoplasmic reticulum stress via the cooperation of cytoplasmic splicing and mRNA translation. Mol. Cell. Biol. 2012, 32, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- Neill, G.; Masson, G.R. A stay of execution: ATF4 regulation and potential outcomes for the integrated stress response. Front. Mol. Neurosci. 2023, 16, 1112253. [Google Scholar] [CrossRef] [PubMed]
- Wek, R.C.; Jiang, H.-Y.; Anthony, T.G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006, 34, 7–11. [Google Scholar] [CrossRef]
- Blais, J.D.; Filipenko, V.; Bi, M.; Harding, H.P.; Ron, D.; Koumenis, C.; Wouters, B.G.; Bell, J.C. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol. Cell. Biol. 2004, 24, 7469–7482. [Google Scholar] [CrossRef]
- Köditz, J.; Nesper, J.; Wottawa, M.; Stiehl, D.P.; Camenisch, G.; Franke, C.; Myllyharju, J.; Wenger, R.H.; Katschinski, D.M. Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood 2007, 110, 3610–3617. [Google Scholar] [CrossRef]
- Han, J.; Back, S.H.; Hur, J.; Lin, Y.H.; Gildersleeve, R.; Shan, J.; Yuan, C.L.; Krokowski, D.; Wang, S.; Hatzoglou, M.; et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013, 15, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Sayers, C.M.; Verginadis, I.I.; Lehman, S.L.; Cheng, Y.; Cerniglia, G.J.; Tuttle, S.W.; Feldman, M.D.; Zhang, P.J.; Fuchs, S.Y.; et al. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. J. Clin. Investig. 2015, 125, 2592–2608. [Google Scholar] [CrossRef] [PubMed]
- Dickhout, J.G.; Carlisle, R.E.; Jerome, D.E.; Mohammed-Ali, Z.; Jiang, H.; Yang, G.; Mani, S.; Garg, S.K.; Banerjee, R.; Kaufman, R.J.; et al. Integrated stress response modulates cellular redox state via induction of cystathionine γ-lyase: Cross-talk between integrated stress response and thiol metabolism. J. Biol. Chem. 2012, 287, 7603–7614. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Cui, H.; Xie, N.; Banerjee, S.; Liu, R.-M.; Dai, H.; Thannickal, V.J.; Liu, G. ATF4 Mediates Mitochondrial Unfolded Protein Response in Alveolar Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2020, 63, 478–489. [Google Scholar] [CrossRef] [PubMed]
- Quirós, P.M.; Prado, M.A.; Zamboni, N.; Amico, D.D.; Williams, R.W.; Finley, D.; Gygi, S.P.; Auwerx, J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017, 216, 2027–2045. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-H.; Ma, X.-J.; Wu, L.-N.; Zhao, Y.-Y.; Zhang, P.-Y.; Zhang, Y.-H.; Shao, M.-W.; Liu, F.; Li, F.; Qin, G.-J. Sirtuin-3 (SIRT3) protects pancreatic β-cells from endoplasmic reticulum (ER) stress-induced apoptosis and dysfunction. Mol. Cell. Biochem. 2016, 420, 95–106. [Google Scholar] [CrossRef]
- Kopp, M.C.; Larburu, N.; Durairaj, V.; Adams, C.J.; Ali, M.M. UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor. Nat. Struct. Mol. Biol. 2019, 26, 1053–1062. [Google Scholar] [CrossRef]
- Tran, H.C.; Van Aken, O. Mitochondrial unfolded protein-related responses across kingdoms: Similar problems, different regulators. Mitochondrion 2020, 53, 166–177. [Google Scholar] [CrossRef]
- Eisner, V.; Picard, M.; Hajnóczky, G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell Biol. 2018, 20, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Münch, C.; Harper, J.W. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 2016, 534, 710–713. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-B.; Wang, R.-X.; Jiang, H.-B.; Zhang, E.-D.; Tan, J.-Q.; Xu, H.-Z.; Zhou, R.-R.; Xia, X.-B. Mitochondrial Ribosomal Protein L10 Associates with Cyclin B1/Cdk1 Activity and Mitochondrial Function. DNA Cell Biol. 2016, 35, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Li, H.; Zhang, H. Abnormal Expression of Mitochondrial Ribosomal Proteins and Their Encoding Genes with Cell Apoptosis and Diseases. Int. J. Mol. Sci. 2020, 21, 8879. [Google Scholar] [CrossRef] [PubMed]
- Papa, L.; Germain, D. SirT3 Regulates the Mitochondrial Unfolded Protein Response. Mol. Cell Biol. 2014, 34, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Borrás, C.; Sastre, J.; García-Sala, D.; Lloret, A.; Pallardó, F.V.; Viña, J. Mitochondria from females exhibit higher antioxidant gene expression and lower oxidative damage than males. Free Radic. Biol. Med. 2003, 34, 546–552. [Google Scholar] [CrossRef]
- Semenova, N.V.; Rychkova, L.V.; Darenskaya, M.A.; Kolesnikov, S.I.; Nikitina, O.A.; Petrova, A.G.; Vyrupaeva, E.V.; Kolesnikova, L.I. Superoxide Dismutase Activity in Male and Female Patients of Different Age with Moderate COVID-19. Bull. Exp. Biol. Med. 2022, 173, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ji, L.-L.; Liu, T.-Y.; Wang, Z.-T. Evaluation of gender-related differences in various oxidative stress enzymes in mice. Chin. J. Physiol. 2011, 54, 385–390. [Google Scholar]
- Sol, E.M.; Wagner, S.A.; Weinert, B.T.; Kumar, A.; Kim, H.S.; Deng, C.X.; Choudhary, C. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS ONE 2012, 7, e50545. [Google Scholar] [CrossRef]
- Kaeberlein, M.; McVey, M.; Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999, 13, 2570–2580. [Google Scholar] [CrossRef]
- Bellizzi, D.; Dato, S.; Cavalcante, P.; Covello, G.; Di Cianni, F.; Passarino, G.; Rose, G.; De Benedictis, G. Characterization of a bidirectional promoter shared between two human genes related to aging: SIRT3 and PSMD13. Genomics 2007, 89, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Jin, J.; Cichewicz, R.H.; Hageman, S.A.; Ellis, T.K.; Xiang, L.; Peng, Q.; Jiang, M.; Arbez, N.; Hotaling, K.; et al. trans-(-)-ε-Viniferin increases mitochondrial sirtuin 3 (SIRT3), activates AMP-activated protein kinase (AMPK), and protects cells in models of Huntington Disease. J. Biol. Chem. 2012, 287, 24460–24472. [Google Scholar] [CrossRef] [PubMed]
- Ying, Y.; Lu, C.; Chen, C.; Liu, Y.; Liu, Y.U.; Ruan, X.; Yang, Y. SIRT3 Regulates Neuronal Excitability of Alzheimer’s Disease Models in an Oxidative Stress-Dependent Manner. Neuromolecular. Med. 2022, 24, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Deng, H.X.; Gius, D.; Schumacker, P.T.; Surmeier, D.J.; Ma, Y.C. Sirt3 protects dopaminergic neurons from mitochondrial oxidative stress. Hum. Mol. Genet. 2017, 26, 1915–1926. [Google Scholar] [CrossRef] [PubMed]
- He, S.; He, C.; Yuan, H.; Xiong, S.; Xiao, Z.; Chen, L. The SIRT 3 expression profile is associated with pathological and clinical outcomes in human breast cancer patients. Cell. Physiol. Biochem. 2014, 34, 2061–2069. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Huang, Z.; Jiang, H.; Shi, F. The sirtuin 3 expression profile is associated with pathological and clinical outcomes in colon cancer patients. Biomed Res. Int. 2014, 2014, 871263. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Fu, L.L.; Wen, X.; Wang, X.Y.; Liu, J.; Cheng, Y.; Huang, J. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 2014, 3, e1047. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Brown, K.; Hirschey, M.D.; Verdin, E.; Chen, D. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metab. 2010, 12, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, N.R.; Gupta, M.; Kim, G.; Rajamohan, S.B.; Isbatan, A.; Gupta, M.P. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Investig. 2009, 119, 2758–2771. [Google Scholar] [CrossRef]
- Pradhan, R.; Kumar, R.; Shekhar, S.; Rai, N.; Ambashtha, A.; Banerjee, J.; Pathak, M.; Dwivedi, S.N.; Dey, S.; Dey, A.B. Longevity and healthy ageing genes FOXO3A and SIRT3: Serum protein marker and new road map to burst oxidative stress by Withania somnifera. Exp. Gerontol. 2017, 95, 9–15. [Google Scholar] [CrossRef]
- Brunelli, E.; Domanico, F.; Russa, L.D.; Pellegrino, D. Sex Differences in Oxidative Stress Biomarkers. Curr. Drug Targets 2014, 15, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Malorni, W.; Straface, E.; Matarrese, P.; Ascione, B.; Coinu, R.; Canu, S.; Galluzzo, P.; Marino, M.; Franconi, F. Redox state and gender differences in vascular smooth muscle cells. FEBS Lett. 2008, 582, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Vassilopoulos, A.; Pennington, J.D.; Andresson, T.; Rees, D.M.; Bosley, A.D.; Fearnley, I.M.; Ham, A.; Flynn, C.R.; Hill, S.; Rose, K.L.; et al. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient-and exercise-induced stress. Antioxid. Redox. Signal. 2014, 21, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Costa-Mattioli, M.; Walter, P. The integrated stress response: From mechanism to disease. Science 2020, 368, eaat5314. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Koo, J.H.; Lee, J.M.; Joo, M.S.; Kim, T.H.; Kim, H.; Jun, D.W.; Kim, S.G. NRF2-mediated SIRT3 induction protects hepatocytes from ER stress-induced liver injury. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22170. [Google Scholar] [CrossRef] [PubMed]
- Durkin, M.E.; Qian, X.; Popescu, N.C.; Lowy, D.R. Isolation of Mouse Embryo Fibroblasts. Bio-protocol 2013, 3, e908. [Google Scholar] [CrossRef] [PubMed]
- Podgorski, I.I.; Pinterić, M.; Marčinko, D.; Popović Hadžija, M.; Filić, V.; Ciganek, I.; Pleše, D.; Balog, T.; Sobočanec, S. Combination of sirtuin 3 and hyperoxia diminishes tumorigenic properties of MDA-MB-231 cells. Life Sci. 2020, 254, 117812. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]

| Pathway | False Discovery Rate |
|---|---|
| Focal adhesion | 2.22 × 10−10 |
| Focal adhesion: PI3K-Akt-mTOR signaling pathway | 4.79 × 10−9 |
| Mechanisms associated with pluripotency | 0.00047 |
| Cholesterol metabolism with Bloch and Kandutsch–Russell pathways | 0.00088 |
| Comprehensive IL-17A signaling | 0.00088 |
| Adipogenesis genes | 0.0029 |
| ESC pluripotency pathways | 0.0031 |
| Mapk signaling pathway | 0.0037 |
| Integrin-mediated cell adhesion | 0.0037 |
| Alpha 6 beta 4 integrin signaling pathway | 0.0111 |
| TGF-beta signaling pathway | 0.0118 |
| Retinol metabolism | 0.0242 |
| Wnt signaling pathway and pluripotency | 0.0318 |
| Omega-9 fatty acid synthesis | 0.0464 |
| Matrix metalloproteinases | 0.0464 |
| Oxidative stress and redox pathway | 0.0464 |
| Male MEF | Female MEF |
|---|---|
| EGF receptor signaling pathway | De novo pyrimidine ribonucleotides biosynthesis |
| Pentose phosphate pathway | De novo purine biosynthesis |
| Wnt signaling pathway | Sterol regulatory element-binding proteins (SREBP) signaling |
| Cadherin signaling pathway | Mitogen activated protein kinase kinase/MAP kinase cascade |
| FGF signaling pathway | Ras pathway |
| Glycolysis | PI3 kinase pathway |
| Hypoxia response via HIF activation | |
| De novo purine biosynthesis | |
| PI3 kinase pathway | |
| JAK/STAT signaling pathway |
| Pathway | Gene | Log2 Fold Change—Male KO MEF | Adjusted p-Value—Female KO MEF | Log2 Fold Change—Female KO MEF | Adjusted p-Value—Female KO MEF |
|---|---|---|---|---|---|
| Prkaa2 (AMPK) | 0.57 | 1.10 × 10−4 | −0.248 | 0.26 | |
| Glycolysis and TCA cycle | Lactate dehydrogenase B (Ldhb) | 2.09 | 5.60 × 10−7 | 0.16 | 0.33 |
| Mitochondrial pyruvate carrier 1 (Mpc1) | 0.37 | 0.02 | 0.15 | 0.82 | |
| Pyruvate dehydrogenase E1 alpha 1 subunit (Pdha1) | 0.14 | 4.40 × 10−4 | 0.18 | 4.40 × 10−6 | |
| Phosphofructokinase-1. Liver-type (Pfkl) | 0.4 | 1.60 × 10−7 | 0.27 | 1.20 × 10−9 | |
| Phosphofructokinase 1 muscle-type (Pfkm) | 0.34 | 3.40 × 10−4 | 0.07 | 0.23 | |
| Phosphofructokinase 2 (Pfkfb2) | 0.48 | 0.012 | −0.11 | 0.86 Ima ima | |
| Pyruvate kinase (Pkm) | −0.11 | 0.016 | 0 | 0.27 | |
| Aldolase A (Aldoa) | 0.44 | 0.694 | −0.15 | 3.00 × 10−4 | |
| Phosphoglycerate kinase (Pgk1) | 0.02 | 0.624 | 0.14 | 5.00 × 10−6 | |
| Isocitrate dehydrogenase (Idh2) | 0.2 | 4.60 × 10−5 | 0.11 | 0.03 | |
| Pyruvate dehydrogenase kinase isoenzyme 1 (Pdk1) | 1.23 | 1.00 × 10−7 | 0.63 | 0.002 | |
| Succinate dehydrogenase complex subunit C (Sdhc) | 0.03 | 0.004 | 0.03 | 0.038 | |
| Alpha-ketoglutarate dehydrogenase (Ogdh) | 0.05 | 0.152 | 0.05 | 0.264 | |
| Succinyl-CoA ligase [ADP-forming] subunit beta (Sucla2) | 0.08 | 0.382 | 0.1 | 0.414 | |
| Succinate dehydrogenase (Sdha) | −0.12 | 0.729 | −0.04 | 0.593 | |
| Fumarate hydratase (Fh1) | 0.03 | 0.926 | 0.12 | 0.363 | |
| Malate dehydrogenase 1 (Mdh1) | −0.02 | 0.528 | 0.16 | 0.191 | |
| Malate dehydrogenase 2 (Mdh2) | −0.04 | 0.870 | 0.01 | 0.666 | |
| Citrate synthase (Cs) | −0.07 | 0.530 | −0.04 | 0.947 | |
| Pyruvate carboxylase (Pcx) | −0.04 | 0.998 | −0.16 | 0.790 | |
| Aconitase (Aco1) | 0.05 | 0.465 | 0.02 | 0.940 | |
| Pentose-phosphate pathway | Glucose-6-Phosphate Dehydrogenase (G6pdx) | 0.22 | 4.10 × 10−5 | 0.21 | 0.124 Ima ima |
| 6-Phosphogluconolactonase (Pgls) | 0.28 | 2.10 × 10−4 | 0.18 | 0.01 | |
| 6-Phosphogluconate Dehydrogenase (Pgd) | 0.17 | 4.90 × 10−4 | −0.02 | 0.947 | |
| Ribulose-5-Phosphate Isomerase (Rpia) | −0.1 | 0.922 | −0.08 | 0.947 | |
| Transketolase (Tkt) | 0.14 | 7.80 × 10−6 | 0.03 | 0.380 | |
| Transaldolase (Taldo1) | −0.07 | 0.718 | −0.08 | 0.79 | |
| Fatty acid metabolism | Acetyl-coA carboxylase (Acaca) | −0.263 | 0.093 | 0.056 | 0.008 |
| Slc27a4 | 0.227 | 4.4 × 10−4 | 0.007 | 0.947 | |
| Fasn (Fatty acid synthase) | −0.134 | 0.011 | 0.039 | 0.940 |
| Gene | Log2 Fold Change—Male KO MEF | Adjusted p-Value—Female KO MEF | Log2 Fold Change—Female KO MEF | Adjusted p-Value—Female KO MEF | Function |
|---|---|---|---|---|---|
| Hspd1 | −0.16 | 3.30 × 10−5 | −0.02 | 0.72 | Mitochondrial chaperones |
| Hspe1 | −0.267 | 2.30 × 10−6 | 0.02 | 0.75 | |
| Hspa9 | 0.071 | 0.021 | −0.121 | 5.5 × 10−4 | |
| Hspa-5 (BiP) | 0.1 | 9.80 × 10−14 | 0.2 | 9.00 × 10−27 | ER chaperones |
| Calr | 0.16 | 1.30 × 10−8 | 0.31 | 4.50 × 10−61 | |
| Pdia4 | 0.3 | 4.20 × 10−15 | 0.26 | 2.10 × 10−15 | |
| Pdia5 | 0.37 | 1.70 × 10−10 | 0.32 | 4.40 × 10−9 | |
| Herpud1 | 0.87 | 1.10 × 10−26 | 0.4 | 0.002 | |
| Derl1 | 0.364 | 3.1 × 10−11 | 0.111 | 0.053 | |
| ClpP | 0.028 | 0.922 | −0.06 | 0.947 | Proteases |
| LonP | 0.295 | 1.8 × 10−13 | −0.19 | 2.0 × 10−4 | |
| Cth | 0.974 | 1.4 × 10−12 | −0.711 | 3.1 × 10−8 | Atf-4 mediated response to hypoxia/oxidative stress |
| Hmox1 | 0.494 | 6.5 × 10−11 | −0.202 | 0.082 | Heme Oxygenase 1, antioxidant enzyme and a known Nrf-2 (Nfe2l2) target |
| Gclc | 0.168 | 0.01 | −0.144 | 0.346 | Glutamate-cysteine ligase; glutathione synthesis |
| Asns | 0.836 | 1.2 × 10−55 | 0.023 | 0.728 | Atf-4 mediated amino acid metabolism and transport |
| Psat1 | 0.279 | 1.2 × 10−19 | −0.052 | 0.941 | Phosphoserine aminotransferase 1, involved in serine biosynthesis |
| Slc3a2 | 0.304 | 2.0 × 10−10 | −0.084 | 0.071 | Amino acid transporter; ER stress regulator [57] |
| Gene | Log2 Fold Change—Male KO MEF | Adjusted p-Value—Female KO MEF | Log2 Fold Change—Female KO MEF | Adjusted p-Value—Female KO MEF | Function |
|---|---|---|---|---|---|
| Mrpl1 | −0.269 | 0.045 | −0.225 | 0.049 | Mitochondrial large unit ribosomal protein |
| Mrpl9 | −0.275 | 0.037 | −6.2 × 10−5 | 0.947 | Mitochondrial large unit ribosomal protein |
| Mrpl19 | −0.068 | 0.003 | 0.235 | 0.993 | Mitochondrial large unit ribosomal protein |
| Mrpl23 | −0.941 | 0.008 | 0.029 | 1 | Mitochondrial large unit ribosomal protein |
| Mrpl27 | −0.334 | 0.037 | −0.075 | 0.917 | Mitochondrial large unit ribosomal protein |
| Mrpl38 | −0.371 | 0.001 | −0.105 | 0.66 | Mitochondrial large unit ribosomal protein |
| Mrpl45 | −0.271 | 0.009 | −0.240 | 0.036 | Mitochondrial large unit ribosomal protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belužić, R.; Šimunić, E.; Podgorski, I.I.; Pinterić, M.; Hadžija, M.P.; Balog, T.; Sobočanec, S. Gene Expression Profiling Reveals Fundamental Sex-Specific Differences in SIRT3-Mediated Redox and Metabolic Signaling in Mouse Embryonic Fibroblasts. Int. J. Mol. Sci. 2024, 25, 3868. https://doi.org/10.3390/ijms25073868
Belužić R, Šimunić E, Podgorski II, Pinterić M, Hadžija MP, Balog T, Sobočanec S. Gene Expression Profiling Reveals Fundamental Sex-Specific Differences in SIRT3-Mediated Redox and Metabolic Signaling in Mouse Embryonic Fibroblasts. International Journal of Molecular Sciences. 2024; 25(7):3868. https://doi.org/10.3390/ijms25073868
Chicago/Turabian StyleBelužić, Robert, Ena Šimunić, Iva I. Podgorski, Marija Pinterić, Marijana Popović Hadžija, Tihomir Balog, and Sandra Sobočanec. 2024. "Gene Expression Profiling Reveals Fundamental Sex-Specific Differences in SIRT3-Mediated Redox and Metabolic Signaling in Mouse Embryonic Fibroblasts" International Journal of Molecular Sciences 25, no. 7: 3868. https://doi.org/10.3390/ijms25073868
APA StyleBelužić, R., Šimunić, E., Podgorski, I. I., Pinterić, M., Hadžija, M. P., Balog, T., & Sobočanec, S. (2024). Gene Expression Profiling Reveals Fundamental Sex-Specific Differences in SIRT3-Mediated Redox and Metabolic Signaling in Mouse Embryonic Fibroblasts. International Journal of Molecular Sciences, 25(7), 3868. https://doi.org/10.3390/ijms25073868







